Abstract
The simultaneous detection of multiple mycotoxins is of great significance for food safety and human health. Herein, a simple, convenient and accurate fluorescent aptasensor was designed based on the dual cross DNA nanostructure for the simultaneous detection of ochratoxin A (OTA) and aflatoxin B1 (AFB1), in which the stable dual cross DNA nanostructure provided an assay platform using the fluorescent dye-labeled aptamers as a sensing element. Owing to the higher affinity of aptamers for their target, the aptamer probes were released from the assay platform in the presence of OTA and AFB1, resulting in an enhanced fluorescence at 570 nm and 670 nm. This “signal-on” fluorescent aptasensor assay system can effectively avoid background signals and minimize false positive. Furthermore, the designed method can realize the simultaneous detection of OTA and AFB1 during the whole experiment. The limits of detection (LOD) were as low as 0.0058 ng/mL for OTA, ranging from 0.01 to 50 ng/mL and 0.046 ng/mL for AFB1, ranging from 0.05 to 100 ng/mL. The proposed fluorescent aptasensor exhibits excellent performance in practical application and provides a novel approach for the simultaneous detection of multiple mycotoxins by simply changing the aptamers.
Graphical abstract
A “signal-on” fluorescent aptasensor assay system based on the stable dual cross DNA nanostructure was successfully developed for simultaneous detection of OTA and AFB1 with lower detection limits in wider linear ranges.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycotoxins are toxic secondary metabolites produced by a certain fungus that colonizes on both crops and food during growth, harvest, storage and processing [1]. These mycotoxins contaminate over 25% harvested crops and cause billions of economic losses around the world each year. In addition, due to their high teratogenicity, mutagenicity and carcinogenicity, mycotoxins present a great potential threat to humans and animal health through the food chain [2, 3]. To date, more than 300 species of mycotoxins have been reported, including Aspergillus, Fusarium and Penicillium spp. The most frequently encountered and dangerous mycotoxins in food products are aflatoxin B1 (AFB1) and ochratoxin A (OTA). It is well known that AFB1 is the most toxic substance. Its toxicity is 10 times that of potassium cyanide (KCN) and 68 times that of arsenic [4]. According to an evaluation by the International Agency of Research on Cancer (IARC), AFB1 has been classified in Group 1 as a potent human carcinogen, and OTA has been categorized as a Group 2B carcinogen [5]. Therefore, the maximum permitted levels for AFB1 and OTA have been set by various countries because of their extensive influence on agricultural products and foods, such as peanuts, cereals, feed, coffee and milk [6].
Accurate, economical and convenient detection of AFB1 and OTA is of great significance for food safety and human health. There are various analytical methods available for the detection of mycotoxins in food and feed. Conventional analytical methods are considered the standard method for mycotoxins due to accuracy and precision, such as high-performance liquid chromatography (HPLC), mass spectrometry (MS), liquid chromatography-mass spectrometry (LCMS) and enzyme-linked immunosorbent assay (ELISA) [7, 8]. However, owing to their high costs, complicated operations and professional skill required, the practical application of these methods is restricted. Moreover, a great deal of false-positive results are common with traditional methods [9]. Currently, more electrochemical [10], colorimetric [11] and fluorometric [12] analysis strategies have been developed with the aim to achieve simple, rapid, efficient and low-cost mycotoxins detection. Among these, the fluorescence method has been largely utilized due to its high sensitivity and instantaneous response [13]. Rasooly et al. presented a portable low-cost fluorescence detector for direct detection of detoxified and active AFB1. Most importantly, this fluorometer combined with Vero cells can be used for quantitative detection of AFB1 activity [14]. Furthermore, in order to improve the specific recognition of biosensors, aptamers as an alternative of antibodies can specifically recognize and bind to a target with the advantages of low cost, simple synthesis, high thermal stability and easy chemical modification [15, 16]. Thus, aptamers are promising sensing elements for the development of prominent biosensors. Fluorescent aptasensors combine the advantages of a fluorescence method with aptamers. Generally, unlabeled fluorescent aptasensors are disturbed by background signals, making it difficult to detect multiple mycotoxins.
DNA nanostructure as a novel nanomaterial can be constructed using economic and programmable single-stranded DNA (ssDNA). A great deal of DNA nanostructures with unique function and flexibility were designed for stable and sensitive biosensors via binding to aptamers [17]. Seyed designed a fluorescent aptasensor based on a DNA pyramid nanostructure and PicoGreen dye for the detection of OTA [18]. The presented aptasensor exhibited high sensitivity and specificity toward OTA and was successfully used in serum and grape juice samples. Yang et al. reported a DNA octahedron fluorescence nanoprobe assembled from eight ssDNAs, which can simultaneous detect and image messenger RNAs (mRNAs) in living cells [19]. However, the construction of complicated DNA nanostructures is difficult and limited by technical expertise. The simple, stable and versatile DNA nanostructures have a promising application in biosensors.
Herein, we developed a labeled fluorescent aptasensor based on the dual cross DNA nanostructure. The developed dual cross DNA nanostructure was composed of aptamer and complementary ssDNA, which provided a sensing platform. The Cy5 and Cy3 were labeled on the aptamers of AFB1 and OTA, respectively, and the complementarity to the ssDNA-modified quencher led to weak fluorescence emission. Owing to the higher affinity of the aptamer for its target, upon addition of AFB1 or OTA, the dual cross DNA nanostructure was disassembled and the aptamer left the quencher, leading to a strong fluorescence emission at specific wavelengths. The presented aptamer probes could effectively reduce the interference of background signals and realize the multiple detections with high sensitivity and selectivity.
Experimental section
Materials
All HPLC-purified DNA sequences were synthesized by Shanghai Sangon. Detail sequences are given in Electronic Supplementary Material Table S1. Aflatoxin B1 (AFB1), aflatoxin B2 (AFB2), ochratoxin A (OTA), ochratoxin B (OTB) and deoxynivalenol (DON) were purchased from Sigma-Aldrich. Fumonisin B1 (FB1) was provided by Acros Organics. Magnesium chloride hexahydrate (MgCl2), sodium chloride (NaCl) and hydrochloric acid (HCl) were purchased from Tianjin Kemiou Chemical Reagent Co., Ltd. Tris(hydroxymethyl)aminomethane (Tris) and DL-dithiothreitol (DTT) were obtained from Shanghai Macklin Biochemical Co., Ltd. Ethylenediaminetetraacetic acid (EDTA) was purchased from Luoyang Chemical Reagent Factory. Agarose, ethidium bromide and DNA loading buffer (6×) were provided by Beijing Solarbio Science & Technology Co., Ltd. Wine and corn were obtained from a local supermarket in Zhengzhou, China. All the chemicals were of analytical grade, and all aqueous solutions were prepared via ultra-pure water (18.2 MΩ).
Preparation of dual cross DNA nanostructure
Before the experiment, all containers were sterilized, and the DNA was dissolved using 20 mM Tris buffer solution. The dual cross DNA nanostructure was prepared based on the complementary pairing via the annealing process. Briefly, 4 μL of OTA-aptamer, AFB1-aptamer, C1, C2 and C3 at a concentration of 10 μM were mixed in the polymerase chain reaction (PCR) tube under shake stirring over 2 min. The mixtures were put into the PCR instrument and heated to 95 °C for 10 min. Then, the DNA was cooled to 4 °C at a rate of 1 °C/min. The DNA nanostructure was characterized using agarose gel electrophoresis (AGE) at 110 V for 40 min (see Electronic Supplementary Material).
Simultaneous determination of OTA and AFB1
The analytical procedures of OTA and AFB1 based on dual cross DNA nanostructure are as follows: In a typical OTA and AFB1 detection experiment, the fluorescent aptasensor (10 μL, 2 μM) was mixed with 10 μL OTA and AFB1 at different concentrations. The mixture was incubated at ambient temperature for 30 min and replenished to 200 μL by Tris-HCl buffer solution. The final concentrations of OTA and AFB1 are given in Electronic Supplementary Material Table S2. The fluorescence was carried out by recording the emission change at 670 nm and 570 nm using 635 nm (Cy5) and 530 nm (Cy3) as the excitation wavelength (Hitachi F-7100 fluorescence spectrophotometer with slits of 5 nm). The quenching efficiencies of aptamer probes were calculated by (F − F0)/F0, in which F0 and F are the fluorescence intensities of Cy5 or Cy3 in the absence and presence of target, respectively. The selectivity of this prepared fluorescent aptasensor for OTA and AFB1 was evaluated by monitoring relative changes in fluorescence at 670 nm and 570 nm in response to 50 ng/mL of other mycotoxins (FB1, DON, AFB2 and OTB) and their mixtures (OTA, OTB, AFB1, AFB2, FB1 and DON). All experiments were conducted at least three times.
OTA and AFB1 detection in real samples
The corn and wine were pretreated for OTA and AFB1 actual detection in real samples [20]. Firstly, the corn was ground into powder (1.0 g) and mixed with 1 mL OTA and AFB1 solution (the concentrations of OTA and AFB1 were 5 ng/mL, 50 ng/mL, 500 ng/mL, respectively). After drying at room temperature, extraction solvent (10 mL, methanol: water, 7:3 [v/v]) was add into the mixture under shake stirring over 30 min. The supernatant was centrifuged at 5000 rmp for 10 min and filtered by a 0.22-μm filter. Then, different concentrations of OTA and AFB1 (0.5 ng/mL, 5 ng/mL, 50 ng/mL) in corn samples were obtained. In addition, 5.0 mL wine was mixed with different concentrations of OTA and AFB1 solution and filtered by a 0.22-μm filter. Different concentrations of OTA and AFB1 (0.1 ng/mL, 1 ng/mL, 10 ng/mL) in wine samples were obtained. The spiked sample can be used for real sample detection. The contaminated feed (corn peanut meal) was also ground into powder (1.0 g). Then, extraction solvent was added under shake stirring over 30 min. The supernatant was treated for AFB1 detection. All results were compared using HPLC.
Results and discussion
Design of the fluorescent aptasensors
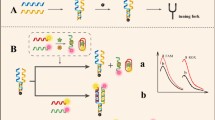
The principle of the proposed fluorescent aptasensor is shown in Scheme 1. The dual cross DNA nanostructure was assembled from OTA aptamer, AFB1 aptamer, C1, C2 and C3 via annealing. The quencher BHQ2 labeled on C1 can complement C3 and C2, which provided a stable sensing platform. The OTA aptamer labeled with Cy3 (OTA APT-Cy3) was partially complementary to C1 (5′ terminal) and C3 (3′ terminal), resulting in a quenched fluorescence of Cy3 due to the fluorescence resonance energy transfer (FRET) between Cy3 and BHQ2. In the presence of OTA, OTA APT-Cy3 binds with OTA preferentially due to the higher affinity with OTA than ordinary DNA duplex, and the fluorescence of Cy3 increases with the presence of OTA. Similarly, the AFB1 aptamer labeled with Cy5 (AFB1 APT-Cy5) was partially complementary to C1 (3′ terminal) and C3 (5′ terminal), leading the fluorescence of Cy5 quenched by BHQ2. Upon addition of AFB1, the fluorescence of Cy5 also increased. Therefore, this “signal-on” fluorescent aptasensor can realize simultaneous detection of OTA and AFB1 and effectively avoid background signal interference.
In addition, 3.0% agarose gel electrophoresis was used to characterize the successful construction of this aptasensor. As shown in Fig. 1, all DNA nanostructures and strands can be displayed in the corresponding lane. The assembled dual cross DNA nanostructure reveals the lowest migration in lane 1, demonstrating the successful formation of the aptasensor. Owning to 36 bases, OTA aptamer shows the fastest move in lane 2. The hairpin structure in AFB1 aptamer provides an insertion site for the dye, causing lane 3 to be bright. The quencher BHQ2 labeled on C1 and C3 causes lane 4 and lane 6 to be very weak. Moreover, the central chain C2 (lane 5) has the largest number of bases with the slowest migration in all ssDNA. Therefore, the designed fluorescent aptasensor was verified to be formed, which provided a stable aptamer probe.
Optimization of detection conditions
For optimal detection performance, the concentrations of dual cross DNA nanostructure and pH were optimized. A very low concentration of dual cross DNA nanostructure gives a little signal enhancement. On the contrary, the background signal has strong interference in a high concentration of dual cross DNA nanostructure. As shown in Fig. 2A, the changes in fluorescence (F − F0)/F0 (F0 and F represent the fluorescence intensity of the aptasensor system in the absence and presence of target at 570 nm [Cy3] and 670 nm [Cy5]) were recorded. The curves gradually increased and tended to be stable with the rise of dual cross DNA nanostructure concentrations. When the concentration was held at 200 nM, the changes increased to its maximum indicating the optimal aptasensor concentration. In addition, the dual cross DNA nanostructure will disintegrate and overcome the electrostatic interaction of the phosphate backbone under acidic or alkaline conditions. The pH condition was optimized and ranged from 7 to 9. It can be seen from Fig. 2B that the fluorescence changes reached a maximum at pH 8.0 due to the weak charge repulsion of ssDNA. Therefore, the fluorescent aptasensor has excellent detection performance due to its stable structure under optimized conditions.
To investigate the feasibility of this fluorescent aptasensor, the effect of OTA and AFB1 on fluorescence was first determined under optimized conditions. Figure 3A displays weak fluorescence due to the FRET between BHQ2-Cy3 and Cy5, and almost unchanged fluorescence at 570 nm and 670 nm with the addition of a buffer solution without target. The fluorescence was enhanced twofold at 570 nm and unchanged at 670 nm in the presence of OTA (Fig. 3B). Similarly, the fluorescence was enhanced 3.3-fold at 670 nm and unchanged at 570 nm in the presence of AFB1 (Fig. 3C). While both OTA and AFB1 were present, the fluorescence of aptasensor had a significant increase at 570 nm and 670 nm owning to the separation of aptamer from the DNA nanostructure by the target (Fig. 3D). These results indicated that the proposed fluorescent aptasensor can be utilized for simultaneous detection of OTA and AFB1.
Simultaneous detection of OTA and AFB1 using fluorescent aptasensors
OTA and AFB1 are two highly toxic mycotoxins that can be determined by fluorescent aptasensors. As depicted in Fig. 4a, the fluorescence intensity at 570 nm was increased with the concentrations of OTA ranging from 0.01 to 50 ng/mL. An excellent linear response was obtained between the logarithm of the OTA concentrations and (F − F0)/F0, in which F0 and F are the emission intensities of Cy3 in the absence and presence of OTA, respectively (Fig. 4b). The corresponding linear equation was calculated to be y = 0.688x + 1.498, with a correlation coefficient of R2 = 0.997. The limit of detection (LOD) was calculated according to: L = 3 × σ/s, where L is the detection limit of target, σ is the standard deviation of blank, and s is the slope of linear correction curve (based on International Union of Pure and Applied Chemistry, IUPAC). The LOD of OTA was calculated to be 0.0058 ng/mL. The limit of quantitation (LOQ) of OTA was determined to be 0.017 ng/mL. Moreover, the fluorescence intensity of aptasensors at 670 nm was increased with the presence of AFB1 (Fig. 4c). The variation curve of (F − F0)/F0 was obtained with the increasing of AFB1 ranging from 0.05 to 100 ng/mL. The linear equation is expressed as follows: y = 1.089x + 1.593, R2 = 0.998 (Fig. 4d). The corresponding LOD was calculated to be 0.046 ng/mL (S/N = 3). The LOQ of AFB1 was determined to be 0.138 ng/mL. To further highlight the performance of this fluorescent aptasensor, a comparison of different analytical methods for OTA and AFB1 is summarized in Table 1. Our proposed fluorescent aptasensor assay system can realize simultaneous detection of OTA and AFB1 with a wider detection range and lower LOD as compared with other analytical methods.
Representative fluorescence emission spectra of the aptasensor at different concentrations of OTA (0.01–50 ng/mL) (a) and AFB1 (0.05–100 ng/mL) (c). The corresponding calibration plot of relative fluorescence intensity toward OTA (b) and AFB1 (d) concentrations. Inset: The linear relationship of relative fluorescence intensity toward the logarithm of OTA and AFB1 concentration
Furthermore, the selectivity of the aptasensor was also evaluated via comparing the fluorescence changes toward different mycotoxins under the same test conditions. Figure 5 shows that the fluorescence scarcely changed in the presence of control mycotoxins, compared to the significant increase of OTA or AFB1 at lower concentrations. Moreover, similar changes in fluorescence between the target and the mixture indicate that OTA and AFB1 do not interact with each other. This is mainly ascribed to the high affinity between the aptamer and its target. All of these results demonstrate that the fluorescent aptasensors have a great potential for the simultaneous determination of OTA and AFB1 in food products.
Practical detections for OTA and AFB1
For practical application, this fluorescent aptasensor was applied to determine the concentrations of OTA and AFB1 in corn and wine samples. According to the linear equation, the concentrations of OTA and AFB1 were found to be close to the spiked amount. The recovery test was calculated by comparing the known amount and found amount, and the recoveries of OTA and AFB1 ranged from 97.2% to 101.8% and 95.8% to 107.8% in the corn samples and from 98.5% to 107.2% and 99.1% to 101.5% in the wine samples, respectively (Table 2). The recoveries are further compared with different analytical methods in Table 1. The certified reference material in feed (corn peanut meal) was also detected using HPLC and the fluorescent aptasensor. The AFB1 in contaminated feed was found to be 18.59 ng/mL using HPLC, and that via the aptasensor was 20.37 ng/mL. These results indicated that the fluorescent aptasensor assay system has obvious advantages over other methods and it can be used as an alternative method for practical application in OTA and AFB1 detection.
Conclusions
In summary, a fluorescent aptasensor was successfully developed based on the dual cross DNA nanostructure modified with Cy3 and Cy5 for the simultaneous detection of OTA and AFB1. The stable dual cross DNA nanostructure was assembled from OTA APT-Cy3, AFB1 APT-Cy5, C1, C2 and C3, which provided an assay platform with weak fluorescence. The fluorescence of this aptasensor will be enhanced in the presence of OTA and AFB1 due to the higher affinity of the aptamer for its target, which results in the aptamer detaching from the DNA nanostructure. This “signal-on” fluorescent aptasensor assay system exhibited sensitive and selective OTA and AFB1 assay and could realize their simultaneous detection. Further, an adequate determination of OTA and AFB1 in corn, wine and feed samples confirmed the applicability of this strategy. The rational design of this fluorescent aptasensor presents a promising method to simultaneously detect multiple mycotoxins and promotes the application of aptamer in sensors.
References
Zhang N, Liu BS, Cui XL, Li YT, Tang J, Wang HX, Zhang D, Li Z. Recent advances in aptasensors for mycotoxin detection: on the surface and in the colloid. Talanta. 2021;223(1):121729. https://doi.org/10.1016/j.talanta.2020.121729.
Jiang Q, Wu JD, Yao K, Yin YL, Max G, Yang CB, Francis L. Paper-based microfluidic device (DON-Chip) for rapid and low-cost deoxynivalenol quantification in food, feed, and feed ingredients. ACS Sens. 2019;4(11):3072–9. https://doi.org/10.1021/acssensors.9b01895.
Li Y, Liu X, Lin Z. Recent developments and applications of surface plasmon resonance biosensors for the detection of mycotoxins in foodstuffs. Food Chem. 2012;132(3):1549–54. https://doi.org/10.1016/j.foodchem.2011.10.109.
Wu H, Wang HY, Wu J, Han GQ, Liu YL, Zou P. A novel fluorescent aptasensor based on exonuclease-assisted triple recycling amplification for sensitive and label-free detection of aflatoxin B1. J Hazard Mater. 2021;415(5):125584. https://doi.org/10.1016/j.jhazmat.2021.125584.
Chauhan R, Singh J, Sachdev T, Basu T, Malhotra B. Recent advances in mycotoxins detection. Biosens Bioelectron. 2016;81(15):532–45. https://doi.org/10.1016/j.bios.2016.03.004.
Wang J, Mukhtar H, Ma L, Pang Q, Wang X. VHH antibodies: reagents for mycotoxin detection in food products. Sensors. 2018;18(2):485. https://doi.org/10.3390/s18020485.
He TT, Zhou T, Wan H, Han QB, Ma YQ, Tan T, Wan YQ. One-step deep eutectic solvent strategy for efficient analysis of aflatoxins in edible oils. J Sci Food Agric. 2020;100(13):4840–8. https://doi.org/10.1002/jsfa.10544.
Jia YM, Wu F, Liu PL, Zhou GH, Yu B, Lou XD, Xia F. A label-free fluorescent aptasensor for the detection of aflatoxin B1 in food samples using AIEgens and graphene oxide. Talanta. 2009;198(1):71–7. https://doi.org/10.1016/j.talanta.2019.01.078.
Sergeyev T, Yarynka D, Piletska E, Linnik R, Zaporozhets O, Brovko O, Piletsky S, El'skaya A. Development of a smartphone-based biomimetic sensor for aflatoxin B1 detection using molecularly imprinted polymer membranes. Talanta. 2019;201:204–10. https://doi.org/10.1016/j.talanta.2019.04.016.
Yang XS, Shi DM, Zhu SM, Wang BJ, Zhang XJ, Wang GF. Portable aptasensor of aflatoxin B1 in bread based on a personal glucose meter and DNA walking machine. ACS Sens. 2018;3(7):1368–75. https://doi.org/10.1021/acssensors.8b00304.
Seok YG, Byun JY, Shim WB, Kim MG. A structure-switchable aptasensor for aflatoxin B1 detection based on assembly of an aptamer/split DNAzyme. Anal Chim Acta. 2015;886(30):182–7. https://doi.org/10.1016/j.aca.2015.05.041.
Tang XQ, Li PW, Zhang Q, Zhang ZW, Zhang W, Jiang J. Time-resolved fluorescence immunochromatographic assay developed using two idiotypic nanobodies for rapid, quantitative, and simultaneous detection of aflatoxin and Zearalenone in maize and its products. Anal Chem. 2017;89(21):11520–8. https://doi.org/10.1021/acs.analchem.7b02794.
Suo ZG, Liu XW, Hou XL, Liu Y, Lu JT, Xing FF, Chen YY, Feng LY. Ratiometric assays for acetylcholinesterase activity and organo-phosphorous pesticide based on superior carbon quantum dots and BLGF-protected gold nanoclusters FRET process. ChemistrySelect. 2020;5(29):9254–60. https://doi.org/10.1002/slct.202002042.
Rasooly R, Do PM, Hernlem BJ. Low cost quantitative digital imaging as an alternative to qualitative in vivo bioassays for analysis of active aflatoxin B1. Biosens Bioelectron. 2016;80:405–10. https://doi.org/10.1016/j.bios.2016.01.087.
Zhou JH, Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat Rev Drug Discov. 2017;16(3):2889–93. https://doi.org/10.1038/nrd.2016.199.
Zhao BJ, Wu P, Zhang H, Cai CX. Designing activatable aptamer probes for simultaneous detection of multiple tumor-related proteins in living cancer cells. Biosens Bioelectron. 2015;68(15):763–70. https://doi.org/10.1016/j.bios.2015.02.004.
Suo ZG, Chen JQ, Hou XL, Hu ZH, Xing FF, Feng LY. Growing prospects of DNA nanomaterials in novel biomedical applications. RSC Adv. 2019;9(29):16479–91. https://doi.org/10.1039/c9ra01261c.
Nameghi MA, Danesh NM, Ramezani M, Hassani FV, Abnous K, Taghdisi SM. A fluorescent aptasensor based on a DNA pyramid nanostructure for ultrasensitive detection of Ochratoxin a. Anal Bioanal Chem. 2016;408(21):5811–8. https://doi.org/10.1007/s00216-016-9693-7.
Zhong L, Cai SX, Huang YQ, Yin LT, Yang YL, Lu CH, Yang HH. A DNA octahedron-based fluorescence nanoprobe for dual tumorrelated mRNAs detection and imaging. Anal Chem. 2018;90(20):12059–66. https://doi.org/10.1021/acs.analchem.8b02847.
Wei M, He X, Xie YL. A novel signal-on fluorescent aptasensor for Ochratoxin a detection based on RecJf exonuclease-induced signal amplification. J Chin Chem Soc. 2020;67(7):1247–53. https://doi.org/10.1002/jccs.201900423.
Wang Y, Hu XF, Pei YF, Sun YN, Wang FY, Song CM, Yin MQ, Deng RG, Li ZX, Zhang GPY. Selection of phage-displayed minotopes of ochratoxin a and its detection in cereal by ELISA. Anal Methods. 2015;7(5):1849–54. https://doi.org/10.1039/c4ay02290d.
AntonellaVatinno A, Palmisano R, Zambonin F, Carlo G. Determination of Ochratoxin a in wine at sub ng/mL levels by solid-phase microextraction coupled to liquid chromatography with fluorescence detection. J Chromatogr A. 2006;1115(1–2):196–201. https://doi.org/10.1016/j.chroma.2006.02.092.
Gke G, Aissa SB, Nemčekov K, Catanante G, Raouafi N, Marty JL. Aptamer-modified pencil graphite electrodes for the impedimetric determination of Ochratoxin a. Food Control. 2020;115:107271. https://doi.org/10.1016/j.foodcont.2020.107271.
Bulbul G, Hayat A, Andreescu S. A generic amplification strategy for electrochemical aptasensors using a non-enzymatic Nanoceria tag. Nanoscale. 2015;7(31):13230–8. https://doi.org/10.1039/c5nr02628h.
Lin CY, Zheng HX, Sun M, Guo YJ, Luo F, Guo LH, Qiu B, Lin ZY, Chen GN. Highly sensitive colorimetric aptasensor for ochratoxin a detection based on enzyme-encapsulated liposome. Anal Chim Acta. 2018;1002(9):90–6. https://doi.org/10.1016/j.aca.2017.11.061.
Yin XT, Wang S, Liu XY, He CM, Tang YL, Li QM, Liu JH, Su HJ, Tan TW, Dong YY. Aptamer-based colorimetric biosensing of Ochratoxin a in fortified white grape wine sample using unmodified gold nanoparticles. Anal Sci. 2017;33(6):659–64. https://doi.org/10.2116/analsci.33.659.
Hao L, Wang W, Shen X, Wang SL, Li Q, An FL, Wu SJ. A fluorescent DNA hydrogel aptasensor based on the self-assembly of rolling circle amplification products for sensitive detection of Ochratoxin a. J Agric Food Chem. 2020;68(1):369–75. https://doi.org/10.1021/acs.jafc.9b06021.
Wu KF, Ma CB, Zhao H, Chen MJ, Deng ZY. Sensitive aptamer-based fluorescene assay for Ochratoxin a based on RNase H signal amplification. Food Chem. 2019;277(30):273–8. https://doi.org/10.1016/j.foodchem.2018.10.130.
Zhao FC, Tian Y, Shen Q, Liu RX, Shi RR, Wang HM, Yang ZY. A novel nanobody and mimotope based immunoassay for rapid analysis of aflatoxin B1. Talanta. 2019;195:55–61. https://doi.org/10.1016/j.talanta.2018.11.013.
Nonaka Y, Saito K, Hanioka N, Narimatsu S, Kataoka H. Determination of aflatoxins in food samples by automated on-line in-tubesolid-phase microextraction coupled with liquid chromatography–massspectrometry. J Chromatogr A. 2009;1216(20):4416–22. https://doi.org/10.1016/j.chroma.2009.03.035.
Lai WQ, Zeng Q, Tang J, Zhang MS, Tang DP (2018) A conventional chemical reaction for use in an unconventional assay: a colorimetric immunoassay for aflatoxin B1 by using enzyme-responsive just-in-time generation of a MnO2 based nanocatalyst. Microchim Acta 185(2):92. https://doi.org/10.1007/s00604-017-2651-z.
Goud KY, Hayat A, Catanante G, Satyanarayana M, Gobi KV, Marty JL. An electrochemical aptasensor based on functionalized graphene oxide assisted electrocatalytic signal amplification of methylene blue for aflatoxin B1 detection. Electrochim Acta. 2017;244(1):96–103. https://doi.org/10.1016/j.electacta.2017.05.089.
Li X, Yang L, Men C, Xie YF, Liu JJ, Zou HY, Li YF, Zhan L, Huang CZ. Photothermal soft nanoballs developed by loading plasmonic Cu2-XSe nanocrystals into liposomes for photothermal immunoassay of aflatoxin B1. Anal Chem. 2019;91(7):4444–50. https://doi.org/10.1021/acs.analchem.8b05031.
Tan HX, Ma L, Guo T, Zhou HY, Chen L, Zhang YH, Dai HJ, Yu Y. A novel fluorescence aptasensor based on mesoporous silica nanoparticles for selective and sensitive detection of aflatoxin B1. Anal Chim Acta. 2019;1068(30):87–95. https://doi.org/10.1016/j.aca.2019.04.014.
Funding
This study was funded by the Key Scientific and Technological Project of Henan Province (212102310001).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 27 kb)
Rights and permissions
About this article
Cite this article
Suo, Z., Liang, X., Jin, H. et al. A signal-enhancement fluorescent aptasensor based on the stable dual cross DNA nanostructure for simultaneous detection of OTA and AFB1. Anal Bioanal Chem 413, 7587–7595 (2021). https://doi.org/10.1007/s00216-021-03723-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03723-8