Abstract
The ldh gene of Corynebacterium glutamicum ATCC 13032 (gene symbol cg3219, encoding a 314 residue NAD+-dependent l-(+)-lactate dehydrogenase, EC 1.1.1.27) was cloned into the expression vector pKK388-1 and over-expressed in an ldhA-null E. coli TG1 strain upon isopropyl-β-D-thiogalactopyranoside (IPTG) induction. The recombinant protein (referred to here as CgLDH) was purified by a combination of dye-ligand and ion-exchange chromatography. Though active in its absence, CgLDH activity is enhanced 17- to 20-fold in the presence of the allosteric activator d-fructose-1,6-bisphosphate (Fru-1,6-P2). Contrary to a previous report, CgLDH has readily measurable reaction rates in both directions, with V max for the reduction of pyruvate being approximately tenfold that of the value for l-lactate oxidation at pH 7.5. No deviation from Michaelis–Menten kinetics was observed in the presence of Fru-1,6-P2, while a sigmoidal response (indicative of positive cooperativity) was seen towards l-lactate without Fru-1,6-P2. Strikingly, when introduced into an lldD − strain of C. glutamicum, constitutively expressed CgLDH enables the organism to grow on l-lactate as the sole carbon source.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corynebacterium glutamicum is an industrially important Gram positive rod-shaped bacterium used in the production of amino acids including l-glutamate and l-lysine (Krämer 2004). It has furthermore been proposed as a source for the organic acids l-lactate and succinate (Okino et al. 2005). The former has long been used in the food and pharmaceutical industries as a preservative and base chemical building block and has more recently become important in the production of the biodegradable plastic poly-lactic acid, or PLA, where optically pure monomers of d- and l-lactate are essential (for a review, see Okano et al. 2010).
The ldh gene in C. glutamicum ATCC 13032 is solely responsible for l-lactate production in that organism, as demonstrated by gene knockout (Inui et al. 2004; Dietrich et al. 2009). Although some work has been carried out on the regulation of ldh expression under certain conditions (Inui et al. 2007; Engels et al. 2008; Dietrich et al. 2009; Toyoda et al. 2009a, b), the encoded enzyme has not been extensively kinetically characterised.
Bacterial lactate dehydrogenases fall into two broad classes—NAD+-linked and NAD+-independent. The first class are cytoplasmic proteins that may be activated by fructose-1,6-bisphosphate (Fru-1,6-P2), are d- or l-lactate-specific and catalyse the conversion of pyruvate to lactate in a reversible or non-reversible manner (Garvie 1980). The second class are all believed to be flavin-dependent, are physiologically required for the oxidation of lactate and are often membrane-bound LDHs that participate in the respiratory chain. C. glutamicum has two such NAD+-independent LDHs (Bott and Niebisch 2003). The protein encoded by lldD (gene symbol cg3227, EC 1.1.2.3) utilises l-lactate, and the second is a d-lactate-specific enzyme encoded by dld (gene symbol cg1027, EC 1.1.2.4). Each has been shown to be necessary for growth of C. glutamicum on l-lactate and d-lactate, respectively, as the sole carbon source (Stansen et al. 2005; Kato et al. 2010). Both are believed to be flavin-dependent and have been kinetically characterised to some extent, the authors reporting sub-millimolar K m values for lactate (LLD: K m 0.51 mM for l-lactate Stansen et al. 2005; DLD: K m 0.61 mM for d-lactate Kato et al. 2010).
The physiological role of the NAD+-dependent LDHs in bacteria is to provide a means for the regeneration of NAD+ during fermentation, when NADH would otherwise accumulate owing to the absence of oxygen. It has recently been reported that the ldh-encoded NAD+-dependent LDH from C. glutamicum ATCC 13032 operates solely in the direction of l-lactate formation (Dietrich et al. 2009). There have been recurrent reports of such irreversible behaviour by bacterial lactate dehydrogenases over many years (Garvie 1980; Freier and Gottschalk 1987; Contag et al. 1990; Wyckoff et al. 1997). Superficially, this implies a thermodynamic paradox that is not usually discussed. Enzymes can only affect the speed of attainment of equilibrium, not the position of equilibrium. How then can they behave irreversibly? However, it is only at equilibrium that the rate of reaction in one direction must unequivocally be balanced by an equal rate of reaction in the opposite direction, and this carries no implication with regard to the relative rates in either direction under initial-rate conditions—i.e. with no products present. Jencks (1975) and Cornish-Bowden (1995) have discussed the feasibility of enzymes acting irreversibly in this sense, and it was this apparent feature of C. glutamicum LDH that directed us to a closer study of the enzyme. In this report, however, we find that there are measurable rates in both directions in the absence of products—rates that are augmented to similar extents by the presence of Fru-1,6-P2. In the absence of this allosteric activator, the reaction with lactate shows strong positive cooperativity (h = 2.2) and S 0.5 of 533 mM, 7 times higher than the K m in its presence.
Furthermore, we demonstrate that, despite the high K m for lactate, constitutive expression of this NAD+-dependent enzyme in a C. glutamicum host deficient in NAD+-independent lactate dehydrogenase enables this (lldD −) strain to grow on l-lactate as the sole carbon source. To our knowledge, this is the first time that complementation of this kind has been seen.
Materials and methods
Bacterial strains, growth conditions, plasmids, oligonucleotides, chemicals and reagents
The bacterial strains, plasmids and oligonucleotides used in this study are listed in Table 1. C. glutamicum strains were grown aerobically (250 r.p.m.) at 34°C in Brain Heart Infusion (BHI) rich medium (Difco) or in CGXII chemically defined medium (Keilhauer et al. 1993) containing 0.2 mg biotin L−1 and either 1% (w/v) glucose, or 1% (w/v) l-lactate. Growth was followed by measuring the OD600 with a Beckman DU 7400 spectrophotometer. Time-course experiments were performed as follows: C. glutamicum strains were grown overnight in BHI medium. Cells were concentrated by centrifugation and re-suspended in CGXII medium at an initial OD600 of 1. Samples were taken every hour, and experiments were done in triplicate. Escherichia coli strains (TG1 and TG1 ∆ldhA) were grown aerobically at 37°C in Luria–Bertani medium (1% tryptone, 1% NaCl, 0.5% yeast extract). Antibiotics used were kanamycin (25 μg mL−1), chloramphenicol (10 μg mL−1) and ampicillin (100 μg mL−1). IPTG was used at a 0.1-mM final concentration. Tryptone and yeast extract were purchased from Formedium, sodium phosphate mono-basic from Aldrich and di-basic from Lancaster Chemicals, with Trizma base being obtained from Sigma, and glycine from Fisher. Riedel-de Haën was the source of ammonium sulphate. Oxidised and reduced dinucleotide coenzymes were sourced from Apollo Scientific and were of at least 98% purity, while sodium l-lactate and d-fructose-1,6-bisphosphate were obtained from Fluka, with pyruvate (sodium salt) coming from Sigma.
DNA isolation, manipulation and transfer
All the molecular biology procedures used in this study were described by Sambrook et al. (1989) and Merkamm and Guyonvarch (2001). C. glutamicum and E. coli cells were transformed by electroporation as described by Bonamy et al. (1990). PCR experiments were carried out with either Taq DNA Polymerase (Promega) or High Fidelity PCR enzyme mix (Fermentas) and a GeneAmp PCR System 2700 thermocycler (Applied Biosystems). Restriction endonucleases were from New England Biolabs, T4 DNA ligase was purchased from Promega, and oligonucleotide primers were synthesised by MWG-Biotech.
Overexpression of the C. glutamicum ldh gene
The open reading frame of ldh was amplified from genomic DNA using oligonucleotides ldh-34 and ldh-36 (Table 1) and digested with BspHI and XmaI. The digestion product was cloned into the NcoI and XmaI sites of the E. coli expression vector pKK388-1 to create pAG1036. The identity of the cloned gene was confirmed by DNA sequencing (MWG-Biotech, Germany). To overexpress the ldh gene in C. glutamicum, pAG1036 was digested with BamHI. The resulting Ptrc-ldh fragment was cloned into the BamHI site of the E. coli–C. glutamicum shuttle vector pCGL482 to create pAG1037. This plasmid was used to express the ldh gene constitutively in C. glutamicum as no regulation by LacI occurs in this background.
Creation of a knockout E. coli expression strain
E. coli genomic DNA was used as the template in two separate PCR reactions to amplify ~500-bp regions flanking the ldhA ORF using the primer combinations ldh-No/ldh-Ni and ldh-Co/ldh-Ci (listed in Table 1). The products were gel purified, combined and used as the template for a crossover PCR reaction using the primer combination ldh-No/ldh-Co. The resultant 1,069-bp construct corresponds to the E. coli genome at the ldhA locus except that the ldhA ORF is replaced by an in-frame 36-bp synthetic ORF encoding a 12-residue peptide (MVINLECEGYCV). This construct was cloned into the NotI and SalI sites of the deletion vector pKOV and was used for the in-frame deletion of the entire ldhA ORF from E. coli TG1 according to the method of Link et al. (Link et al. 1997).
Construction of a C. glutamicum lldD mutant
Disruption of the C. glutamicum ldh gene was previously described by Dietrich et al. (2009). To disrupt the chromosomal lldD gene (cg3227), an internal lldD fragment was amplified by PCR using primers Lld-1 and Lld-2. Chromosomal C. glutamicum DNA was used as template. PCR products were purified using the High Pure PCR Product Purification Kit (Roche Diagnostics). The purified 356-bp PCR fragment was cloned into the pCR2.1-TOPO cloning vector to create pAG1001. Plasmid pAG1001 was introduced into C. glutamicum by electroporation. Kanamycin-resistant transformants were the result of chromosomal integration by single crossover events, as verified by Southern blotting. Inactivation of the lldD gene was confirmed by the absence of LLD activity (Kato et al. 2010) in the membrane fraction of the RES::pAG1001 strain.
Overexpression of CgLDH
pAG1036 was introduced into the E. coli strain TG1ΔldhA and cultured on LB agar containing 100 μg mL−1 ampicillin. A single colony was used to inoculate 5 mL sterile LB medium (1% tryptone, 1% NaCl, 0.5% yeast extract) containing 100 μg mL−1 ampicillin, which was grown overnight at 37°C. One millilitre of this pre-culture was used to inoculate 500 mL fresh growth medium, and this was grown at 30°C to an OD600 of ~0.6–0.8 before being induced with 1 mM isopropyl-β-D-thio-galactopyranoside (IPTG; Melford Laboratories, UK). Cells were harvested following growth overnight at 30°C.
Purification of CgLDH
Cells overexpressing CgLDH were harvested by centrifugation and broken in 10 mM sodium phosphate buffer, pH 7.5 using a Sonicator® Ultrasonic Processor XL (Misonix, Farmingdale NY). The lysed suspension was then centrifuged at 23,500×g for 20 min at 4°C and the supernatant precipitated with 70% ammonium sulphate. The resultant pellet was resuspended in 10 mM sodium phosphate buffer, pH 7.0 (Buffer A) and dialysed overnight against three changes of 500 mL of the same buffer before being applied to a Blue Sepharose dye-affinity chromatography column. The flow-through (having CgLDH activity) was loaded onto a Q-Sepharose™ Fast Flow anion-exchange column equilibrated with Buffer A. Non-binding proteins were removed from the column using Buffer A, and CgLDH was eluted with a 0–0.5 M NaCl gradient in the same buffer. After precipitation in 70% saturated ammonium sulphate followed by dialysis into Buffer A, protein was applied to a second dye-affinity chromatography column (Procion Red HE-3B immobilised on Sepharose CL-6B). Purified CgLDH (from the flow-through of the Procion Red column) was stored at 4°C in 70% ammonium sulphate.
Determination of the pH profile of CgLDH
Assays were carried out at 25°C in a mixed buffer consisting of sodium phosphate, Trizma and glycine, each at a final concentration of 50 mM. The pH was adjusted to the desired value with NaOH or HCl following the addition of pyruvate or l-lactate. For reactions in the direction of pyruvate formation, l-lactate concentration was fixed at 200 mM and NAD+ at 1.0 mM, while reactions in the opposite direction were carried out in the presence of fixed amounts of 2.5 mM pyruvate and 0.3 mM NADH.
Determination of kinetic parameters
Assays were carried out at 25°C in 0.1 M sodium phosphate buffer (pH 7.5), and changes in NADH concentration were monitored via the change in absorbance at 340 nm in a 1-cm cuvette using a Cary 50 UV–Vis spectrophotometer (Varian Inc., Palo Alto CA). The absorbance change was converted into units of enzyme activity (μmol min−1 mg−1) taking the extinction coefficient of NADH to be 6.22 mM−1 cm−1. Assays performed at high NADH concentrations were monitored at 370 or 380 nm, and the rates adjusted by factors of 2.27 or 4.75, respectively, to take account of the lower extinction coefficient of NADH at these wavelengths. Reactions were usually started by the addition of 10 μL of coenzyme, bringing the total reaction volume to 1 mL. The concentration of purified enzyme was determined spectrophotometrically at 280 nm using a calculated extinction coefficient of 32,890 cm−1 M−1 (ProtParam tool Gasteiger et al. 2003).
The kinetic parameters were determined from initial-rate measurements by varying the concentration of one substrate over a wide range while maintaining the other at near-saturating levels. For reactions using lactate as a substrate, l-lactate concentration was varied from 1 to 250 mM at a fixed concentration of 1 mM NAD+, and NAD+ concentration was varied from 0.007 to 3.0 mM with 200 mM l-lactate. In reactions where pyruvate was a substrate, pyruvate was varied from 0.15 to 13.5 mM at a fixed concentration of 0.6 mM NADH, and NADH was varied from 0.006 to 0.6 mM in 2.5 mM pyruvate. In all cases, the concentration of Fru-1,6-P2 in the reaction cuvette was 2.0 mM. Unless otherwise stated, 10 μL enzyme (in 0.01 M sodium phosphate buffer, pH 7.0) and 10 μL of 200 mM Fru-1,6-P2 (in water) were combined in the cuvette before the addition of buffer/substrate solution, prior to the addition of the coenzyme to start the reaction.
Assays without the activator Fru-1,6-P2 were usually carried out over an extended substrate range in order to obtain reliable initial-rate measurements that could be used to extract the kinetic parameters. Fixed substrate concentrations were the same as for reactions containing Fru-1,6-P2.
Time course of enzyme activation by d-fructose-1,6-bisphosphate
CgLDH was added to a reaction mixture consisting of 0.1 M sodium phosphate, pH 7.5, and 2 mM Fru-1,6-P2 containing either 200 mM l-lactate or 2.5 mM pyruvate, thermo-equilibrated at 25°C. Aliquots of 990 μL were withdrawn immediately following enzyme addition and mixing, and at time intervals thereafter, and transferred to cuvettes containing 10 μL of the appropriate coenzyme for assay (final concentrations of 1 and 0.6 mM for NAD+ and NADH, respectively). Enzyme activity was also measured prior to the addition of Fru-1,6-P2 to the assay buffer in order to determine non-activated rates.
Results
Cloning of the C. glutamicum ldh ORF and E. coli TG1 ΔldhA preparation
Automated DNA sequencing (carried out by MWG-Biotech) confirmed that the cloned open reading frame was 100% identical to the reported sequence of the ldh gene from Corynebacterium glutamicum ATCC 13032 (gene locus cg3219). It has been inserted into the bacterial expression vector pKK388-1 under the control of a lac-inducible trc promoter (to give the plasmid pAG1036), so that translation begins from the native start codon of the C. glutamicum ldh gene.
The in-frame deletion of the d-lactate-specific ldhA gene from E. coli TG1 has been confirmed by colony PCR using the ldhA-No and ldhA-Co primers, where a PCR product of 1,069 bp is indicative of gene deletion. Using the same PCR conditions, a product of 2,020 bp is expected if the gene has not been replaced. Of 36 colonies chosen at random following the knockout experiment, 11 gave the shorter PCR product (result not shown). A ΔldhA clone and a wild-type E. coli TG1 clone were chosen for further analysis, in which cell-free extracts from overnight cultures of each in LB medium were assayed spectrophotometrically for LDH activity. Both clones showed an LDH-independent decrease in NADH absorption in the absence of pyruvate. This background was identical in the ΔldhA strain in the presence of 2.5 mM pyruvate (indicating a successful gene deletion), while the activity of the wild-type strain with pyruvate was more than double its background rate. The assays were carried out at 25°C in 0.1 M sodium phosphate buffer, pH 7.5, in the presence of 0.3 mM NADH and 2 mM Fru-1,6-P2.
Overexpression and purification of CgLDH
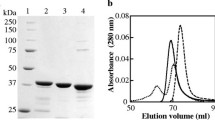
The aforementioned purification procedure yielded approximately 3 mg of pure CgLDH as judged by SDS–PAGE (result not shown) from 500 mL of bacterial culture. This represented a yield of about 10% of the total LDH activity in the crude extract.
Kinetic parameters of CgLDH
The kinetic parameters of CgLDH at pH 7.5 are summarised in Table 2. Over the broad ranges of substrate concentration tested (90- to 430-fold, depending on the substrate), CgLDH gave straightforward Michaelis–Menten kinetics in the presence of Fru-1,6-P2 (see Fig. 1). Some substrate inhibition was seen at higher concentrations for NADH, pyruvate and lactate, and so the data points for calculating the kinetic parameters were chosen to avoid this inhibition. The effect of Fru-1,6-P2 is pronounced, resulting in an increase in activity of 17-fold at 2.5 mM pyruvate, and 20-fold at 200 mM l-lactate. Aside from generating a rate increase, the activation of CgLDH by Fru-1,6-P2 brings about a dramatic change in behaviour towards l-lactate—from apparent positive cooperativity in its absence (h = 2.2), with a very high S 0.5, to classical Michaelis–Menten behaviour when it is present. This cooperative behaviour may account for the failure of Dietrich et al. (2009) to detect activity towards l-lactate, as no mention is made in their publication of adding Fru-1,6-P2 to reactions in this direction.
Graphs of CgLDH activity with increasing substrate concentration for all four substrates, demonstrating a good fit in all cases to the Michaelis–Menten equation with 2 mM fructose-1,6-bisphosphate in the reaction mixture (open circles). The quality of the fit for lactate (R 2 = 0.998) and pyruvate (R 2 = 0.993) is not so apparent owing to the abscissa scale needed to visualise data generated without the effector for these substrates. In the absence of fructose-1,6-bisphosphate (closed circles), lower reaction rates are observed with all substrates, and sigmoidal behaviour towards l-lactate is seen. Reactions were carried out in 0.1 M sodium phosphate buffer, pH 7.5, at 25°C as described in the “Methods” section. Nonlinear regression fit to rectangular hyperbolae and to the Hill equation was performed using SigmaPlot 8.0
pH activity profiles of CgLDH
CgLDH activity has been determined in both directions in the presence of Fru-1,6-P2; over the pH range 6.0–9.5 for the oxidation of l-lactate and from pH 5.5–9.0 for the reduction of pyruvate, both at half-unit intervals (Fig. 2). For the reduction of pyruvate under our assay conditions, the smooth curves indicate a pH optimum between pH 6.5 and 7.0, in agreement with the previous report (Dietrich et al. 2009). The pH optimum for l-lactate oxidation is higher—between 8.0 and 8.5. Fixed substrate concentrations were near-saturating, and a mixed buffer was used to avoid buffer-specific effects.
Graphs of CgLDH activity at different pH values. With lactate as the substrate, initial concentrations were 200 mM l-lactate and 1 mM NAD+. Substrate concentrations for the reverse reaction were 2.5 mM pyruvate and 0.3 mM NADH. Both sets of reactions were carried out in a mixed buffer at 25°C in the presence of 2 mM fructose-1,6-bisphosphate as described in the “Methods” section
Time course of activation by d-fructose-1,6-bisphosphate
When CgLDH is introduced to buffer containing l-lactate or pyruvate and Fru-1,6-P2 at the final reaction concentrations (enzyme never encounters more than 2 mM Fru-1,6-P2), full activation of the enzyme takes 10–15 min (at 25°C) depending on the direction of the reaction. For the reduction of pyruvate under the conditions tested, the activated rate is approximately 424 μmol min−1 mg−1 (compared to a non-activated rate of 93 μmol min−1 mg−1) and T½ for the activation is ~2.7 min. For lactate oxidation, the rate of the activated enzyme is approximately 40 μmol min−1 mg−1, and the T½ for activation is ~0.3 min (the rate is 2.6 μmol min−1 mg−1 without Fru-1,6-P2).
Effect of ldh over-expression on C. glutamicum growth
In order to test the effect of ldh over-expression on l-(+)-lactate consumption, strains RES167 (wild type), RES167::pMM29 (ldh minus strain) and RES167::pAG1001 (lldD minus strain) were transformed either with pCGL482 (control) or pAG1037 (ldh over-expression). The transformed strains were cultivated on minimal medium with either 1% glucose or 1% l-(+)-lactate as carbon sources. The LDH and LLD activities were measured for all strains in both conditions (Table 3). Overexpression of ldh from pAG1037 and the absence of lldD expression in the lldD − strain were confirmed. As seen in Fig. 3, all the strains exhibited quite similar growth behaviour on glucose as expected. On l-(+)-lactate, strains WT (control), WT (ldh over-expr.), ldh − (control), ldh − (ldh over-expr.) grew and lldD − (control) did not, as expected (Fig. 3). Surprisingly, lldD − (ldh over-expr.) grew perfectly. To ensure that this phenotypic complementation was not due to an unexpected additional mutation, ten lldD − (ldh over-expr.) clones obtained from two independent transformation experiments were tested. All exhibited the same growth behaviour on l-lactate. This clearly indicates that constitutive overexpression of ldh complements an lldD deficiency. This also indicates that the CgLDH enzyme can function in vivo to convert lactate to pyruvate. In addition, this experiment revealed that a yet unidentified l-lactate transporter should exist in C. glutamicum, as l-lactate should be in the protonated form at pH 7 and is unlikely to enter cells freely in that form, and as no protein with consistent similarity to the well-characterised l-lactate transporters LldP and GlcA from E. coli (Núñez et al. 2001) was found in the C. glutamicum genome.
Growth of C. glutamicum on glucose or l-lactate. Wild-type (open squares), ldh − (open triangles) and lldD − (closed circles) strains of C. glutamicum transformed with a plasmid for the constitutive expression of CgLDH or an empty-vector control were grown in Erlenmeyer flasks from 0D600 1.0 on minimal medium containing either glucose or l-lactate as the sole carbon source. Comparison of panels c and d (l-lactate cultures) clearly shows that plasmid-borne constitutive expression of NAD+-dependent CgLDH is a functional replacement for NAD+-independent LLD (encoded by lldD), enabling the lldD − strain to grow on l-lactate. Graphs shown are the result of three independent experiments
Discussion
Fructose-1,6-bisphosphate-activated l-(+)-LDHs are usually reported to be unidirectional, often requiring a high concentration of l-lactate in highly alkaline conditions, or use of an NAD+ analogue to detect even low levels of activity in this direction (Garvie 1980; Bryant 1991). Many such LDHs also show no activity, even with pyruvate, in the absence of Fru-1,6-P2 (Garvie 1980; Sommer et al. 1985; Williams and Andrews 1986; Wyckoff et al. 1997). CgLDH, on the other hand, is active in both directions in the absence of Fru-1,6-P2, and each direction of reaction shows a similar increase in rate when 2 mM Fru-1,6-P2 is present. This holds not only for recombinant CgLDH purified from E. coli, but also for CgLDH extracted from C. glutamicum when the protocol for activity determination that has been developed in this study is applied. A lag in Fru-1,6-P2 activation is avoided if the enzyme is pre-incubated with this ligand—a scenario observed previously for the LDHs from Staphylococcus epidermidis and Streptococcus lactis (Götz and Schleifer 1978; Hardman et al. 1985).
We have found four other instances in the literature where Fru-1,6-P2-activated l-(+)-LDHs have been shown to be reversible (Crow and Pritchard 1977; Taguchi et al. 1985; Wrba et al. 1990; Özkan et al. 2004). The first is LDH from Streptococcus lactis where the authors mention K m values but no rates of reaction. The other three enzymes are from thermophilic organisms—Thermus caldophilus, Thermotoga maritima and Clostridium thermocellum, respectively, the first two of which are reported to have measurable rates in each direction that are augmented by the presence of Fru-1,6-P2. The third group reported ‘no marked catalytic activity’ with lactate as substrate when assaying their enzyme in the absence of Fru-1,6-P2, although it is not clear at what temperature or pH they carried out their kinetic analysis to determine K m and V max values that are broadly similar to those found here for CgLDH at 25°C. Another difference is that C. thermocellum LDH shows positive cooperativity with l-lactate (in the presence of Fru-1,6-P2), whereas we show here that while C. glutamicum LDH is cooperative with this substrate in the absence of Fru-1,6-P2 (Hill coefficient of 2.2), it displays straightforward Michaelis–Menten kinetics when the ligand is present. Interestingly, the T. caldophilus and C. thermocellum LDHs displayed a severe substrate inhibition by pyruvate at concentrations above 1 mM in the presence of Fru-1,6-P2—a phenomenon not seen for CgLDH (where only moderate inhibition is seen at higher concentrations).
The K m of CgLDH for pyruvate of 0.85 mM in comparison to a K m for l-lactate of 73.9 mM suggests that the enzyme functions in vivo to convert pyruvate to lactate. Nevertheless, we have clearly demonstrated in this study that CgLDH can also function in vivo in the reverse direction when constitutively overexpressed. We conducted a pH survey for the activity of CgLDH which shows that the pH optimum for lactate formation is between 6.5 and 7.0. The pH optimum for the conversion of l-lactate to pyruvate is higher (between 8.0 and 8.5), but there is still considerable activity at neutral pH owing to a shoulder in the profile. The intracellular pH of C. glutamicum has been measured to vary between pH 7.7–8.3 during batch- and fed-batch culture (Leyval et al. 1997), a pH range compatible with a CgLDH reverse activity.
References
Bonamy C, Guyonvarch A, Reyes O, David F, Leblon G (1990) Interspecies electro-transformation in corynebacteria. FEMS Microbiol Lett 54:263–269
Bott M, Niebisch A (2003) The respiratory chain of Corynebacterium glutamicum. J Biotechnol 104:129–153
Brosius J (1988) A survey of molecular cloning vectors and their uses. In: Rodriguez RL, Denhardt DT (eds) Vectors. Butterworth, Boston, Mass, pp 205–225
Bryant FO (1991) Characterization of the fructose 1,6-bisphosphate-activated, l-(+)-lactate dehydrogenase from Thermoanaerobacter ethanolicus. J Enzyme Inhib 5:235–248
Contag PR, Williams MG, Rogers P (1990) Cloning of a lactate dehydrogenase gene from Clostridium acetobutylicum B643 and expression in Escherichia coli. Appl Environ Microbiol 56:3760–3765
Cornish-Bowden A (1995) Fundamentals of enzyme kinetics. Portland Press Ltd, Portland
Crow VL, Pritchard GG (1977) Fructose 1,6-diphosphate activated l-lactate dehydrogenase from Streptococcus lactis: kinetic properties and factors affecting activation. J Bacteriol 131:82–91
Dietrich C, Nato A, Bost B, Le Marechal P, Guyonvarch A (2009) Regulation of ldh expression during biotin-limited growth of Corynebacterium glutamicum. Microbiology 155:1360–1375
Dusch N, Pühler A, Kalinowski J (1999) Expression of the Corynebacterium glutamicum panD gene encoding l-aspartate-α-decarboxylase leads to pantothenate overproduction in Escherichia coli. Appl Environ Microbiol 65:1530–1539
Engels V, Lindner SN, Wendisch VF (2008) The global repressor SugR controls expression of genes of glycolysis and of the l-lactate dehydrogenase LdhA in Corynebacterium glutamicum. J Bacteriol 190:8033–8044
Freier D, Gottschalk G (1987) l-(+)-lactate dehydrogenase of Clostridium acetobutylicum is activated by fructose-1,6-bisphosphate. FEMS Microbiol Lett 43:229–233
Garvie EI (1980) Bacterial lactate dehydrogenases. Microbiol Rev 44:106–139
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Götz F, Schleifer KH (1978) Biochemical properties and the physiological role of the fructose-1,6-bisphosphate activated l-lactate dehydrogenase from Staphylococcus epidermidis. Eur J Biochem 90:555–561
Hardman MJ, Crow VL, Cruickshank DS, Pritchard GG (1985) Kinetics of activation of l-lactate dehydrogenase from Streptococcus lactis by fructose 1,6-bisphosphate. Eur J Biochem 146:179–183
Inui M, Murakami S, Okino S, Kawaguchi H, Vertes AA, Yukawa H (2004) Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196
Inui M, Suda M, Okino S, Nonaka H, Puskas LG, Vertes AA, Yukawa H (2007) Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 153:2491–2504
Jencks WP (1975) Binding energy, specificity, and enzymic catalysis: the Circe effect. Adv Enzymol Relat Areas Mol Biol 43:219–410
Kato O, Youn JW, Stansen KC, Matsui D, Oikawa T, Wendisch VF (2010) Quinone-dependent d-lactate dehydrogenase Dld (Cg1027) is essential for growth of Corynebacterium glutamicum on d-lactate. BMC Microbiol 10:321
Keilhauer C, Eggeling L, Sahm H (1993) Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J Bacteriol 175:5595–5603
Krämer R (2004) Production of amino acids: physiological and genetic approaches. Food Biotechnol 18:171–216
Leyval D, Debay F, Engasser JM, Goergen JL (1997) Flow cytometry for the intracellular pH measurement of glutamate producing Corynebacterium glutamicum. J Microbiol Meth 29:121–127
Link AJ, Phillips D, Church GM (1997) Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol 179:6228–6237
Merkamm M, Guyonvarch A (2001) Cloning of the sodA gene from Corynebacterium melassecola and role of superoxide dismutase in cellular viability. J Bacteriol 183:1284–1295
Núñez MF, Kwon O, Wilson TH, Aguilar J, Baldoma L, Lin ECC (2001) Transport of l-lactate, d-lactate, and glycolate by the LldP and GlcA membrane carriers of Escherichia coli. Biochem Biophys Res Commun 290:824–829
Okano K, Tanaka T, Ogino C, Fukuda H, Kondo A (2010) Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl Microbiol Biotechnol 85:413–423
Okino S, Inui M, Yukawa H (2005) Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol 68:475–480
Özkan M, Yilmaz EI, Lynd LR, Ozcengiz G (2004) Cloning and expression of the Clostridium thermocellum l-lactate dehydrogenase gene in Escherichia coli and enzyme characterization. Can J Microbiol 50:845–851
Salim K, Haedens V, Content J, Leblon G, Huygen K (1997) Heterologous expression of the Mycobacterium tuberculosis gene encoding Antigen 85A in C. glutamicum. Appl Environ Microbiol 63:4392–4400
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Sommer P, Klein JP, Scholler M, Frank RM (1985) Lactate dehydrogenase from Streptococcus mutans: purification, characterization, and crossed antigenicity with lactate dehydrogenases from Lactobacillus casei, Actinomyces viscosus, and Streptococcus sanguis. Infect Immun 47:489–495
Stansen C, Uy D, Delaunay S, Eggeling L, Goergen JL, Wendisch VF (2005) Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl Environ Microbiol 71:5920–5928
Taguchi H, Machida M, Matsuzawa H, Ohta T (1985) Allosteric and kinetic properties of l-lactate dehydrogenase from Thermus caldophilus GK24, an extremely thermophilic bacterium. Agric Biol Chem 49:359–365
Toyoda K, Teramoto H, Inui M, Yukawa H (2009a) The ldhA gene, encoding fermentative l-lactate dehydrogenase of Corynebacterium glutamicum, is under the control of positive feedback regulation mediated by LldR. J Bacteriol 191:4251–4258
Toyoda K, Teramoto H, Inui M, Yukawa H (2009b) Molecular mechanism of SugR-mediated sugar-dependent expression of the ldhA gene encoding l-lactate dehydrogenase in Corynebacterium glutamicum. Appl Microbiol Biotechnol 83:315–327
Wilkinson GN (1961) Statistical estimations in enzyme kinetics. Biochem J 80:324–332
Williams RA, Andrews P (1986) Purification of the fructose 1,6-bisphosphate-dependent lactate dehydrogenase from Streptococcus uberis and an investigation of its existence in different forms. Biochem J 236:721–727
Wrba A, Jaenicke R, Huber R, Stetter KO (1990) Lactate dehydrogenase from the extreme thermophile Thermotoga maritima. Eur J Biochem 188:195–201
Wyckoff HA, Chow J, Whitehead TR, Cotta MA (1997) Cloning, sequence, and expression of the l-(+)-lactate dehydrogenase of Streptococcus bovis. Curr Microbiol 34:367–373
Acknowledgments
PCE and MS wish to acknowledge the support of Science Foundation Ireland through a Fellowship grant to PCE and also the Irish Higher Education Authority’s support via its Programme for Research in Third-Level Institutions which has funded the Conway Institute.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Wolfgang Buckel.
Rights and permissions
About this article
Cite this article
Sharkey, M.A., Maher, M.A., Guyonvarch, A. et al. Kinetic characterisation of recombinant Corynebacterium glutamicum NAD+-dependent LDH over-expressed in E. coli and its rescue of an lldD − phenotype in C. glutamicum: the issue of reversibility re-examined. Arch Microbiol 193, 731–740 (2011). https://doi.org/10.1007/s00203-011-0711-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-011-0711-z