Abstract
Powdery mildew is one of the most devastating diseases of wheat in areas with cool and maritime climates. Chinese wheat landrace Baihulu confers a high level of resistance against a wide range of Blumeria graminis DC f. sp. tritici (Bgt) races, especially those currently prevailing in Shaanxi. The objectives of this study were to determine the chromosome bin location of the mlbhl gene from Baihulu and its allelism with Pm24. To investigate the inheritance of powdery mildew resistance and detect adjacent molecular markers, we constructed a segregating population of 301 F2 plants and corresponding F2:3 families derived from Baihulu/Shaanyou 225. Genetic analysis revealed that a single dominant gene was responsible for seedling stage powdery mildew resistance in Baihulu. A genetic map comprising Xgwm106, Xgwm337, Xgwm1675, Xgwm603, Xgwm789, Xbarc229, Xgpw4503, Xcfd72, Xcfd83, Xcfd59, Xcfd19, and mlbhl spanned 28.2 cM on chromosome 1D. Xgwm603/Xgwm789 and Xbarc229 were flanking markers tightly linked to mlbhl at genetic distances of 1.5 and 1.0 cM, respectively. The mlbhl locus was located in chromosome bin 1DS 0.59–1.00 delimited by the SSR markers Xgwm337 and Xbarc229. When tested with a differential array of 23 Bgt isolates Baihulu displayed a response pattern that was clearly distinguishable from that of Chiyacao and varieties or lines possessing documented Pm genes. Allelism analysis indicated that mlbhl is a new gene, either allelic or closely linked with Pm24. The new gene was designated Pm24b.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Powdery mildew of common wheat (Triticum aestivum L. em Thell.), caused by Blumeria graminis (DC.) E. O. Speer f. sp. tritici, is a serious fungal disease in the wheat growing areas of the world. Deployment of resistant cultivars provides an effective approach for disease control, eliminates the use of fungicides and minimizes crop losses. To date, more than 60 genes conferring resistance to powdery mildew have been identified at 41 gene loci (Pm1-Pm45) (http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2011.pdf) in wheat and its wild relatives. Five of these loci (Pm1a-Pm1e, Pm3a-Pm3k, Pm4a-Pm4d, Pm5a-Pm5e and Pm8/Pm17) have multiple alleles (Hsam and Zeller 1997; Huang et al. 2003; Singrün et al. 2003; Yahiaoui et al. 2009; Schmolke et al. 2012). A significant problem is that powdery mildew resistance genes are frequently overcome by virulent races within a short time period after introduction to agriculture. Consequently, it is necessary to search for new powdery mildew resistance genes/alleles and to pyramid several resistance genes in a single cultivar using linked markers, to achieve effective resistance.
The most widely used resistance sources in wheat are the wild relatives. Introgressions from relatives may involve large alien segments that are barriers to recombination. Wheat landraces, on the other hand, are valuable sources of resistance genes, especially in regard to ease of transfer in breeding programs. Three formally designated powdery mildew resistance genes/alleles (Pm5d, Pm5e and Pm24) (Huang et al. 2003; Nematollahi et al. 2008; Huang and Röder 2011) have been identified from Chinese wheat landraces. Indeed, there are numerous unidentified genes for resistance to a wide range of diseases in wheat landraces (Zeven 1998; Newton et al. 2010).
As part of a program to identify powdery mildew resistance in Chinese wheat landraces, we assembled a collection of more than 1,000 accessions, which were screened with mixed Bgt isolates or by exposure to natural infection in the field. A more detailed analysis of selected landraces revealed that some might possess novel genes or alleles that have not been identified. Baihulu is one of them. A previous study located powdery mildew resistance gene mlbhl on chromosome 1D in Baihulu and showed that it was linked to marker Xgwm337 (Zhao et al. 2010). The major objectives of this study were to (1) map gene mlbhl by means of molecular markers and (2) investigate allelism between mlbhl and Pm24.
Materials and methods
Plant materials
The mapping population derived from a cross between Baihulu and Shaanyou 225 comprised 301 F2 plants and the derived F3 families. Fifteen plants of each F3 family were tested to identify the genotypes of corresponding F2 plants. Shaanyou 225 and Chancellor which carry no known powdery mildew resistance genes were used as susceptible controls in the disease reaction tests. Baihulu served as a resistant parent and resistant control.
Chinese Spring (CS) and chromosome 1D deletion lines (Table 1) kindly provided by Drs Takashi Endo and Shuhei Nasuda, Kyoto University, Japan, were used for chromosomal arm assignment and bin mapping of the molecular markers. A set of 40 differential wheat genotypes (Table 2) possessing known powdery mildew resistance genes were evaluated for response to 23 Bgt isolates. Chinese wheat landrace Chiyacao carrying resistance gene Pm24, together with Baihulu and a total of 560 F2 plants from the cross Baihulu/Chiyacao were employed to test the allelism of mlbhl and Pm24.
B. graminis f. sp. tritici pathotypes
Twenty-three Bgt differential isolates were used to postulate and differentiate resistance genes mlbhl and Pm24. The Bgt isolates were collected from different areas of China and purified at least twice from single colonies. They were continuously maintained in isolation on seedlings of Chancellor at the institute of Plant Protection, CAAS, Beijing. Isolate E09 was used in host genetic studies as it is avirulent on both Chiyacao and Baihulu. The parents, F2 population, and F2:3 progenies from Baihulu/Shaanyou 225 were inoculated with isolate E09, which was also used to evaluate the F2 population from Baihulu/Chiyacao.
Scoring for powdery mildew response
Seedlings were grown in the greenhouse at 16–20 °C and inoculated when they were at the 2- to 3-leaf stage. Seedlings were inoculated by dusting conidia from infected seedlings of Chancellor. Approximately 12 days later, infection types were scored when the control varieties Shaanyou 225 and Chancellor were fully infected. Infection types (IT) were scored on a scale of zero to four (Sheng 1988). For analysis two main classes of host reactions were distinguished as resistant (IT = 0, 0;, 1 and 2) and susceptible (IT = 3 and 4).
Molecular marker analysis
Genomic DNA of parents and progenies were extracted from seedling leaves according to the CTAB protocol (Saghai-Maroof et al. 1984). Molecular markers were evaluated by bulked segregant analysis (BSA) (Michelmore et al. 1991) to identify markers linked to the powdery mildew resistance gene in Baihulu. Resistant and susceptible bulks were made by pooling equal amounts of DNA from ten resistant and ten susceptible F2 plants, respectively. Wheat SSR markers located on chromosome 1D were selected from primer sets of the GWM (Röder et al. 1998), WMC (Somers et al. 2004), BARC (Song et al. 2005), CFD (Guyomarc’h et al. 2002), and GPW series (http://wheat.pw.usda.gov/ggpages/SSRclub/GeneticPhysical/). Previously published wheat genetic maps of EST-SSR markers (CWEM set) (Peng and Lapitan 2005) and EST-STS markers (MAG set) (Xue et al. 2008) assigned to chromosome 1DS were also screened for polymorphisms between the parents and bulks. Polymorphic markers were further checked for linkage to mlbhl, using the entire F2:3 mapping population. Primer sequences were obtained from the GrainGenes database (http://wheat.pw.usda.gov/GG2/index.shtml). Unpublished primer sequences Xgwm603 and Xgwm789 were kindly provided by Dr. M. Röder, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Gatersleben, Germany. Xgwm1675 and Xgwm1291 were kindly provided by Dr. M. Ganal, TraitGenetics GmbH, Gatersleben, Germany.
The PCR were done in total volumes of 10 μL, including 1× PCR reaction buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.25 U Taq DNA polymerase, 0.5 μM of each primer, and 40–100 ng total DNA. PCR was performed in an S1000 thermocycler (Bio-Rad, California, USA) using the following program: initial denaturation at 94 °C for 3 min, followed by 35 cycles of 30 s at 94 °C, 45 s at 50–60 °C (based on the primer information from the GrainGenes database), 50 s at 72 °C, and a final extension step of 10 min at 72 °C before cooling to 4 °C. PCR products were resolved in 8 % non-denaturing polyacrylamide gel (37.5 acrylamide: 1 bisacrylamide) electrophoresis and visualized by silver staining.
Physical bin mapping
The chromosome bin assignments of markers linked to the resistance gene on chromosome arm 1DS were obtained using CS and CS chromosome 1D deletion lines produced by Endo and Gill (1996).
Data analysis
Chi-squared tests for goodness of fit were used to test for deviation of observed segregation ratios from theoretic Mendelian ratios by SAS 8.0 software. Linkages between markers and the resistance gene were established using JoinMap 4.0 (http://www.kyazma.nl/index.php/mc.JoinMap/sc.General/), with a LOD threshold of 3.0. The genetic map was drawn with the software Mapdraw V2.1 (Liu and Meng 2003).
Results
Inheritance of powdery mildew resistance in Baihulu
Seedlings of 301 F2 plants and the derived F2:3 families produced from the cross Baihulu/Shaanyou 225, and parents were inoculated with Bgt isolate E09. Shaanyou 225 was susceptible with IT 4, whereas Baihulu was highly resistant with IT 0, 0; and 1 (necrotic or small pustules). F2 segregation of 223:78 was consistent with a 3:1 ratio (\( \chi^{2} = 0.134,P = 0.714, \, \chi^{2}_{0.05, \, 1} = 3.841 \)). Similarly, the observed ratio of the F2:3 families (73 homozygous resistant:150 segregating:78 homozygous susceptible) fitted the expected 1:2:1 ratio (\( \chi^{2} = 0.169,P = 0.919, \, \chi^{2}_{0.05, 2} = 5.991 \)). The combined F2 and F3 family data indicated that resistance in Baihulu was conferred by a single dominant gene.
Molecular marker analysis and chromosome bin location
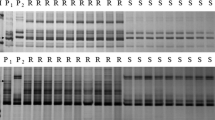
To determine the chromosomal bin location of mlbhl, resistant and susceptible DNA bulks as well as the parents were initially employed in BSA using 61 SSR markers located on chromosome 1D. Markers Xgwm106, Xgwm337, Xgwm1675, Xgwm603, Xgwm789, Xbarc229, Xgpw4503, Xcfd72, Xcfd83, Xcfd59 and Xcfd19 were polymorphic between the parents and bulks and showed expected patterns for linked markers after being tested on individual F2 plants. All were inherited co-dominantly (Fig. 1). A linkage map of gene mlbhl and closely linked markers with a total map length of 28.2 cM are shown in Fig. 2. Gene mlbhl was flanked by loci Xgwm603/Xgwm789 and Xbarc229 at genetic distances of 1.5 and 1.0 cM, respectively. Xgwm1291, that co-segregated with Pm24 in earlier work (Huang and Röder 2011), was not polymorphic between Baihulu and Shaanyou 225. None of the eight EST-based markers previously mapped on 1DS showed linkage with mlbhl.
Polyacrylamide gel electrophoresis of PCR products amplified with mlbhl linked polymorphic markers Xgwm337, Xgwm603, Xgwm789, Xbarc229, Xcfd83 and Xcfd72 in the F2 population of Baihulu/Shaanyou 225. 1 Baihulu, 2 Shaanyou 225, 3 resistant bulk, 4 susceptible bulk, 5–10 homozygous susceptible individuals, 11–16 heterozygous resistant individuals, 17–22 homozygous resistant individuals. Arrows indicate the polymorphic amplification products. M D2000 ladder
CS and CS deletion lines of chromosome 1D were used to locate flanking markers Xgwm337 and Xbarc229, and therefore mlbhl, to a specific segment of the chromosome arm. Fragments of both Xgwm337 and Xbarc229 were missing in 1DS-01 and 1DS-03, and Xgwm337 was also missing in 1DS-05, indicating that Xgwm337 was located in the distal deletion bin 1DS-05 (0.70–1.00), whereas Xbarc229 was in bin 1DS-01 (0.59–0.70) (Fig. 3); thus mlbhl was placed in bin 0.59–1.00. A comparison with published genetic maps (Huang and Röder 2011) suggested that mlbhl was located in the same or a similar chromosomal region to Pm24 (Fig. 2).
The relationship of Pm24 and mlbhl
A comparison of the responses of Baihulu and Chiyacao and lines possessing other identified resistance genes when inoculated with the 23 differential isolates is shown in Table 2. Baihulu was resistant to 14 isolates and susceptible to nine. The responses of Baihulu and Chiyacao differed for six isolates: five avirulent to the former and one avirulent to the latter. Thus the resistance gene in Baihulu was different from Pm24, as well as being different from all other genes represented in Table 2.
All 560 F2 plants from Baihulu/Chiyacao, inoculated with isolate E09 were resistant. On the basis of these results, and the fact that the molecular maps for the present work and for Pm24 (Huang and Röder 2011) are very similar, we propose that the resistance gene in Baihulu be designated Pm24b and that the former Pm24 be designated Pm24a.
Discussion
Many landraces have tolerances to biotic and abiotic stresses (Zeven 1998). Wheat landraces are abundant in China. Useful traits can be easily transferred from landraces to elite common wheat cultivars. To date, three powdery mildew resistance genes have been identified in Chinese wheat landraces, viz. Pm24 (Chiyacao), Pm5d (CI 10904), and Pm5e (Fuzhuang 30) (Huang et al. 2000b, 2003; Nematollahi et al. 2008). In addition to formally named powdery mildew resistance genes, other Chinese wheat landraces were reported to have powdery mildew resistance genes, temporarily designated as mlxbd (Xiaobaidong) and PmH (Hongquanmang) (Huang et al. 2000a; Zhou et al. 2005). Further resistant genotypes were identified among 3,000 landraces, some of which are immune or highly resistant, such as Youbailan, Bensanyuehuang, Mangmai, Baiyouyantiao, Shangeda, Hongtutou, Hongkemai, and Mazhamai (Duan unpublished; Sheng et al. 1992; Xue et al. 2009). Gene Pm24b named in this study was present in a largely unexploited reservoir of Chinese wheat landraces. This new gene is highly effective in the field in Shaanxi. Despite its inferior agronomic traits (e.g. low grain yield, lodging), Baihulu can serve as a valuable resistance resource for common wheat improvement.
So far, Pm24 is the only known wheat powdery mildew resistance locus that was located on chromosome arm 1DS. It was initially mapped in the vicinity of the centromere (Huang et al. 2000b), but was later assigned to bin 1DS5-0.70–1.00 (Huang and Röder 2011). The markers flanking Pm24a were Xgwm789/Xgwm603 and Xbarc229 at 2.4 and 3.6 cM, respectively. It was also near Xgwm337 and co-segregated with Xgwm1291 (Huang and Röder 2011). In our research a genetic map containing 11 markers linked to Pm24b was constructed and markers Xgwm337 and Xbarc229 flanked it at genetic distances of 3.7 and 1.0 cM, respectively. According to Sourdille et al. (2004) these markers are close to the 1D centromere, but in the present study the Pm24b region was shown to be in deletion bin 1DS-0.59–1.00 (Fig. 3). Therefore, Pm24 must be physically located in the distal region of chromosome 1DS.
We investigated the response spectra of 40 wheat genotypes including a set of differentials carrying known resistance genes. The response arrays of Baihulu and Chiyacao were different from all other reference sources and from each other. The lack of segregation in the intercross, and the very similar map locations of both genes, indicated they are allelic or very closely linked. The present study provides a springboard for fine mapping of Pm24b and for its use in wheat breeding programs using the closely linked markers to combine it with other resistance genes as a means of achieving durable resistance.
References
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307
Guyomarc’h H, Sourdille P, Charmet G, Edwards K, Bernard M (2002) Characterisation of polymorphic microsatellite markers from Aegilops tauschii and transferability to the D-genome of bread wheat. Theor Appl Genet 104:1164–1172
Hsam SLK, Zeller FJ (1997) Evidence of allelism between genes Pm8 and Pm17 and chromosomal location of powdery mildew and leaf rust resistance genes in the common wheat cultivar ‘Amigo’. Plant Breed 116:119–122
Huang XQ, Röder MS (2011) High-density genetic and physical bin mapping of wheat chromosome 1D reveals that the powdery mildew resistance gene Pm24 is located in a highly recombinogenic region. Genetica 139:1179–1187
Huang XQ, Hsam SLK, Zeller FJ (2000a) Chromosomal location of powdery mildew resistance genes in Chinese wheat (Triticum aestivum L. em. Thell.) landraces Xiaobaidong and Fuzhuang 30. J Genet Breed 54:311–317
Huang XQ, Hsam SLK, Zeller FJ, Wenzel G, Mohler V (2000b) Molecular mapping of the wheat powdery mildew resistance gene Pm24 and marker validation for molecular breeding. Theor Appl Genet 101:407–414
Huang XQ, Wang LX, Xu MX, Röder MS (2003) Microsatellite mapping of the powdery mildew resistance gene Pm5e in common wheat (Triticum aestivum L.). Theor Appl Genet 106:858–865
Liu R, Meng J (2003) MapDraw: a Microsoft Excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas 25:317–321
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Nematollahi G, Mohler V, Wenzel G, Zeller FJ, Hsam SLK (2008) Microsatellite mapping of powdery mildew resistance allele Pm5d from common wheat line IGV1-455. Euphytica 159:307–313
Newton AC, Akar T, Baresel JP, Bebeli PJ, Bettencourt E, Bladenopoulos KV, Czembor JH, Fasoula DA, Katsiotis A, Koutis K, Koutsika-Sotiriou M, Kovacs G, Larsson H, de Carvalho MAAP, Rubiales D, Russell J, Dos Santos TMM, Patto MCV (2010) Cereal landraces for sustainable agriculture. A review. Agron Sustain Dev 30:237–269
Peng JH, Lapitan NLV (2005) Characterization of EST-derived microsatellites in the wheat genome and development of eSSR markers. Funct Integr Genomics 5:80–96
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Schmolke M, Mohler V, Hartl L, Zeller FJ, Hsam SLK (2012) A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum). Mol Breed 29:449–456
Sheng BQ (1988) Grades of resistance to powdery mildew classified by different phenotypes of response in the seedling stage of wheat. Plant Prot 1:49
Sheng BQ, Duan XY, Zhou YL, Wang JX (1992) Cluster of powdery mildew resistance genes carried in some Chinese wheat landraces. Crop Genet Resour 4:33–35
Singrün C, Hsam SLK, Hartl L, Zeller FJ, Mohler V (2003) Powdery mildew resistance gene Pm22 in cultivar Virest is a member of the complex Pm1 locus in common wheat (Triticum aestivum L. em Thell.). Theor Appl Genet 106:1420–1424
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109:1105–1114
Song QJ, Shi JR, Singh S, Fickus EW, Costa JM, Lewis J, Gill BS, Ward R, Cregan PB (2005) Development and mapping of microsatellite (SSR) markers in wheat. Theor Appl Genet 110:550–560
Sourdille P, Singh S, Cadalen T, Brown-Guedira GL, Gay G, Qi L, Gill BS, Dufour P, Murigneux A, Bernard M (2004) Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct Integr Genomics 4:12–25
Xue S, Zhang Z, Lin F, Kong Z, Cao Y, Li C, Yi H, Mei M, Zhu H, Wu J, Xu H, Zhao D, Tian D, Zhang C, Ma Z (2008) A high-density intervarietal map of the wheat genome enriched with markers derived from expressed sequence tags. Theor Appl Genet 117:181–189
Xue F, Duan XY, Zhou YL, Ji WQ (2009) Postulation of powdery mildew resistant genes carried in some Chinese wheat landraces and the genetic diversity analysis. J Triticeae Crop 29:228–235
Yahiaoui N, Kaur N, Keller B (2009) Independent evolution of functional Pm3 resistance genes in wild tetraploid wheat and domesticated bread wheat. Plant J 57:846–856
Zeven AC (1998) Landraces: a review of definitions and classifications. Euphytica 104:127–139
Zhao NJ, Xue F, Wang CY, Han JR, JI WQ, Zhen L (2010) SSR analysis of powdery mildew resistance gene in Chinese wheat landrace Baihulu. J Triticeae Crop 30:411–414
Zhou R, Zhu Z, Kong X, Huo N, Tian Q, Li P, Jin C, Dong Y, Jia J (2005) Development of wheat near-isogenic lines for powdery mildew resistance. Theor Appl Genet 110:640–648
Acknowledgments
This research was partially supported by National 863 Program (2011AA100103) and National 973 Program (2009CB118301) of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by F. Ordon.
F. Xue and C. Wang equally contributed to this article.
Rights and permissions
About this article
Cite this article
Xue, F., Wang, C., Li, C. et al. Molecular mapping of a powdery mildew resistance gene in common wheat landrace Baihulu and its allelism with Pm24 . Theor Appl Genet 125, 1425–1432 (2012). https://doi.org/10.1007/s00122-012-1923-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-012-1923-6