Abstract
The powdery mildew resistance allele Pm5d in the backcross-derived wheat lines IGV1-455 (CI10904/7*Prins) and IGV1-556 (CI10904/7*Starke) shows a wide spectrum of resistance and virulent pathotypes have not yet been detected in Germany. Although this allele may be distinguished from the other documented Pm5 alleles by employing a differential set of Blumeria graminis tritici isolates, the use of linked molecular markers could enhance selection, especially for gene pyramiding. Pm5d was genetically mapped relative to six microsatellite markers in the distal part of chromosome 7BL using 82 F3 families of the cross Chinese Spring × IGV1-455. Microsatellite-based deletion line mapping placed Pm5d in the terminal 14% of chromosome 7BL. The closely linked microsatellite markers Xgwm577 and Xwmc581 showed useful variation for distinguishing the different Pm5 alleles except the ones originating from Chinese wheat germplasm. Their use, however, would be limited to particular crosses because they are not functional markers. The occurrence of resistance genes closely linked to the Pm5 locus is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Powdery mildew caused by Blumeria graminis f.sp. tritici is one of the most destructive foliar diseases of common wheat worldwide and is particularly prevalent in areas with cool or maritime climates. Breeding of resistant cultivars is the most economical and environmentally sound method to decrease the use of fungicides and to reduce crop losses due to this disease. Currently, 38 major host resistance genes are documented (Miranda et al. 2006; McIntosh personal communication). Five of these resistance gene loci (Pm1, Pm3, Pm4, Pm5, and Pm8) have more than one resistance allele making a total of 54 named Pm resistance genes.
The availability of different resistance genes allows their pyramiding into the same genotypes as a means of delaying a breakdown of resistance. Molecular markers tightly linked to disease resistance genes allow selection for resistance in the absence of the pathogen. Various molecular marker types have been used for mapping powdery mildew resistance genes in wheat (Huang and Röder 2004). Microsatellites, also termed simple sequence repeats (SSR), reveal much higher polymorphisms in wheat than any other marker system and are now being preferentially used for genetic mapping. Linkage to microsatellites was recently described for Pm33 (Zhu et al. 2005), Pm34 (Miranda et al. 2006), and several tentatively designated genes, e.g. MlZec1 (Mohler et al. 2005), PmY201 and PmY212 (Sun et al. 2006).
Among the 38 powdery mildew resistance loci, Pm5 was located on the long arm of chromosome 7B (Law and Wolfe 1966; Lebsock and Briggle 1974). Pm5 is widespread in cultivars and landraces of China and Europe (Huang et al. 1997; Zeller et al. 1998). Eight recessive resistance alleles at, or near, the Pm5 locus have been reported (Hsam et al. 2001; Huang et al. 2000a, 2002), of which Pm5e was mapped relative to microsatellite markers (Huang et al. 2003). Line IGV1-455 carrying Pm5d provides resistance to all powdery mildew races in Germany and is, therefore, a valuable resistance source for the enhancement of wheat germplasm (Hsam et al. 2001). The major objectives of the present study were to construct a genetic linkage map around the powdery mildew resistance gene Pm5d using microsatellite markers, to determine the location of the resistance gene on the physical map of wheat chromosome 7B and to identify microsatellite markers which could be useful for marker-assisted selection (MAS).
Materials and methods
Plant materials
A total of 82 F3 families originating from a cross between the powdery mildew-susceptible cultivar Chinese Spring (CS) and the resistant line IGV1-455 was used for linkage analysis of molecular markers and powdery mildew resistance allele Pm5d. Wheat genotypes Hope (Pm5a), Kormoran (Pm5b), T. sphaerococcum cv. Kolandi (Pm5c), IGV1-455 (Pm5d), IGV1-556 (Pm5d), Fuzhuang 30 (Pm5e), Xiaobaidong (Mlxbd, a recessive Pm gene on 7BL), Prins (nearly isogenic to IGV1-455), CS, and another 22 wheat genotypes (Table 2) were used for determining allele sizes of closely linked microsatellite markers. The backcross-derived wheat lines IGV1-455 (CI10904/7*Prins) and IGV1-556 (CI10904/7*Starke) were kindly provided by Dr. J. MacKey (Uppsala, Sweden). CS deletion lines 7BS-1, 7BL-14, 7BL-2, 7BL-9, 7BL-7, 7BL-10, 7BL-3, kindly provided by J. Raupp (Wheat Genetics Resource Centre, Kansas State University, USA), were used for physical mapping of powdery mildew resistance allele Pm5d. The fraction length (FL) value of a given deletion line identifies the breakpoint in the deleted chromosome and the length of the remaining chromosome arm from the centromere relative to the length of the complete arm (Endo and Gill 1996). Assignment of markers to bins was described in Werner et al. (1992).
Powdery mildew reaction tests
A set of up to 13 Blumeria graminis tritici (Bgt) pathotypes was used to test some of the cultivars and lines used in the present study (Table 1). These Bgt isolates possess the ability to differentiate known Pm5 alleles (Hsam et al. 2001). To follow segregation of Pm5d in the mapping population, Bgt isolates 10 and 14, both avirulent to IGV1-455 and virulent to CS, were used. A minimum of 15 plants of each F3 family was tested to identify the genotype of each corresponding F2 plant. Reaction tests were conducted on agar-mounted detached primary leaf segments. The methods of inoculation, conditions of incubation and disease assessment were according to Hsam et al. (2001). χ2 tests for goodness-of-fit were used to test for deviations of observed and expected segregation ratios.
Molecular mapping techniques
Nuclear DNA extraction from primary leaves followed the procedure described by Huang et al. (2000b). For genetic mapping of Pm5d, eight microsatellite markers from the distal half of chromosome 7BL (Xgwm146, Xgwm611, Xgwm577, Xwmc70, Xwmc526, Xwmc581, Xwmc613 and Xbarc1073) were chosen from the GrainGenes database (http://wheat.pw.usda.gov). Microsatellite analysis was carried out according to Huang et al. (2000c). A partial linkage map around Pm5d was computed with the program JoinMap® 3.0 (Van Ooijen and Voorrips 2001). Map distances were calculated using the Kosambi function (Kosambi 1944), which assumes cross-over interference. Charts of genetic linkage maps were drawn with the computer program MapChart 2.1 (Voorrips 2002).
Results
Powdery mildew reaction tests
The disease response patterns of 21 wheat cultivars and lines possessing either single resistance genes or gene combinations are given in Table 1. All wheats carrying a Pm5 allele as the only resistance factor were resistant to Bgt pathotypes 10 and 14; however, a differential response to other isolates, especially 2, 5, 9, 13, 71, 77 and E, allowed identification of each allele by a unique reaction pattern. Lines IGV1-455 and IGV1-556 carrying Pm5d were resistant to all isolates. All other entries with known resistance genes and gene combinations showed differential responses. Cultivars Borneo, Centrum, Enorm, Greif and Triso were known to carry alleles at the Pm5 locus (Anonymous 2006), but the specific alleles were undetermined. Clearly, none carried Pm5d. Segregation of F3 lines in the CS × IGV1-455 mapping population with isolates 10 and 14 showed 26 homozygous resistant, 38 segregating and 18 homozygous susceptible lines (\( \chi ^{{\text{2}}}_{{{\text{1:2:1}}}} \) = 2.00, P > 0.3), indicating variation at a single locus.
Genetic and physical mapping
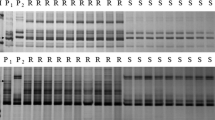
The powdery mildew resistance gene in line IGV1-455 was previously identified and designated Pm5d (Hsam et al. 2001). Eight microsatellite markers from the distal half of chromosome 7BL were assayed on DNA from the nearly isogenic lines (NILs), IGV1-455 and Prins, and the parental lines of the mapping population. Microsatellite markers Xbarc1073, Xgwm146, Xgwm577, Xgwm611, Xwmc526, Xwmc581 and Xwmc613 showed polymorphism between IGV1-455 and Prins, whereas Xwmc70 was null-allelic for both. In the parental survey, polymorphism was revealed for Xgwm146, Xgwm577, Xgwm611, Xwmc70, Xwmc581 and Xwmc613. These six markers were used to generate a genetic map of the target region in the cross CS × IGV1-455. Xgwm146, Xwmc581 and Xwmc613 showed co-dominant inheritance, whereas the other three were dominant in repulsion phase. SSR marker Xwmc581 was duplicated in both parental lines and showed fragment sizes of 125 bp and 141 bp in CS and 127 bp and 145 bp in IGV1-455. These duplicated bands were inherited together. Due to extreme stutter combined with the small size difference between the marker alleles of the parents, Xwmc613 could be scored only as a dominant marker linked in repulsion with Pm5d. The genetic map of the distal part of wheat chromosome 7BL spanned a distance of 32.5 cM (Fig. 1). Pm5d was flanked by loci Xgwm611 and Xgwm577 with linkage distances of 2.1 cM and 2.0 cM, respectively. A set of seven CS deletion lines, each line carrying a different deletion of chromosome 7B, was used for targeting resistance allele Pm5d to an individual breakpoint interval. While deletion line 7BS-1 showed amplification for the six markers linked to the disease resistance locus, no PCR products were obtained from the DNA of any of the other lines with 7BL deletions. This indicated that Pm5 is located distal to breakpoint 0.86 of deletion line 7BL-3 (Fig. 1).
Allelic sizes of linked microsatellite markers
None of the three closely linked markers could be used singly, or in combination, to distinguish between the different resistance alleles at the Pm5 locus (Table 2). Wheat lines IGV1-455, IGV1-456 (Pm5d), Fuzhuang 30 (Pm5e) and Xiaobaidong (Mlxbd), all of Chinese origin, shared common marker haplotypes in the region of interest. Alleles of Xgwm577 distinguished wheat lines carrying resistance alleles Pm5a, Pm5b or Pm5c. The marker size of wheat cultivar Spica was the same as cv. Hope confirming the likely presence of Pm5a previously indicated from the disease response pattern. Likewise, Dream had the same allele as the Pm5b reference line Kormoran. Chinese wheats with Pm5 alleles shared a null allele in common with the German cv. Kolibri, carrying Pm3d. However, the marker sizes of susceptible CS and Prins and another 14 wheats with divergent Pm genes were clearly different from lines carrying Pm5 alleles. Based on Xgwm577 marker alleles, Pm5a was postulated for cv. Borneo, and Pm5b was postulated for Greif, Enorm, Centrum and Triso.
In addition to IGV1-455 and CS, Xwmc581 was also duplicated in wheat genotypes Hope, Spica and Prins and the Chinese wheats with resistance alleles at the Pm5 locus. In the set of wheats tested, the 145 bp allele was unique to Chinese wheats possessing Pm5 resistance alleles. Among Pm5a-carrying wheats, a 113 bp allele was present in Borneo distinguishing it from Hope and Spica. In addition, the non-Pm5 cv. Contra had the same allele. All cultivars previously classified as carrying Pm5b had the same Xwmc581 allele, but it was also found in T. sphaerococcum cv. Kolandi with gene Pm5c. Thus Xwmc581 is only of limited use for MAS of all Pm5 alleles.
SSR marker Xgwm611 was not suitable for MAS of Pm5 because the same marker alleles, or null alleles, were present in many genotypes irrespective of powdery mildew response.
Discussion
The majority of mapped powdery mildew resistance genes in wheat are located at the chromosome ends; for example, resistance gene mlRD30 is distal to RFLP marker Xmwg2062 (Singrün et al. 2004) which maps to the terminal 1% of chromosome 7AL (Hohmann et al. 1994). This is also evident for Pm5 since it was located in the most terminal bin on the deletion map of chromosome 7BL.
All the named Pm5 alleles can be distinguished based on their differential reactions (Table 1). In addition, their chromosomal location on wheat chromosme 7BL as well as allelism tests in the F2 and F3 generations indicated that Pm5a to Pm5d are alleles (Hsam et al. 2001). However, genuine allelism of some of the named Pm5 genes appears doubtful. Huang et al. (2000a) observed one susceptible and two segregating lines among the 61 F3 families of a cross between Hope (Pm5a) and Fuzhuang 30 (Pm5e), indicating that Pm5e is a closely linked resistance gene rather than a Pm5 allele. Likewise, one susceptible and two segregating families were found in the F3 progeny of the cross Xiaobaidong (Mlxbd) × Selpek (Pm5a), whereas no susceptible lines were detected in 277 F2 progenies of the cross between Xiaobaidong and Fuzhuang 30 (Huang et al. 2000a). These results suggest that genes at, or near, the Pm5 locus may be clustered as also reported for powdery mildew resistance genes on chromosomes 1A (Pm3, Pm25, and Pm genes from wheat lines Abessi, N324 and GUS122 (Shi et al. 1998; Yahiaoui et al. 2006)) and 7A (Pm1, Pm9, mlRD30, resistance genes in hexaploid germplasms NC96BGTA4 and NC99BGTAG11, and Mlm2033 and Mlm80 in einkorn (Singrün et al. 2004; Srnić et al. 2005; Yao et al. 2007)).
The different resistance alleles at the Pm5 locus could be distinguished from the susceptible allele through the combined use of the closely linked SSR markers Xgwm577 and Xwmc581. Xgwm577 showed specific alleles for Pm5a, Pm5b and Pm5c, whereas a null allele was found for Pm5d, Pm5e and Mlxbd. Null markers are not as efficient as amplifiable markers in MAS because they cannot be screened in heterozygotes and therefore require at least two generations for detecting plants carrying target alleles (Ishii and Yonezawa 2007). To circumvent this disadvantage, Xwmc581 can be used in MAS for any of the three Pm5 alleles of Chinese origin.
Based on Xgwm577 alleles, cultivars Greif, Enorm, Centrum and Triso were considered likely to carry Pm5b and Borneo possibly possessed Pm5a. To our knowledge, only resistance alleles Pm5a and Pm5b have been distributed in European wheat germplasm. Based on the present reaction tests, specific Pm5 alleles in complex Pm genotypes were difficult to predict. However, Bgt isolate E showing virulence for Pm3b, Pm5b and Pm6, but avirulence for Pm5a, supported the postulation of Pm5b in cultivars Enorm and Greif (Table 1). This was not possible for Borneo, Centrum and Triso since Pm4b which is common to these lines conferred resistance to Bgt isolate E.
In conclusion, the closely linked markers will assist gene postulation in complex Pm genotypes and permit MAS of resistance alleles at, or near, the Pm5 locus in those crosses or populations that have appropriate polymorphisms.
References
Anonymous (2006) Beschreibende Sortenliste, Deutscher Landwirtschaftsverlag GmbH, Hannover, Germany
Endo TR, Gill BS (1996) The deletion stocks of common wheat. J Hered 87:295–307
Hohmann U, Endo TR, Gill KS, Gill BS (1994) Comparison of genetic and physical maps of group-7 chromosomes from Triticum aestivum L. Mol Gen Genet 245:644–653
Hsam SLK, Huang XQ, Zeller FJ (2001) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em. Thell.). 6. Alleles at the Pm5 locus. Theor Appl Genet 102:127–133
Huang XQ, Röder MS (2004) Molecular mapping of powdery mildew resistance genes in wheat: a review. Euphytica 137:203–223
Huang XQ, Hsam SLK, Zeller FJ (1997) Identification of powdery mildew resistance genes in common wheat (Triticum aestivum L.). IX. Cultivars, landraces and breeding lines grown in China. Plant Breed 116:233–238
Huang XQ, Hsam SLK, Zeller FJ (2000a) Chromosomal location of powdery mildew resistance genes in Chinese wheat (Triticum aestivum L. em. Thell.) landraces Xiaobaidong and Fuzhuang 30. J Genet Breed 54:311–317
Huang XQ, Zeller FJ, Hsam SLK et al (2000b) Chromosomal location of AFLP markers in common wheat utilizing nulli-tetrasomic stocks. Genome 43:298–305
Huang XQ, Hsam SLK, Zeller FJ et al (2000c) Molecular mapping of the wheat powdery mildew resistance gene Pm24 and marker validation for molecular breeding. Theor Appl Genet 101:407–414
Huang XQ, Hsam SLK, Zeller FJ (2002) Chromosomal location of genes for resistance to powdery mildew in Chinese wheat lines Jieyan 94-1-1 and Siyan 94-1-2. Hereditas 136:212–218
Huang XQ, Wang LX, Xu MX, Röder MS (2003) Microsatellite mapping of the powdery mildew resistance gene Pm5e in common wheat (Triticum aestivum L.). Theor Appl Genet 106:858–865
Ishii T, Yonezawa K (2007) Optimization of the marker-based procedures for pyramiding genes from multiple donor lines: I schedule of crossing between the donor lines. Crop Sci 47:537–546
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Law CN, Wolfe MS (1966) Location of genetic factors for mildew resistance and ear emergence time on chromosome 7B of wheat. Can J Genet Cytol 8:462–470
Lebsock KL, Briggle LW (1974) Gene Pm5 for resistance to Erysiphe graminis f. sp. tritici in Hope wheat. Crop Sci 14:561–563
Miranda LM, Murphy JP, Marshall D, Leath S (2006) Pm34: a new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L). Theor Appl Genet 113:1497–1504
Mohler V, Zeller FJ, Wenzel G, Hsam SLK (2005) Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 9. Gene MlZec1 from the Triticum dicoccoides-derived wheat line Zecoi-1. Euphytica 142:161–167
Shi AN, Leath S, Murphy JP (1998) A major gene for powdery mildew resistance transferred to common wheat from wild einkorn wheat. Phytopathology 88:144–147
Singrün C, Hsam SLK, Zeller FJ et al (2004) Localization of a novel recessive powdery mildew resistance gene from common wheat line RD30 in the terminal region of chromosome 7AL. Theor Appl Genet 109:210–214
Srnić G, Murphy JP, Lyerly JH et al (2005) Inheritance and chromosomal assignment of powdery mildew resistance genes in two winter wheat germplasm lines. Crop Sci 45:1578–1586
Sun XL, Liu D, Zhang HQ et al (2006) Identification and mapping of two new genes conferring resistance to powdery mildew from Aegilops tauschii (Coss.) Schmal. J Integr Plant Biol 48:1204–1209
Van Ooijen JW, Voorrips RE (2001) Joinmap 30, software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Werner JE, Endo TR, Gill BS (1992) Toward a cytogenetically based physical map of the wheat genome. Proc Natl Acad Sci USA 89:11307–11311
Yahiaoui N, Brunner S, Keller B (2006) Rapid generation of new powdery mildew resistance genes after wheat domestication. Plant J 47:85–98
Yao G, Zhang JL, Yang LL et al (2007) Genetic mapping of two powdery mildew resistance genes in einkorn (Triticum monococcum L.) accessions. Theor Appl Genet 114:351–358
Zeller FJ, Huang XQ, Paderina EV et al (1998) Identification of powdery mildew resistance genes in common wheat (Triticum aestivum L. em. Thell.). XII. Cultivars and landraces grown in Mediterranean countries. Plant Genet Resour Newsl 116:5–8
Zhu ZD, Zhou RH, Kong XY et al (2005) Microsatellite markers linked to 2 powdery mildew resistance genes introgressed from Triticum carthlicum accession PS5 into common wheat. Genome 48:585–590
Acknowledgments
The authors would like to thank Prof. Bob McIntosh for substantially improving the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ghazaleh Nematollahi and Volker Mohler equally contributed to this work.
Rights and permissions
About this article
Cite this article
Nematollahi, G., Mohler, V., Wenzel, G. et al. Microsatellite mapping of powdery mildew resistance allele Pm5d from common wheat line IGV1-455. Euphytica 159, 307–313 (2008). https://doi.org/10.1007/s10681-007-9494-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-007-9494-3