Abstract
This paper reports on the transcriptional regulation mechanism of the Corynebacterium glutamicum ldhA gene encoding l-lactate dehydrogenase responsible for production of l-lactate. DNA affinity purification allowed us to identify SugR, a global repressor of genes involved in sugar uptake and glycolysis, as a protein binding to the ldhA promoter region. Whereas ldhA gene expression and ldhA promoter activity were completely repressed during growth of wild-type cells in the absence of sugar, no such repression was observed in sugR mutant cells, indicating that SugR represses ldhA transcription. Electrophoretic mobility shift assays and DNase I footprint analyses revealed that two direct repeats, centered at position-17 with respect to the transcriptional start point, are required for SugR binding to the ldhA promoter and that fructose-1-phosphate (F-1-P) is the strongest negative effector of repressor activity of SugR. Furthermore, the ldhA promoter activity during growth on either fructose or sucrose, under which F-1-P is generated, is higher than on glucose, supporting the results of the in vitro binding assays. Thus, C. glutamicum ldhA is repressed by SugR in the absence of sugar and the expression level is dependent on the extent of derepression, which varies in response to sugars provided.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the absence of an external electron acceptor, facultative aerobic bacteria induce fermentative pathways to maintain the cytosolic NADH to NAD+ ratio. Fermentation of sugars to lactate is widespread. A variety of bacteria produce lactate during anaerobic growth; in some cases, the l-isomer is made; in others, d-lactate is the product. Escherichia coli produces d-lactate (Clark 1989), while Bacillus subtilis and many lactobacilli produce l-lactate (Cruz Ramos et al. 2000). Lactate dehydrogenase (LDH) is a cytoplasmic NAD-linked enzyme that converts pyruvate to lactate. During fermentation, LDH is the key enzyme involved in reoxidation of the NADH formed during glycolysis.
Expression of LDH-encoding genes in E. coli and B. subtilis is under the control of multiple transcriptional regulators. In E. coli, fermentative LDH, encoded by the ldhA gene, is induced in anaerobically grown cultures (Clark 1989; Mat-Jan et al. 1989). Expression of the ldhA gene is affected by both the redox sensor system ArcAB and the carbohydrate metabolism regulators, CsrAB and Mlc (Jiang et al. 2001). However, direct binding of the regulators to the ldhA promoter region and the regulatory mechanisms involved have not been investigated. In B. subtilis, the LDH-encoding gene, ldh, is induced together with the alsSD genes, encoding acetolactate synthase and acetolactate decarboxylase, respectively, by ArfM (Cruz Ramos et al. 2000; Marino et al. 2001). ArfM is activated via the redox-sensing regulators, ResDE and Fnr, in response to anaerobiosis (Marino et al. 2001). The induction of the genes is repressed in the presence of nitrate by YdiH (Rex; Cruz Ramos et al. 2000; Larsson et al. 2005; Reents et al. 2006). YdiH is a homolog of Rex, a redox-sensing transcriptional regulator that responds to cellular NADH to NAD+ ratio in Streptomyces coelicolor A3(2) (Brekasis and Paget 2003; Schau et al. 2004).
Corynebacterium glutamicum is a nonpathogenic high-GC Gram-positive bacterium which is widely used in the industrial production of amino acids, e.g., l-glutamate and l-lysine (Kinoshita et al. 1957; Hermann 2003). In C. glutamicum, the ldhA gene encodes LDH responsible for production of l-lactate (Inui et al. 2004). We have previously shown that both the expression level of the ldhA gene and LDH activity are increased in response to low oxygen tension at the onset of the stationary phase of growth on glucose and remain high under oxygen deprivation (Inui et al. 2007). However, the regulatory mechanism of ldhA expression in C. glutamicum has remained elusive.
In this study, SugR, a repressor of genes involved in the phosphoenolpyruvate:sugar phosphotransferase system (PTS; Engels and Wendisch 2007; Gaigalat et al. 2007; Tanaka et al. 2008), was identified as a protein binding to the promoter region of the C. glutamicum ldhA gene. By measuring both ldhA mRNA levels and LDH activity in the sugR mutant and the wild-type strain, we show that SugR represses ldhA expression. This is consistent with an independent recent study identifying the ldhA promoter as one of the targets of SugR binding by ChIP-chip analysis (Engels et al. 2008). Moreover, we here report detailed analyses of SugR binding by DNase I footprinting analyses and electrophoretic mobility shift assays (EMSAs) using various mutated ldhA promoter fragments. Analyses of ldhA expression pattern in response to sugars by lacZ reporter assays revealed the sugar-dependent regulation mechanism mediated by SugR.
Materials and methods
Bacterial strains and plasmids
The strains and plasmids used in this study are listed in Table 1.
Culture conditions
For genetic manipulation, E. coli strains were grown at 37°C in Luria-Bertani medium. C. glutamicum strains were grown at 33°C in nutrient-rich medium (A medium; Inui et al. 2007) with 4% glucose. When appropriate, the media were supplemented with antibiotics. The final antibiotic concentrations for E. coli were 50 μg of ampicillin per milliliter and 50 μg of kanamycin per milliliter; for C. glutamicum, kanamycin (50 μg ml−1) was used. Growth experiments of C. glutamicum were performed using A medium containing 1% glucose or acetate as described previously (Toyoda et al. 2008).
DNA manipulations
The oligonucleotides used in this study (Table 2) were obtained from Gene design (Osaka, Japan). Plasmid DNA was isolated with the QIAprep spin miniprep kit (Qiagen, Hilden, Germany). Chromosomal DNA was isolated from C. glutamicum with Genomic prep (GE Healthcare Bioscience, Piscataway, NJ, USA), modified by using 4 mg ml−1 lysozyme at 37°C for 2 h. Restriction endonucleases were purchased from Takara (Osaka, Japan) and used according to the manufacturer’s instructions. Polymerase chain reaction (PCR) was performed using a GeneAmp PCR system (Applied Biosystems, Foster City, CA, USA) using LA Taq polymerase (Takara) or Phusion polymerase (New England Biolabs). The resulting PCR fragments were purified with the QIAquick PCR purification kit (Qiagen). E. coli was transformed by the CaCl2 procedure, as described by Sambrook et al. (1989). C. glutamicum was transformed by electroporation as described previously (Vertès et al. 1993). The nucleotide sequence of cloned DNA fragments was determined with Genetic analyzer 3130xl (Applied Biosystems).
DNA affinity purification
DNA affinity purification of proteins binding to the ldhA promoter region was performed as described previously (Toyoda et al. 2008). The ldhA promoter region between −177 and +89 relative to the transcriptional start point (TSP) of the ldhA gene was generated by PCR using primer pldhA RV-bio, which was tagged with biotin, and primer pldhA FW (Table 2). The Dynabeads (Dynal) coupled with the ldhA promoter fragment were equilibrated with 600-μl binding buffer (50 mM Tris–HCl, pH 7.5, 1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 100 mM NaCl, 0.05% Triton-X, and 10% (vol/vol) glycerol) and incubated with cell extract for 2 h at room temperature with shaking. Unbound and nonspecifically bound proteins were removed by magnetic separation with a magnet particle concentrator (Dynal) and several washes with 600 μl of binding buffer with 120 mM NaCl. The proteins bound to the coupled Dynabeads were eluted with 0.2 and 1 M NaCl (40 μl each). Eluted fractions were collected and subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the technique of Laemmli (1970). Subsequently, the samples were electrophoretically transferred onto a Sequi-Blot polyvinylidene fluoride membrane (Bio-Rad Laboratories) using the TransBlot SD cell (Bio-Rad Laboratories). Proteins on the blot were detected by Coomassie brilliant blue staining. The N-terminal amino acid sequence of the protein bands was analyzed by Edman sequence analysis (APRO Life Science Institute, Inc, Tokushima, Japan).
Quantitative RT-PCR
Total RNA was extracted from C. glutamicum cells using the Qiagen RNeasy Mini kit (Qiagen) as described previously (Toyoda et al. 2008). Isolated RNA samples were checked for purity by both agarose gel electrophoresis and spectrophotometric analysis and stored at −80°C. Quantitative reverse-transcriptase PCR (qRT-PCR) was performed using an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems) and Power SYBR Green PCR Master Mix with MuLV Reverse Transcriptase and RNase Inhibitor of GeneAmp RNA PCR kit (Applied Biosystems) as described previously (Toyoda et al. 2008). Specific primers (Table 2) were designed using the Primer Express Software version 3.0 (Applied Biosystems). The specificity of the amplicons was checked by qRT-PCR dissociation curve analysis.
LDH assay
C. glutamicum cells were harvested by centrifugation and washed with 50 mM Tris–HCl buffer, pH 7.5, and resuspended with buffer containing 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 1 mM EDTA, 1 mM DTT, and 10% (vol/vol) of glycerol. The cells were mechanically disrupted using zirconia/silica beads as described previously (Toyoda et al. 2008), and the supernatant after centrifugation was used as crude extracts for the enzyme assay. The activity of LDH was determined as described previously (Bunch et al. 1997).
Construction of ldhA promoter-lacZ fusions
DNA fragments containing the ldhA promoter region were generated by PCR using primers pldhA FW1 and pldhA RVNaeI and cloned in front of the lacZ gene in pCRA741 (Inui et al. 2007), yielding pCRC614. Direction and sequence of the inserted fragment were confirmed by sequencing. The plasmid pCRC614 was isolated as nonmethylated DNA from E. coli JM110 for efficient gene introduction into C. glutamicum R chromosome (Vertès et al. 1993) and was subsequently integrated into the chromosome of the wild-type C. glutamicum R and sugR mutant KT1 (Toyoda et al. 2008) by markerless gene insertion methods, as described previously (Inui et al. 2007), resulting in strains KT14 and KT15, respectively. The sequence of the ldhA promoter in PldhA-lacZ fusion inserted in the chromosome was confirmed by direct sequencing of PCR fragments which were amplified using primer specific for lacZ and primer pldhA FW1 and genomic DNA, extracted from the mutants obtained, as a template.
β-galactosidase assay
β-galactosidase activity in C. glutamicum was determined as described previously (Toyoda et al. 2008). Cells permeabilized with toluene were incubated with o-nitrophenyl-β-galactoside, and activity was measured in Miller units as previously described (Miller 1972). Assays were carried out in triplicate for each sample, and results are presented as mean ± standard deviations.
Electrophoretic mobility shift assay
Cy3-labeled DNA fragments used in EMSAs were generated by PCR and purified with PCR purification kit (Qiagen). The ldhA promoter fragments P1, P2, and P3 were generated using primers pldhA FW1, pldhA FW2, and pldhA FW3, respectively, together with primer pldhA RV Cy3. DNA fragment P4 was generated using a pair of primers pldhA RV1 and pldhA FW Cy3. DNA fragments DR-I and DR-II, covering the direct repeats in the ldhA promoter, were generated using primer pairs pldhA RV2/pldhA FW Cy3 and pldhA FW4/pldhA RV Cy3, respectively. DNA fragments containing mutations in the direct repeat in the DR-I were generated using primer pairs pldhA RV2 mut1/pldhA FW Cy3 to pldhA RV2 mut4/pldhA FW Cy3. DNA fragments containing mutations in the direct repeat in the DR-II were generated with primer pairs pldhA FW4 mut1/pldhA RV Cy3 to pldhA FW4 mut4/pldhA RV Cy3. EMSA with His-tagged SugR was performed as described previously (Toyoda et al. 2008). The resulting DNA–protein complexes were loaded onto a 5% polyacrylamide gel. DNA and DNA–protein complexes were visualized by a Typhoon TRIO Variable Mode Imager (GE Healthcare Bioscience). To test for possible effectors, the protein was incubated with glucose-6-phosphate (G-6-P), fructose-1-phosphate (F-1-P), fructose-6-phosphate (F-6-P), and fructose-1,6-bisphosphate (F-1,6-P) in the binding buffer for 15 min before labeled DNA was added and the mixture was incubated for an additional 25 min at room temperature.
DNase I footprinting
DNase I footprinting was performed as described previously (Toyoda et al. 2009). Labeled DNA probes were obtained by amplification with 5′-IRD700-labeled oligonucleotides. Primer pair pldhA IRD FW/pldhA RV3 or pldhA IRD RV/pldhA FW8 was used to generate the coding- or noncoding-strand-labeled DNA fragment. DNA fragment containing the ldhA promoter region was amplified by PCR using primer pair pldhA RV3/pldhA FW8 and cloned into pGEM T-easy, yielding pCRC615. The DNA sequencing reactions were set up using the same IRD-700-labeled primers and pCRC615 as a template and a DYEnamic direct cycle sequencing kit with 7-deaza-dGTP (GE healthcare Bioscience). To test for possible effectors, the protein was preincubated with metabolites as described in the part of electrophoretic mobility shift assay; then, the IRD700-labeled probe was added and the mixture was treated as described above.
Results
Identification of proteins binding to the ldhA promoter of C. glutamicum
To obtain transcriptional regulators binding to the ldhA promoter region, we performed DNA affinity purification with magnetic streptavidin beads as described previously (Toyoda et al. 2008). A 266-bp biotinylated promoter region between position +89 and −177 relative to the TSP of the ldhA gene (Inui et al. 2007) was linked to streptavidin-coated magnetic beads and incubated with cell extracts from wild-type C. glutamicum R grown in minimal medium containing glucose as the sole carbon source. We identified five proteins by comparing the N-terminal amino acid sequences of the corresponding bands on SDS-PAGE (Fig. 1) with those of all open reading frames of C. glutamicum R (Yukawa et al. 2007). The proteins identified included three transcriptional regulators: two DeoR-type transcriptional regulators (CgR0187 and CgR1761) and a GntR-type transcriptional regulator (CgR2816). CgR1761 corresponded to SugR, a repressor of genes involved in PTS in C. glutamicum (Engels and Wendisch 2007; Gaigalat et al. 2007; Tanaka et al. 2008). The protein encoded by cgR_2816 corresponded to LldR, which has been identified as a repressor of the l-lactate utilization operon (Georgi et al. 2008). The protein encoded by cgR_0187 was a transcriptional regulator which has not yet been characterized to date. In this study, we investigated the role of SugR in regulation of ldhA expression.
SDS-PAGE of C. glutamicum proteins binding to the ldhA promoter region. Cell extracts of C. glutamicum R grown in minimal medium containing 1% glucose was incubated with DNA affinity beads and bound proteins were eluted with 1 M NaCl. The N-terminal amino acid sequence of the protein bands A to E was analyzed; A, putative gluconate kinase (CgR2896); B, DeoR-type transcriptional regulator (CgR0187) and 50S ribosomal protein L2 (CgR0611); C, SugR (CgR1761); D, ROK family sugar kinase (CgR1739); E, LldR (CgR2816). A molecular mass standard is shown to the left of the panel
Inactivation of sugR upregulates ldhA expression
SugR is a global regulator involved in sugar-dependent gene expression (Engels and Wendisch 2007; Gaigalat et al. 2007; Tanaka et al. 2008; Toyoda et al. 2008). Hence, we compared ldhA expression of the sugR mutant strain KT1 to that of the wild-type strain during growth on either glucose or acetate. Total RNA was extracted from cells grown in nutrient-rich medium (A medium) containing glucose or acetate at the mid-exponential (3 h) and at the onset of the stationary phase (6 h), and the relative mRNA levels of ldhA were determined by qRT-PCR (Fig. 2a). As previously shown (Toyoda et al. 2008), there was no marked difference in growth rate on either glucose or acetate between the KT1 and the wild-type strain. Wild-type ldhA mRNA levels on glucose were comparable to those on acetate at the exponential phase (Fig. 2a). Whereas the ldhA mRNA levels increased nearly 30-fold at the onset of the stationary phase on glucose, they did not show any changes on acetate (Fig. 2a). Strain KT1 ldhA mRNA levels were more than fivefold higher than those in the wild-type strain under all the conditions tested, except at the onset of the stationary phase on glucose, where the ldhA mRNA levels in strain KT1 were only slightly higher. The increase in the level of the ldhA mRNA in the sugR mutant was most noticeable at the onset of the stationary phase on acetate.
Effect of deletion of the sugR gene on ldhA mRNA level (a) and LDH activity (b). a The level of ldhA mRNA in the wild-type R (white) and the sugR mutant KT1 (black) during growth in nutrient-rich A medium with 1% glucose (Glc) or acetate (Ace) at the exponential (3 h) and the stationary (6 h) phases was analyzed by qRT-PCR, and the levels relative to that in the wild type grown in the presence of glucose at 3 h were determined. Mean values from three independent cultures are shown with standard deviations. b LDH activity in the wild type (white) and the sugR mutant KT1 (black) during growth under the same conditions as described for ldhA mRNA level. Mean values from at least three independent cultures are shown with standard deviations
The activity of NAD-dependent LDH in the sugR mutant KT1 grown in A medium containing either glucose or acetate was compared with that in the wild-type strain (Fig. 2b). The LDH activity was barely detectable in the wild type at the exponential phase of growth on either glucose or acetate. The wild-type LDH activity on glucose increased nearly 15-fold at the onset of the stationary phase, whereas that on acetate did not change. As observed with the level of ldhA mRNA, the LDH activity of strain KT1 was higher than that of the wild type under all the conditions examined. These results indicated that ldhA expression is dependent on the growth phase and carbon source used and that SugR has a negative effect on ldhA expression not only during the exponential phase of growth on glucose but throughout growth on acetate.
Inactivation of sugR upregulates ldhA promoter activity
To investigate the promoter activity of the ldhA gene, we integrated the ldhA promoter-lacZ translational fusion (PldhA-lacZ) into the chromosome of wild-type C. glutamicum R and sugR mutant strain KT1. The resulting strains were designated KT14 and KT15, respectively. We measured β-galactosidase activity to examine the PldhA-lacZ expression in strains KT14 and KT15 grown for 10 h in A medium containing either glucose or acetate (Fig. 3). In the presence of glucose, almost no β-galactosidase activity was detected in strain KT14 at the exponential phase, while the activity was markedly higher at the onset of the stationary phase (6 h; Fig. 3a). In the presence of acetate, no β-galactosidase activity was detected in strain KT14 throughout the cultivation period (Fig. 3b). Strain KT15 exhibited significantly higher β-galactosidase activity than strain KT14 throughout the growth on glucose. In the presence of acetate, the β-galactosidase activity of strain KT15 increased with growth of cells (Fig. 3). The results were consistent with the ldhA mRNA level and LDH activity and demonstrated that ldhA expression in cells grown on acetate is completely repressed by SugR.
Binding of SugR to the ldhA promoter region
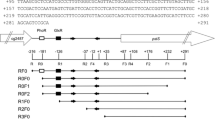
To verify the binding of SugR to the ldhA promoter region, we performed EMSAs with a DNA fragment containing the region between positions −102 and +91 relative to the TSP of ldhA and the recombinant SugR protein. The results of EMSAs showed that SugR specifically bound to the ldhA promoter (Fig. 4). To identify the SugR binding site in the ldhA promoter region, we performed EMSAs with various lengths of the ldhA promoter fragments (Fig. 4). SugR was able to bind to fragment P2 but unable to bind to fragments P3 and P4, indicating that the region between positions −31 and −11 relative to the TSP of the ldhA gene contains SugR binding site (Fig. 4).
Binding of SugR to the ldhA promoter region. a DNA fragments (P1–P4) used in the electrophoretic mobility shift assays are indicated with bars. The DNA fragments binding and not binding to SugR are indicated with black and gray bars, respectively. The ldhA gene is indicated with the box arrow and the position with respect to the transcriptional start point (+1) of ldhA is indicated. b EMSA using the ldhA promoter fragments and SugR protein. The DNA fragments used in the EMSAs are indicated at the top of the gels. Each well contained 10 nM of DNA fragment. Lanes 1 to 4 show EMSAs using 0, 100, 200, and 400 nM of SugR protein, respectively. Lane 5 in the panel of the fragment P1 shows an EMSA using 800 nM of SugR and the fragment P1. The free DNA probe and SugR–DNA complexes are indicated with white and black arrowheads, respectively. Nonspecific bands are indicated with asterisks
To determine the binding sites of SugR, DNase I footprinting assays were performed. Each of the strands of a DNA fragment encompassing the region from positions −203 to +146 with respect to the TSP was labeled, incubated with an increasing quantity of SugR, and then hydrolyzed by DNase I. The results showed that the protected region on the coding strand extended from positions −38 to +26 relative to the TSP of the ldhA gene while protected region on the noncoding strand extended from −41 to +23 (Fig. 5). In the protected region, we found two 10-bp direct repeats (5′-CGGNCATAATCGGNCATAAT-3′) between positions −26 and −7 (Fig. 6). Relevance of the motif to the SugR binding was tested by mutational analyses using EMSAs (Fig. 6). Mutations in GGNCAT of the direct repeats almost completely inhibited SugR binding to DNA (Fig. 6). In contrast, mutations in AAT of the direct repeats and three bases outside the direct repeat had almost no effect on binding of SugR. These results showed that CGGNCAT of the two direct repeats is essential for the binding of SugR to the ldhA promoter. Whereas DNA fragments containing the two direct repeats generated two SugR–DNA complexes upon binding (Fig. 4, fragment P2), deletion of one of the direct repeats resulted in single shifted band (Fig. 6, fragments DR-I and DR-II). This suggested that multiple SugR proteins bind to the direct repeats.
DNase I footprinting analysis of the interaction between SugR and the ldhA promoter regions examined on the coding and the noncoding strands. A DNA fragment (10 nM) was incubated with increasing amounts of SugR: lanes 1 and 5, no protein; lane 2, 200 nM; lane 3, 400 nM; lane 4, 800 nM. Protected regions are indicated by bars and hypersensitive sites are indicated by arrows. Positions with respect to the TSP of ldhA are indicated. The DNA sequencing reactions were set up using the same labeled primer and plasmid pCRC615
SugR binds to the two direct repeats in the ldhA promoter. a Nucleotide sequence of the two direct repeats is shown. Arrows indicate the direct repeats. The position with respect to the TSP of the ldhA gene is shown above the sequence. DNA fragments, DR-I and DR-II, used in EMSAs, are shown with bold bars. b Mutations introduced in the DR-I and DR-II fragments are listed below the wild-type sequence. The direct repeats included in the fragments are indicated with bold letters and arrows. c EMSA using the DR-I, DR-II, and their derivatives containing mutations and SugR protein. The DNA fragments used in the EMSAs are indicated at the top of the gels. Each well contained 10 nM of DNA fragment. Lanes labeled with a minus sign contains no SugR and those labeled with a plus sign contains 800 nM of SugR
Fructose-1-phosphate and fructose-1,6-bisphosphate have negative effects on binding of SugR to the ldhA promoter
As binding of SugR to the target promoters is inhibited by sugar metabolites (Engels and Wendisch 2007; Gaigalat et al. 2007; Toyoda et al. 2008), we investigated effects of the metabolites on SugR binding to the ldhA promoter by EMSAs (Fig. 7). F-1-P inhibited binding of SugR at 40 μM. F-1,6-P had an inhibitory effect on SugR binding above 20 mM. F-6-P and G-6-P, at up to 40 mM, had no effect on binding of SugR.
Effects of sugar intermediate concentrations on SugR binding to the ldhA promoter. EMSA using 10-nM DNA fragment containing the ldhA promoter (fragment DR-II in Fig. 6) and 400 nM SugR in the presence of a fructose-1-phosphate (F-1-P), b fructose-6-phosphate (F-6-P), c glucose-6-phosphate (G-6-P), and d fructose-1,6-bisphosphate (F-1,6-P) was conducted. The free DNA probe and SugR–DNA complexes are indicated with white and black arrowheads, respectively
DNase I footprinting experiments were then used to confirm these results. SugR was preincubated with different concentrations of F-1-P or other molecules to be tested prior to addition of the DNA-labeled probe. These experiments showed that SugR lost its ability to protect its target sequence with increasing F-1-P concentration (Fig. 8), whereas high concentrations of F-6-P or G-6-P did not interfere with the binding activity of SugR. The results provided confirmation that F-1-P acts as the negative effector molecule on SugR binding to the ldhA promoter.
Effect of fructose-1-phosphate (F-1-P), glucose-6-phosphate (G-6-P), and fructose-6-phosphate (F-6-P) on SugR binding to the ldhA promoter region. The experiment was performed as described in the legend for Fig. 5 for DNase I footprinting on the labeled coding and noncoding strands. Varying amounts of the metabolites to be tested were incubated with SugR before addition of labeled DNA probe. Lanes 1 and 15, no protein; lane 2, 200 nM of SugR; lanes 3–14, 400 nM of SugR; lanes 4–10, with 0.025, 0.05, 01, 0.25, 0.5, 1.0, and 2.5 mM of F-1-P; lanes 11 and 12, with 5 and 20 mM of G-6-P; lanes 13 and 14, with 5 and 20 mM of F-6-P. Protected regions are indicated by bars. Positions with respect to the TSP of ldhA are indicated
ldhA promoter activity was enhanced in the presence of fructose and sucrose
The strongest effector of SugR, F-1-P, is generated upon uptake of fructose by PtsF, fructose-specific component of PTS, during growth on fructose and sucrose but is not generated during glucose metabolism. To gain insight into effects of the sugar metabolites on SugR activity in vivo, PldhA-lacZ expression in the genetic background of the wild-type strain was examined during growth on fructose and sucrose. The β-galactosidase activity of the fructose-grown cells was higher than that of the glucose-grown cells at the exponential phase. The activity of both the fructose- and glucose-grown cells similarly increased at the onset of the stationary phase, and subsequently the activity of the fructose-grown cells decreased to the same level as that of the glucose-grown cells at the stationary phase. The PldhA-lacZ expression level of the sucrose-grown cells was comparable to that of the fructose-grown cells throughout the cultivation period. The enhanced promoter activity in fructose- or sucrose-grown cells was comparable with that in the sugR mutant strain KT15 grown on glucose throughout the growth (Figs. 9 and 3). These results suggest that repressor activity of SugR is completely lost due to F-1-P generated during the growth in the presence of fructose or sucrose but not in the presence of glucose.
Discussion
In this study, we identified SugR as a protein binding to the ldhA promoter and examined the involvement of the protein in the regulation of ldhA expression in response to sugar. Analyses of the LDH activity, the ldhA mRNA level, and the ldhA promoter-lacZ fusion revealed that ldhA expression is markedly induced at the onset of the stationary phase only in the presence of sugar, while the induction is completely repressed in the absence of sugar (Fig. 3). The results indicate that the presence of sugar is essential for the induction of ldhA expression, although, as previously shown (Inui et al. 2007), the expression is also controlled by oxygen tension. Our observation that upregulation of the ldhA mRNA level by disruption of the sugR gene in the absence of sugar is consistent with recent studies of SugR regulon defined by transcriptome analyses (Gaigalat et al. 2007) and by ChIP-chip analyses (Engels et al. 2008). These findings indicate that SugR acts as a transcriptional repressor of ldhA, in addition to PTS sugar uptake genes and glycolytic genes (Engels and Wendisch 2007; Gaigalat et al. 2007; Engels et al. 2008; Tanaka et al. 2008; Toyoda et al. 2008), and that the presence of sugar alleviates the repression to induce the target genes coordinately (Fig. 10). The coordinated regulation will allow the cells to efficiently utilize sugar in response to environmental conditions. As demonstrated in this study, detailed characteristics of SugR binding to the ldhA promoter in vitro and expression patterns of the ldhA promoter-lacZ fusion in vivo provided further experimental evidence to elucidate the sugar response mechanism of ldhA transcription mediated by SugR.
Model of regulation of sugar uptake (PtsG and PtsF), GAPDH (GapA), and LDH (LdhA) by SugR. Uptake of glucose and fructose and the glycolytic pathway are depicted. Bold blunted lines indicate repression of gene expression by SugR. Dashed curved lines with bars indicate negative effect of sugar phosphates on repressor activity of SugR
Repressor activity of SugR is controlled by sugar phosphates, including F-1-P and F-1,6-P, while effects of the molecules on binding of SugR are different among the target promoters studied. As in the case of the gapA promoter, F-1-P and F-1,6-P have a negative effect on SugR binding to the ldhA promoter, and the effect of F-1-P is much stronger than that of F-1,6-P (Fig. 7; Toyoda et al. 2008). F-6-P and G-6-P have no effect on SugR binding to the promoters of gapA and ldhA (Fig. 7; Toyoda et al. 2008). In addition to F-1-P and F-1,6-P, G-6-P also has a negative effect on SugR binding to the intergenic region between ptsI and ptsF operon (Gaigalat et al. 2007). As for the ptsG promoter, only F-6-P inhibits SugR binding (Engels and Wendisch 2007). Although cause of the difference in the effects is not clear, the distinct effects may modulate sugar-dependent global regulation mediated by SugR. F-1-P is generated upon uptake of fructose by PtsF, fructose-specific component of PTS. F-1-P is also generated during growth on sucrose in C. glutamicum because C. glutamicum requires PtsF to metabolize sucrose completely due to the absence of fructokinase activity to phosphorylate fructose liberated from sucrose (Dominguez and Lindley 1996). On the other hand, glucose is phosphorylated to G-6-P upon uptake by PtsG and subsequently metabolized through glycolysis to F-6-P and F-1,6-P. The ldhA promoter activity during growth on either fructose or sucrose was higher than that on glucose (Fig. 9), supporting the result that F-1-P acts as the strongest negative effector of SugR in vitro. During growth on fructose, the ldhA promoter is active even at the exponential phase where almost no promoter activity is detected during growth on glucose (Fig. 9). l-lactate production has been observed even under fully aerobic conditions during growth on fructose in C. glutamicum (Péquignot et al. 1997; Dominguez et al. 1998; Kiefer et al. 2002). This could be attributed in part to inactivation of repressor activity of SugR by F-1-P. In many bacteria, expression of fermentative LDH is induced under oxygen limitation in the absence of external electron acceptor. Although sugars are essential for growth under fermentative conditions, studies on sugar response of ldhA expression are limited. In E. coli, carbohydrate metabolism regulators, CsrAB and Mlc, are indirectly involved in regulation of ldhA (Jiang et al. 2001), although direct regulators are not known so far. In Streptococcus thermophilus and Streptococcus bovis, CcpA, a global regulator involved in catabolite repression in low-GC Gram-positive bacteria, is involved in upregulation of ldh in response to sugar provided (van den Bogaard et al. 2000; Asanuma et al. 2004).
DNase I footprinting analyses and EMSAs show that the 10-bp direct repeats, located between positions −7 and −26 with respect to the TSP, are required and enough for SugR binding to the ldhA promoter region (Figs. 4, 5, and 6). Based on the consensus sequence, TC(A/G)(A/G)ACA, proposed by comparing the promoter sequences of the target genes, two putative SugR binding sites are suggested to be located in the ldhA promoter region between positions −35 and −42 and between −21 and −28 with respect to the TSP (Engels et al. 2008). However, SugR did not bind to the distal site (between positions −35 and −42), while the proximal site is included in the direct repeats. Each half site of the direct repeats (CGGNCATAAT) was able to bind to SugR (Fig. 6). In addition, a part of the direct repeats (ATCGGGCATAAT), which was not proposed to be the binding site by Engels et al. (2008), is perfectly conserved in the upstream region of ldhA in C. glutamicum ATCC 13032 (NC_003450), Corynebacterium efficiens YS-314 (NC_004369), and Corynebacterium diphtheriae NCTC 13129 (NC_002935), indicating the importance of the sequence in control of the promoter activity in these organisms. The mutational analyses using EMSA show that the sequence, GGNCAT, in the direct repeats is essential for SugR binding (Fig. 6). This is consistent with the consensus sequence proposed (Engels et al. 2008). The location of the SugR binding sites indicates that SugR represses ldhA expression by preventing RNA polymerase from binding to the promoter region. It should be noted that the SugR binding site is not conserved in the ldhA promoter region of Corynebacterium jeikeium K411 (NC_007164). This might be linked to the absence of components of PTS in its genome (Tauch et al. 2005).
To avoid wasting NADH by LDH, ldhA expression should be controlled in response not only to sugar by SugR but also to redox balance in the cell by unknown transcriptional regulators. This might be supported by the fact that increase in the ldhA promoter activity at the onset of the stationary phase was observed in the sugR mutant as well as in the wild type (Fig. 3). In B. subtilis, transcriptional regulators controlling anaerobic respiration and fermentation (ResDE, Fnr, ArfM, and Rex) are involved in ldh expression (Cruz Ramos et al. 2000; Larsson et al. 2005; Wang et al. 2008). In E. coli, a redox sensor system, ArcAB, is also involved in ldhA expression (Jiang et al. 2001). Although open reading frames encoding putative transcriptional regulators and two-component systems are found in the genome of C. glutamicum, those involved in a redox-sensing mechanism have not been reported so far. In addition to SugR, we obtained two other transcriptional regulators, CgR0187 and CgR2816 (Fig. 1), but these regulators have no relevance to the redox-sensing systems of E. coli and B. subtilis. Independently, GlxR, a CRP-type transcriptional regulator controlling various physiological reactions (Kim et al. 2004; Letek et al. 2006; Jungwirth et al. 2008), was recently shown to bind to the ldhA promoter region at position −62 with respect to the TSP in vitro (Kohl et al. 2008). It is suggested that GlxR plays a positive role on the ldhA expression due to the location of the binding site (Kohl et al. 2008). These findings indicate that regulation of ldhA is under the control of complex transcriptional regulatory network.
References
Asanuma N, Yoshii T, Hino T (2004) Molecular characterization of CcpA and involvement of this protein in transcriptional regulation of lactate dehydrogenase and pyruvate formate-lyase in the ruminal bacterium Streptococcus bovis. Appl Environ Microbiol 70:5244–5251
Brekasis D, Paget M (2003) A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2). EMBO J 22:4856–4865
Bunch PK, Mat-Jan F, Lee N, Clark DP (1997) The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143:187–195
Clark DP (1989) The fermentation pathways of Escherichia coli. FEMS Microbiol Rev 63:223–234
Cruz Ramos H, Hoffmann T, Marino M, Nedjari H, Presecan-Siedel E, Dreesen O, Glaser P, Jahn D (2000) Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression. J Bacteriol 182:3072–3080
Dominguez H, Lindley ND (1996) Complete sucrose metabolism requires fructose phosphotransferase activity in Corynebacterium glutamicum to ensure phosphorylation of liberated fructose. Appl Environ Microbiol 62:3878–3880
Dominguez H, Rollin C, Guyonvarch A, Guerquin-Kern JL, Cocaign-Bousquet M, Lindley ND (1998) Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur J Biochem 254:96–102
Engels V, Wendisch VF (2007) The DeoR-Type regulator SugR represses expression of ptsG in Corynebacterium glutamicum. J Bacteriol 189:2955–2966
Engels V, Lindner S, Wendisch VF (2008) The global repressor SugR controls expression of genes of glycolysis and of the l-lactate dehydrogenase LdhA in Corynebacterium glutamicum. J Bacteriol 190:8033–8044
Gaigalat L, Schlueter JP, Hartmann M, Mormann S, Tauch A, Pühler A, Kalinowski J (2007) The DeoR-type transcriptional regulator SugR acts as a repressor for genes encoding the phosphoenolpyruvate:sugar phosphotransferase system (PTS) in Corynebacterium glutamicum. BMC Mol Biol 8:104
Georgi T, Engels V, Wendisch VF (2008) Regulation of l-lactate utilization by the FadR-type regulator LldR of Corynebacterium glutamicum. J Bacteriol 190:963–971
Hermann T (2003) Industrial production of amino acids by coryneform bacteria. J Biotechnol 104:155–172
Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H (2004) Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol 7:182–196
Inui M, Suda M, Okino S, Nonaka H, Puskás LG, Vertès AA, Yukawa H (2007) Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 153:2491–2504
Jiang GR, Nikolova S, Clark DP (2001) Regulation of the ldhA gene, encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 147:2437–2446
Jungwirth B, Emer D, Brune I, Hansmeier N, Pühler A, Eikmanns BJ, Tauch A (2008) Triple transcriptional control of the resuscitation promoting factor 2 (rpf2) gene of Corynebacterium glutamicum by the regulators of acetate metabolism RamA and RamB and the cAMP-dependent regulator GlxR. FEMS Microbiol Lett 281:190–197
Kiefer P, Heinzle E, Wittmann C (2002) Influence of glucose, fructose and sucrose as carbon sources on kinetics and stoichiometry of lysine production by Corynebacterium glutamicum. J Ind Microbiol Biotechnol 28:338–343
Kim HJ, Kim TH, Kim Y, Lee HS (2004) Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J Bacteriol 186:3453–3460
Kinoshita S, Udaka S, Shimono M (1957) Studies on the amino acid fermentation. Part 1. Production of l-glutamic acid by various microorganisms. J Gen Appl Microbiol 3:193–205
Kohl TA, Baumbach J, Jungwirth B, Pühler A, Tauch A (2008) The GlxR regulon of the amino acid producer Corynebacterium glutamicum: in silico and in vitro detection of DNA binding sites of a global transcription regulator. J Biotechnol 135:340–350
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Larsson JT, Rogstam A, von Wachenfeldt C (2005) Coordinated patterns of cytochrome bd and lactate dehydrogenase expression in Bacillus subtilis. Microbiology 151:3323–3335
Letek M, Valbuena N, Ramos A, Ordóñez E, Gil JA, Mateos LM (2006) Characterization and use of catabolite-repressed promoters from gluconate genes in Corynebacterium glutamicum. J Bacteriol 188:409–423
Marino M, Ramos HC, Hoffmann T, Glaser P, Jahn D (2001) Modulation of anaerobic energy metabolism of Bacillus subtilis by arfM (ywiD). J Bacteriol 183:6815–6821
Mat-Jan F, Alam KY, Clark DP (1989) Mutants of Escherichia coli deficient in the fermentative lactate dehydrogenase. J Bacteriol 171:342–348
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Péquignot C, Dussap CG, Gros JB (1997) Intra- and extracellular concentrations of glutamate, lactate and acetate during growth of Corynebacterium glutamicum on different media. J Ind Microbiol Biotechnol 18:312–318
Reents H, Munch R, Dammeyer T, Jahn D, Härtig E (2006) The Fnr Regulon of Bacillus subtilis. J Bacteriol 188:1103–1112
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schau M, Chen Y, Hulett FM (2004) Bacillus subtilis YdiH is a direct negative regulator of the cydABCD operon. J Bacteriol 186:4585–4595
Studier FW, Moffatt BA (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130
Tanaka Y, Teramoto H, Inui M, Yukawa H (2008) Regulation of expression of general components of the phosphoenolpyruvate: carbohydrate phosphotransferase system (PTS) by the global regulator SugR in Corynebacterium glutamicum. Appl Microbiol Biotechnol 78:309–318
Tauch A, Kaiser O, Hain T, Goesmann A, Weisshaar B, Albersmeier A, Bekel T, Bischoff N, Brune I, Chakraborty T, Kalinowski J, Meyer F, Rupp O, Schneiker S, Viehoever P, Pühler A (2005) Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J Bacteriol 187:4671–4682
Toyoda K, Teramoto H, Inui M, Yukawa H (2008) Expression of the gapA gene encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum is regulated by the global regulator SugR. Appl Microbiol Biotechnol 81:291–301
Toyoda K, Teramoto H, Inui M, Yukawa H (2009) Involvement of the LuxR-type transcriptional regulator, RamA, in regulation of expression of the gapA gene encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum. J Bacteriol 191:968–977
van den Bogaard PTC, Kleerebezem M, Kuipers OP, de Vos WM (2000) Control of lactose transport, β-galactosidase activity, and glycolysis by CcpA in Streptococcus thermophilus: evidence for carbon catabolite repression by a non-phosphoenolpyruvate-dependent phosphotransferase system sugar. J Bacteriol 182:5982–5989
Vertès AA, Inui M, Kobayashi M, Kurusu Y, Yukawa H (1993) Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res Microbiol 144:181–185
Wang E, Bauer MC, Rogstam A, Linse S, Logan DT, von Wachenfeldt C (2008) Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol Microbiol 69:466–478
Yukawa H, Omumasaba CA, Nonaka H, Kós P, Okai N, Suzuki N, Suda M, Tsuge Y, Watanabe J, Ikeda Y, Vertès AA, Inui M (2007) Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 153:1042–1058
Acknowledgements
We thank Crispinus A. Omumasaba (RITE) for critical reading of the manuscript. This work was partially supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Toyoda, K., Teramoto, H., Inui, M. et al. Molecular mechanism of SugR-mediated sugar-dependent expression of the ldhA gene encoding l-lactate dehydrogenase in Corynebacterium glutamicum . Appl Microbiol Biotechnol 83, 315–327 (2009). https://doi.org/10.1007/s00253-009-1887-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-1887-x