Abstract
Auxin concentrations in plants are tightly regulated through both biosynthesis and degradation. In the past few years, much progress was made in the area of auxin metabolism. Genetic and biochemical studies in Arabidopsis unequivocally established a complete tryptophan (Trp)-dependent two-step auxin biosynthesis pathway in which Trp is first converted into indole-3-pyruvate (IPA) by the TAA family of aminotransferases and subsequently indole-3-acetic acid (IAA) is produced from IPA by the YUC family of flavin monooxygenases. The TAA/YUC pathway is highly conserved in the plant kingdom and is probably the main auxin biosynthesis pathway in plants. Recent work also demonstrated that oxidative degradation of auxin plays an essential role in maintaining auxin homeostasis and in regulating plant development. In this chapter, we discuss the recent advancements in auxin biosynthesis and catabolism.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Auxin is an essential hormone for many aspects of plant growth and development (Zhao 2010). Plants have evolved a sophisticated network to control auxin levels with spatial and temporal precision in response to environmental cues and developmental signals. Indole-3-acetic acid (IAA), the main natural auxin in plants, can be produced from de novo biosynthesis. Free IAA, which is the presumed active form of auxin, can also be released from IAA conjugates including IAA esters, IAA-saccharides, and IAA-amino acids. A third probable route for producing IAA is to convert indole-3-butyric acid (IBA) to IAA using enzymes similar to those used in β-oxidation of fatty acids. When auxin levels need to be lowered, plants employ several mechanisms to deactivate IAA. IAA can be quickly converted into the presumed inactive forms by reaction of the carboxyl group of IAA with amino acids, sugars, and other small molecules. The IAA conjugates may serve as a first step for the eventual complete degradation of IAA. IAA is also inactivated by oxidation of the indole ring of IAA. For example, IAA can be converted to 2-oxindole-3-acetic acid (OxIAA). In this chapter, we discuss the progress made in the area of auxin biosynthesis and metabolism in the past few years.
2 De Novo Auxin Biosynthesis
De novo auxin biosynthesis is broadly divided into two categories: Tryptophan (Trp) dependent and Trp independent. Trp-independent auxin biosynthesis pathway was proposed two decades ago based on results from feeding plants with labeled Trp and Trp biosynthetic intermediates and from studies on Trp-deficient mutants (Wright et al. 1991; Normanly et al. 1993). However, the molecular mechanisms and genes for the Trp-independent pathway are not known. Therefore, the Trp-independent pathway will not be discussed further in this chapter.
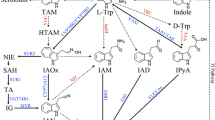
Trp has long been known as a precursor for the production of IAA in plants. Feeding plants with labeled Trp yields labeled IAA, indicating that plants have the enzymes to convert Trp to IAA (Wright et al. 1991; Normanly et al. 1993). Many biosynthetic pathways have been elucidated using analytic biochemistry techniques in combination with labeled precursors and intermediates. For example, the biosynthetic routes for brassinolide and ethylene have been established long before the biosynthetic genes have been identified (Yang and Hoffmann 1984; Sakurai and Fujioka 1993). However, the classic feeding and analytic biochemical approaches failed to identify the key components for Trp-dependent plant auxin biosynthesis pathways. There are several reasons for this apparent failure. First, Trp is a precursor for many metabolites (Fig. 2.1). Trp is a precursor for indole-3-pyruvate (IPA), tryptamine (TAM), indole-3-acetaldoxime (IAOx), indole-3-acetamide (IAM), indole-3-acetonitrile (IAN), and indole-3-acetaldehyde (IAAld) (Fig. 2.1). Arabidopsis and many other plants have the capacity to produce all of the above-mentioned intermediates (Fig. 2.1) at a given developmental stage (Ouyang et al. 1999; Sugawara et al. 2009). Some of the intermediates such as IAN exist in very high concentrations (Fig. 2.1) (Sugawara et al. 2009). Such a complex profile of Trp metabolism makes it difficult to identify Trp-dependent IAA synthesis intermediates. Second, some of the intermediates are intrinsically unstable in vitro and can be nonenzymatically converted to other compounds during the experimental process, therefore complicating the analysis of metabolic profiling. For example, IPA is readily converted nonenzymatically into IAA in vitro (Bentley et al. 1956). Third, most of the Trp metabolic intermediates display auxin activities during in vitro bioassays (Fig. 2.2). In the presence of IAM in growth media, light-grown Arabidopsis seedlings have long hypocotyls and epinastic cotyledons (Fig. 2.2). The IAM-induced phenotypes are identical to those observed in auxin overproduction mutants (Boerjan et al. 1995; Romano et al. 1995; Zhao et al. 2001). Therefore, the phenotypes of plants grown on IAM media are likely caused by overaccumulation of IAA in plants. Arabidopsis seedlings grown on IAN-containing media produce more adventitious roots and have short primary roots (Normanly et al. 1997) (Fig. 2.2). The IAN-induced phenotypes are very similar to those observed in plants grown on IAA-containing media, suggesting that IAN is probably converted to IAA in plants (Fig. 2.2). Indeed, a genetic screen for mutants insensitive to IAN identified Arabidopsis nitrilase genes that encode enzymes for the hydrolysis IAN to IAA (Bartel and Fink 1994; Normanly et al. 1997). Interestingly, treatments with IAN or IAM cause auxin overaccumulation in plants and high-auxin phenotypes. However, the IAM-induced phenotypes are dramatically different from those caused by IAN (Fig. 2.2). It is speculated that both IAM and IAN need to be metabolized into IAA to show auxin activities as the observed phenotypic differences may be simply caused by different tissue specificities of the hydrolytic enzymes for IAM and IAN. Although it is very clear that Trp metabolic intermediates can be converted to IAA in plants, it is difficult to determine how important their contribution to the total IAA pool under natural conditions is. The fact that plants produce a large number of Trp metabolic intermediates (Fig. 2.1) and that some of the Trp metabolites have auxin activities when added to growth media (Fig. 2.2) made it very difficult to dissect Trp-dependent auxin biosynthesis pathways using classic analytic biochemistry techniques alone.
Selected tryptophan metabolic intermediates. Arabidopsis plants produce all of the intermediates shown in the figure. The numbers in parenthesis refer to the actual concentrations in ng/g fresh weight. IAA indole-3-acetic acid, IAAld indole-3-acetaldehyde, IAOx indole-3-acetaldoxime, IAM indole-3-acetamide, IAN indole-3-acetonitrile, IPA indole-3-pyruvate, TAM tryptamine, and Trp tryptophan
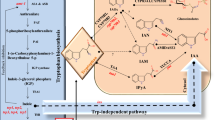
The main criterion for determining whether a Trp metabolite is important for de novo auxin biosynthesis is to use the “deletion test.” If the intermediate is important for auxin biosynthesis, we expect that plants show dramatic developmental defects similar to those observed in mutants defective in auxin transport or signaling if the plants lose the ability to make the intermediate. Recent results from a combination of analytic biochemical studies and Arabidopsis genetics research have established that the main auxin biosynthesis pathway in Arabidopsis is a simple two-step pathway that converts Trp to IAA (Fig. 2.3). The pathway is highly conserved throughout the plant kingdom.
2.1 The TAA/YUC Pathway as the Main Auxin Biosynthesis Pathway
The YUCCA flavin-containing monooxygenases catalyze the rate-limiting step
The YUCCA (YUC) flavin-containing monooxygenase (FMO) gene was identified as a key auxin biosynthesis gene a decade ago from an activation-tagging screen for long hypocotyl mutants in Arabidopsis (Zhao et al. 2001). The dominant yucca (later renamed as yuc1D) mutant was caused by the insertion of four copies of the CaMV 35S transcriptional enhancer downstream of the YUC gene (Zhao et al. 2001). The enhancers greatly increase the YUC expression levels, resulting in dramatic developmental defects. Physiological and molecular studies demonstrated that yuc1D is an auxin overproduction mutant (Zhao et al. 2001). Direct auxin measurements show that yuc1D contains 50 % more free IAA than wild-type Arabidopsis plants. Moreover, the auxin reporter DR5-GUS is greatly upregulated in yuc1D further supporting that yuc1D is an auxin overproduction mutant. It was suggested that YUC flavin-containing monooxygenases catalyze a rate-limiting step in auxin biosynthesis (Zhao et al. 2001).
YUC was later found to be a member of a gene family with 11 genes in the Arabidopsis genome. The founding member was renamed as YUC1. Overexpression of any of the YUC family members leads to auxin overproduction phenotypes in Arabidopsis, suggesting that all of the YUC genes participate in auxin biosynthesis (Cheng et al. 2006, 2007). The YUC genes have overlapping functions and inactivation of a single YUC gene does not cause any obvious developmental defects (Cheng et al. 2006, 2007). The observed genetic redundancy among YUC genes may provide an explanation for why YUC genes had not been discovered previously by forward loss-of-function genetic screens. Detailed analyses of various yuc mutant combinations have demonstrated that YUC genes are essential for almost all of the major developmental processes including embryogenesis, seedling growth, vascular initiation and patterning, flower development, and plant architecture (Cheng et al. 2006, 2007). For example, the yuc1 yuc4 double mutants do not make tertiary veins in rosette leaves and fail to make continuous vascular boundless in flowers. Overall yuc1 yuc4 flowers contain fewer floral organs and are completely sterile. A key piece of evidence that demonstrates the roles of YUC genes in auxin biosynthesis is the genetic rescue of yuc1 yuc4 mutants with the bacterial auxin biosynthesis gene iaaM under the control of the YUC1 promoter (Cheng et al. 2006, 2007).
The biochemical mechanisms of YUC-mediated auxin biosynthesis have been solved recently (Mashiguchi et al. 2011; Dai et al. 2013). YUC enzymes use NADPH and molecular oxygen to catalyze the oxidative decarboxylation of IPA to generate IAA (Fig. 2.3). On the basis of sequence homology to the mammalian microsome FMOs, it is expected that YUCs use a flavin (FAD or FMN) as a cofactor. Expressed in and purified from E. coli, the Arabidopsis YUC6 displayed a bright yellowish colour, suggesting that YUC6 contains a flavin cofactor. HPLC and other experiments demonstrate that the cofactor in YUC6 is FAD, not FMN (Dai et al. 2013). The YUC6-catalyzed conversion of IPA to IAA can be divided into three consecutive chemical steps: (1) reduction of FAD to FADH2 using electrons from NADPH; (2) binding of molecular oxygen to FADH2 to form the C4a-(hydro)peroxyl flavin intermediate; (3) the reaction of the C4a intermediate with IPA to produce IAA from decarboxylation of IPA (Fig. 2.3) (Dai et al. 2013). Interestingly, the reduction of YUC6 by NADPH takes place regardless of the presence of IPA. IPA also does not affect the rate of YUC6 reduction. The kinetic pattern and rate of the formation of the C4a intermediate is also not affected by IPA (Dai et al. 2013). However, the decomposition of C4a intermediate is greatly accelerated by IPA (Dai et al. 2013). The oxidized YUC6, reduced YUC6, and the C4a intermediate display distinct spectroscopic properties and can be monitored spectroscopically. The oxidized YUC6 shows two peaks at 376 and 448 nm in the UV-visible spectrum, while reduction of YUC6 causes the disappearance of the 448 nm peak. The YUC6 C4a-(hydro)peroxyl flavin intermediate has a maximum absorbance at 381 nm in a UV-visible spectrum (Dai et al. 2013). The FAD cofactor in YUC6 provides a convenient handle to follow the progression of the YUC-catalyzed reactions.
Besides IPA as a substrate, YUC6 can also catalyze the decarboxylation of phenyl-pyruvate (PPA) to produce phenyl-acetic acid (PAA), suggesting that YUC enzymes do not have strict substrate specificities (Dai et al. 2013). It is not known whether the YUC6-catalyzed conversion of PPA to PAA has any physiological significance. However, it is known that PAA displays auxin activities when added into growth media. Both YUC enzymes and mammalian FMOs share sequence homologies and form the C4a-(hydro)peroxyl flavin intermediate. Mammalian FMOs are mainly known for their ability to oxygenate soft nucleophiles such as nitrogen- or sulfur-containing molecules, whereas YUCs such as YUC6 oxygenate electrophilic substrates such as IPA and PPA (Ziegler 1988, 2002; Dai et al. 2013). However, mammalian FMOs recently have been shown to use electrophilic substrates as well and YUCs were previously shown to oxygenate soft nucleophiles in vitro (Zhao et al. 2001; Kim et al. 2007; Lai et al. 2010). The stability of the C4a intermediate is also quite different for YUCs and mammalian FMOs. The YUC6 intermediate has a half-life of about 20 s, whereas that of some FMOs from mammalian cells is more than 30 min (Ziegler 1988, 2002; Dai et al. 2013). It is important to use both in vitro enzymatic assays and in vivo genetic evidence to determine the physiological functions of flavin-containing monooxygenases.
In the presence of excess PPA or IPA, some uncoupled YUC6 reactions still take place and produce hydrogen peroxide. The uncouple ratio is about 4 % (Dai et al. 2013). It is not clear whether the uncoupled reaction plays any physiological role. It is conceivable that H2O2 produced from the uncoupled reaction may participate in deactivating YUC enzymes, providing an intrinsic mechanism for turning off auxin biosynthesis.
Genetic, physiological, and biochemical studies have unambiguously demonstrated that the YUC family of flavin-containing monooxygenases plays a key role in auxin biosynthesis. Genetic evidence suggests that the conversion of IPA to IAA is the rate-limiting and the committed step for IAA biosynthesis.
Tryptophan Aminotransferase of Arabidopsis (TAA) family of aminotransferases plays a key role in auxin biosynthesis
Three groups independently discovered that TAA1, the founding member of a large family of aminotransferases, is an important auxin biosynthesis enzyme (Stepanova et al. 2008; Tao et al. 2008; Yamada et al. 2009). Mutations in TAA1, which is also called SAV3, WEI8, and TIR2, alter shade-avoidance responses, cause resistance to ethylene and to the auxin transport inhibitor NPA (Stepanova et al. 2008; Tao et al. 2008; Yamada et al. 2009). Although inactivation of TAA1 along does not cause dramatic developmental phenotypes, simultaneously disruption of TAA1 and its close homolog TAR2 leads to defects in vascular pattern formation and in flower development in Arabidopsis. The taa mutants produce less free IAA compared to wild-type plants (Stepanova et al. 2008; Tao et al. 2008; Yamada et al. 2009).
TAA1 and its related proteins catalyze the transfer of the amino group from Trp to pyruvate or to α-ketoglutarate to produce IPA and Ala or Glu (Fig. 2.3) in vitro. Therefore, it is important to keep in mind that TAA genes not only produce IPA but also affect the homeostasis of other α-keto acids and other amino acids. It is not clear which α-keto acid is the preferred in vivo acceptor of the amino group from Trp.
TAAs and YUCs were previously placed in two separate pathways (Zhao et al. 2001; Stepanova et al. 2008; Tao et al. 2008). But several recent genetic studies have demonstrated that YUCs and TAAs participate in the same pathway (Mashiguchi et al. 2011; Stepanova et al. 2011; Won et al. 2011). The yuc mutants and taa mutants share many similarities. For example, yuc1 yuc2 yuc4 yuc6 quadruple mutants have dramatic vascular and floral defects, which are also observed in taa1 tar2 double mutants (Cheng et al. 2006; Stepanova et al. 2008). In fact, all of the characteristics of taa mutants can be phenocopied by inactivating certain combinations of YUC genes (Won et al. 2011). Overexpression of YUC genes leads to auxin overproduction phenotypes, which are dependent on the presence of functional TAA genes (Won et al. 2011). Furthermore, taa mutants are partially IPA deficient, whereas yuc mutants accumulate IPA, suggesting that TAA genes participate in IPA production and that YUCs use IPA as a substrate (Mashiguchi et al. 2011; Won et al. 2011). Finally, recent biochemical studies on the catalytic mechanisms of YUC flavin monooxygenases provide the final proof of the TAA/YUC two-step pathway as the main auxin biosynthesis pathway (Mashiguchi et al. 2011; Dai et al. 2013; Zhao 2013).
The TAA/YUC pathway is widely distributed throughout the plant kingdom. YUC genes from maize (Gallavotti et al. 2008), rice (Woo et al. 2007; Yamamoto et al. 2007; Fujino et al. 2008; Abu-Zaitoon et al. 2012), tomato (Exposito-Rodriguez et al. 2011), petunia (Tobena-Santamaria et al. 2002), strawberry (Liu et al. 2012), and other species (Kim et al. 2012; Cheol Park et al. 2013) have been functionally characterized and they all participate in auxin biosynthesis. The TAA genes in maize have also been shown to participate in auxin biosynthesis (Phillips et al. 2011). The committed step for auxin biosynthesis is catalyzed by the YUC flavoproteins. Thus the YUC-catalyzed reaction has to be tightly controlled. It has been shown that YUC genes are only expressed in discrete groups of cells (Cheng et al. 2006, 2007). Such tight control of YUC transcription provides a mechanism for temporal and spatial regulation of auxin production.
2.2 Other Trp-Dependent Auxin Biosynthesis Pathways
Trp is metabolized into several other indolic compounds (Fig. 2.1), some of which show auxin activities when applied to plants (Fig. 2.2). The physiological roles of the indolic compounds other than IPA in auxin biosynthesis are still ambiguous. That a compound can be metabolized into IAA both in vitro and in vivo does not mean that the compound is actually an important contributor to auxin biosynthesis in plants. Further genetic analysis of the genes responsible for generating the Trp metabolic intermediates (Fig. 2.1) is needed to assess the roles of the compounds in auxin biosynthesis.
IAM pathway
Arabidopsis and maize have detectable amount of IAM (Sugawara et al. 2009), which is the key intermediate in the bacterial auxin biosynthesis pathway characterized in Agrobacterium and Pseudomonas two decades ago (Yamada et al. 1985; Romano et al. 1995). In plant pathogenic bacteria, Trp is oxidized by the iaaM Trp-2-monooxygenase to IAM that is subsequently hydrolyzed by iaaH to produce IAA. Unlike the bacterial IAM pathway, the genes and enzymes responsible for producing IAM in plants have not been identified. It appears that plants do not have genes with high sequence homology to the bacterial iaaM gene. Therefore, IAM may be synthesized using a different mechanism. It is possible that IAM may be synthesized from IAA as a way to control free IAA levels. Conversion of IAA to IAM may be accomplished using mechanisms similar to glutamine biosynthesis.
Hydrolysis of IAM occurs in plants as feeding plants with IAM leads to elevated auxin levels and “high-auxin” phenotypes (Fig. 2.2). It is proposed that a group of hydrolases, which are homologous to the bacterial hydrolase iaaH, plays a role in converting IAM to IAA (Pollmann et al. 2006; Hoffmann et al. 2010). It is still inconclusive whether IAM is an important auxin biosynthesis intermediate in plants because IAM-deficient mutants have not been identified.
TAM pathway
Tryptamine is presumably produced by Trp decarboxylase, but the enzymes responsible for the reaction in Arabidopsis have not been characterized. Sequence homology-based prediction may not lead to the correct identification of the genes. TAM was a proposed substrate for the YUC flavin monooxygenases (Zhao et al. 2001; Kim et al. 2012), which have now been shown to catalyze the conversion of IPA to IAA in vitro and in vivo. However, all of the flavin-containing monooxygenases form the C4a-(hydro)peroxyl flavin intermediates, which are the catalytically active intermediates. The C4a intermediate can do both nucleophilic and electrophilic reactions, depending on the reaction conditions. For example, mammalian FMOs have long been recognized for their roles in xenobiotic metabolism by reacting with soft nucleophiles such as nitrogen-containing compounds (Ziegler 2002). It has also been shown that Human FMOs can catalyze a Baeyer–Villiger type reaction, in which the C4a intermediate reacts with an electrophilic carbonyl carbon (Lai et al. 2010). To date, it has not been ruled out that TAM is an important intermediate in auxin biosynthesis; however, biosynthesis and metabolism of TAM are not well understood.
IAN pathway
IAN is very abundant compared to other Trp metabolites (Fig. 2.1). IAN stimulates adventitious root development and inhibits primary root elongation (Fig. 2.2). The conversion of IAN to IAA is catalyzed by nitrilases. Inactivation of nitrilase genes leads to resistance to exogenous IAN, but the nitrilase mutants do not display obvious developmental defects observed in known auxin signaling and transport mutants (Bartel and Fink 1994; Normanly et al. 1997). Arabidopsis genome contains four copies of the nitrilase gene. The developmental consequences of disrupting all four nitrilase genes have not been investigated, partially due to the fact that two of the copies are immediately adjacent to each other on the same chromosome. Therefore, it is still an open question whether IAN plays a significant role in auxin biosynthesis.
The routes for IAN production are not well understood either. It has been reported that metabolism of indolic glucosinolate yields IAN (de Vos et al. 2008). However, maize does not produce glucosinolates, but still produces IAN, suggesting that other routes can produce IAN. It has been suggested that IAN may also be produced from other indolic compounds such as IAOx (Sugawara et al. 2009).
IAAld pathway
IAAld was previously proposed as an intermediate in the IPA pathway for auxin biosynthesis (Zhao 2010). In plants, it is now known that IAAld is not an intermediate in the IPA pathway (Mashiguchi et al. 2011; Won et al. 2011) as IPA is converted to IAA by the YUC flavin-containing monooxygenases without producing IAAld (Dai et al. 2013). In some IAA-producing bacteria, IAAld is produced from IPA by IPA decarboxylases (Carreno-Lopez et al. 2000). IAAld can be further oxidized into IAA by aldehyde oxidases. In Arabidopsis, genes homologous to the bacterial IPA decarboxylases appear not to play a role in auxin biosynthesis. Inactivation of Arabidopsis aldehyde oxidases does not disturb auxin homeostasis, suggesting that it is very likely that IAAld does not contribute significantly to de novo auxin biosynthesis (Mashiguchi et al. 2011). However, IAAld can also be oxidized by aldehyde dehydrogenases, which have not been characterized in Arabidopsis.
IAOx pathway
IAOx has only been detected in Arabidopsis and related species (Mashiguchi et al. 2011). Monocots such as rice and maize do not have detectable levels of IAOx (Mashiguchi et al. 2011). In Arabidopsis, CYP79B2 and CYP79B3 convert Trp directly to IAOx (Hull et al. 2000; Zhao et al. 2002). Overexpression of CYP79B2 in Arabidopsis leads to long hypocotyl and epinastic cotyledons, a phenotype that is also observed in YUC overexpression lines, suggesting that IAOx can be a precursor for IAA biosynthesis (Zhao et al. 2002). IAOx is also a precursor for indolic glucosinolate biosynthesis (Boerjan et al. 1995; Bak and Feyereisen 2001). When the genes encoding glucosinolate biosynthesis enzymes are mutated, more IAOx is fluxed into IAA biosynthesis, causing auxin overproduction phenotypes (Boerjan et al. 1995; Bak and Feyereisen 2001). For example, the sur1 and sur2 mutants that are defective in glucosinolate biosynthesis overproduce auxin, which leads to the development of long hypocotyls and numerous adventitious roots.
It appears that CYP79B2 and B3 are the only genes responsible for producing IAOx in Arabidopsis. The cyp79b2 cyp79b3 double mutants appear to completely abolish the biosynthesis of IAOx and the double mutants have no detectable amount of IAOx (Sugawara et al. 2009). The double mutants have subtle growth defects when grown at high temperature, but have no obvious phenotypes under normal growth conditions (Zhao et al. 2002). Therefore, it is believed that IAOx is not an essential intermediate for auxin biosynthesis. Nor is IAOx a universal intermediate for auxin biosynthesis.
In summary, after three decades molecular genetics studies in Arabidopsis, the picture of de novo auxin biosynthesis has become clearer. The two-step Trp-dependent pathway catalyzed by TAAs and YUCs is the main auxin biosynthesis pathway that plays essential roles in almost all of the main developmental processes. In retrospect, Arabidopsis probably is not the best model for auxin biosynthesis studies for two main reasons. First, the Arabidopsis glucosinolate biosynthesis pathway really complicated the analyses of IAA biosynthesis because the glucosinolate biosynthesis intermediate IAOx can be converted into IAA. The aforementioned glucosinolate mutants such as sur1 and sur2 had dramatic auxin overproduction phenotypes (Boerjan et al. 1995; Bak and Feyereisen 2001). Second, the genetic redundancy within YUCs and TAAs in Arabidopsis made it difficult for loss-of-function studies. The single Arabidopsis yuc or taa mutants do not show dramatic auxin phenotypes. Only the multiple mutants of taa or yuc display dramatic developmental defects (Cheng et al. 2006, 2007; Stepanova et al. 2008). In contrast, some monocots such as maize offer a relatively simpler system for analyzing auxin biosynthesis. Maize does not produce indolic glucosinolate (Sugawara et al. 2009). Furthermore, inactivation of a single YUC gene or TAA gene in maize leads to dramatic developmental phenotypes, whereas inactivation of at least two YUC genes or TAA genes in Arabidopsis is needed to cause main developmental defects (Gallavotti et al. 2008; Phillips et al. 2011).
3 IAA Production from Non-De Novo Pathways
Besides de novo in loco synthesis and transportation from neighboring cells, free IAA can also be made available by releasing IAA from its conjugated forms or from indole butyric acid (IBA) (Woodward and Bartel 2005). In fact, the majority of IAA in plants exists in the conjugated forms, which are proposed to serve as a storage pool. It is known that IAA can be conjugated via ester bonds with simple alcohols and with sugars such as glucose and myo-inositol or via amide bonds with amino acids, peptides, or proteins. Free IAA can be produced when the conjugates are hydrolyzed. Hydrolysis of conjugates provides plants with a potentially faster way to modulate free IAA levels than de novo biosynthesis. For example, in the germinating seeds of maize, large amount of IAA is released from the endosperm from its ester form to support the growth of developing seedlings (Bialek et al. 1992). Some of the enzymes responsible for hydrolyzing IAA-sugar or IAA-amino acids have been characterized and they show different substrate specificities and are developmentally regulated (Bartel and Fink 1995; Davies et al. 1999; Lasswell et al. 2000; LeClere et al. 2002; Rampey et al. 2004).
3.1 Conversion of IBA to IAA
IBA has long been used in agriculture for promoting root initiation/growth from plant cuttings. Arabidopsis plants accumulate detectable amount of IBA. However, it is not understood how IBA is synthesized in plants. IBA is known to inhibit primary root elongation and stimulate lateral root formation. Genetic screens for Arabidopsis mutants resistant to exogenous IBA have identified many loci (ibr, IBA resistant). The majority of the ibr loci encode enzymes related to β-oxidation of fatty acids or biogenesis of peroxisome, where β-oxidation takes place. The genetic data suggest that the observed auxin activities of IBA depend on the conversion of IBA to IAA (Zolman et al. 2000; Strader et al. 2010). However, it has not been completely ruled out that IBA itself has some biological activities (Simon et al. 2013).
The physiological roles of IBA-derived IAA are difficult to determine because the enzymes responsible for IBA to IAA conversion may also participate in other pathways such as fatty acid metabolism. Recent characterization of mutations resistant to IBA leads to the discovery that disruption of ENOYL-COA HYDRATASE2 (ECH2) gene causes defects in IBA responsiveness, but appears not to affect sugar and fatty acid metabolism. Further analysis of ech2 and other ibr mutants demonstrated that IBA-derived IAA plays important roles in root hair development and cotyledon cell expansion (Strader et al. 2010, 2011).
3.2 Release of Free IAA from IAA Conjugates
IAA conjugates with ester-link to sugars and small alcohols or amide-link to amino acids and peptides have been identified in plants. The various conjugates may serve as a storage form of IAA and can release free IAA when needed. The most studied case of releasing free IAA from conjugates is the hydrolysis of IAA-amino acid conjugates. Among the 20 potential amino acid conjugates, 19 (except IAA-Arg) were tested for their ability to release free IAA in a bioassay based on root elongation in Arabidopsis (LeClere et al. 2002). It was shown that IAA-Ala, -Leu, -Phe, -Asn, -Gln, -Glu, -Gly, -Met, -Ser, -Thr, and -Tyr inhibited root elongation by more than 50 % at 40 μM, suggesting that these amino acid conjugates can be hydrolyzed to release free IAA. In contrast, IAA-Asp, -Cys, -His, -Ile, -Lys, -Pro, -Trp, and -Val appeared not a source for free IAA (LeClere et al. 2002). Genetic screens for Arabidopsis mutants resistant to IAA-Leu and IAA-Ala identified a family of hydrolases including I AA– L eu R esistant 1 (ILR1), I AA– A la r esistant (IAR3), and the IL R1- l ike protein (ILL2) responsible for releasing free IAA from the IAA-amino acid conjugates (Davies et al. 1999; Lasswell et al. 2000; LeClere et al. 2002; Rampey et al. 2004). The ilr1 iar3 ill2 triple mutants are resistant to several IAA-amino acid conjugates and have shorter hypocotyl and fewer lateral roots than wild-type plants, suggesting that releasing free IAA from conjugates plays important roles in IAA homeostasis and plant development (Rampey et al. 2004).
4 Deactivation of IAA
The active form of IAA is believed to be free IAA. The carboxyl group in IAA is essential for its auxin activities. IAA is inactivated by complete oxidation, a process that is still not well understood. IAA can also be taken out of action by forming various conjugates with alcohols, sugars, and amino acids (Woodward and Bartel 2005).
4.1 Synthesis of IAA Conjugates
Great progresses have been made in recent years towards understanding the enzymes responsible for synthesizing IAA esters and amide conjugates. In maize, synthesis of IAA-ester with sugar starts with the formation of IAA-glucose that is preceded by activation of glucose by the formation of glucose-UDP that is then joined with IAA. IAA-glucose is further converted to other IAA-sugar ester conjugates that are mostly believed to be a storage form of IAA (Michalczuk and Bandurski 1982; Leznicki and Bandurski 1988a, b). The formation of methyl IAA by the IAMT1 methyl transferase has been implicated in regulating leaf development in Arabidopsis (Qin et al. 2005).
In Arabidopsis, 20 amidosynthases encoded by the large Gretchen Hagen 3 (GH3) family of genes conjugate IAA as well as some other plant hormones such as jasmonic acid and salicylic acid with amino acids to form amide conjugates (Hagen et al. 1991; Liu et al. 1994; Staswick et al. 2005). GH3 genes are among the early-induced genes by auxin treatments (Hagen et al. 1991). Originally discovered as being able to adenylate IAA in vitro, GH3 amidosynthases are later shown to be responsible for synthesizing IAA-amino acid conjugates. The adenylyl-IAA serves as the activated intermediate and readily reacts with some amino acids (Staswick et al. 2005). Some of the IAA-amino acid conjugates can be hydrolyzed to release free IAA, while some of the conjugates appear non-hydrolyzable in vivo (LeClere et al. 2002). The latter group of IAA-conjugates may serve as a way to inactivate IAA. For example, once IAA-Asp is formed, it would not be hydrolyzed and the conjugated IAA is consequently permanently deactivated. IAA-Asp is also known as a target for oxidative degradation. GH3 proteins have also been shown to play roles in response to environmental stimuli such as light and wounding processes, possibly through the regulation the formation of IAA, jasmonic acid, and/or salicylic acid conjugates (Woodward and Bartel 2005). Interestingly, some of the IAA conjugates possesses antagonist effects against IAA. Externally applied IAA-Trp effectively antagonizes the inhibitory effects of IAA treatment in Arabidopsis roots (Staswick 2009a, b). IAA-peptide and IAA-protein conjugates have also been discovered (Walz et al. 2002), indicating that IAA may serve as a small molecular tag but their functions are still unclear.
4.2 IAA Degradation via Oxidation
IAA starts the oxidative degradation either with decarboxylation on the side chain or with oxidation of the indole ring. Very little is known regarding oxidative degradation of IAA. It has been reported that peroxidase may be involved in the oxidative decarboxylation of IAA (Normanly 2010). Oxidative intermediates including OxIAA have been discovered in plants (Reinecke and Bandurski 1983; Ostin et al. 1998; Kai et al. 2007; Peer et al. 2013). In Arabidopsis, other IAA metabolites such as N-(6-hydroxyindol-3-ylacetyl)-phenylalanine (6-OH-IAA-Phe), N-(6-hydroxyindol-3-ylacetyl)-valine (6-OH-IAA-Val), and 1-O-(2-oxoindol-3-ylacetyl)-beta-d-glucopyranose (OxIAA-Glc) have been observed with OxIAA-Glc being the main oxidative product. Recently, it was reported that in Arabidopsis roots, OxIAA is the major catabolic product of IAA (Pencik et al. 2013). Because OxIAA has little auxinic effects, irreversible oxidation of IAA into OxIAA effectively removes the IAA from the auxin pool. Another recent discovery in rice shed light on the genes underlying the conversion of IAA to OxIAA (Zhao et al. 2013). Rice plants with a mutation in the Dioxygenase for Auxin Oxidation (DAO) gene have elevated free IAA levels in anthers and ovaries and are defective in anther dehiscence, pollen fertility, and seed development (Zhao et al. 2013). The dao mutants also do not have detectable level of oxIAA, and the purified DAO protein expressed in E. coli could convert IAA to oxIAA in vitro (Zhao et al. 2013). The new findings mark the beginning of understanding the molecular and genetic mechanisms underlying IAA oxidation and the roles of oxidative degradation of IAA in auxin homeostasis.
5 Conclusions
Plants employ many ways to control auxin levels thus to ensure proper growth and development. The major advancement in our understanding of auxin homeostasis in the past few years is the elucidation of a complete two-step Trp-dependent auxin biosynthesis pathway where Trp is converted to IPA by the TAA family of amino transferases and the YUC flavin monooxygenases catalyzes the production of IAA using IPA as a substrate. However, there are still some gaps in our understanding of both auxin biosynthesis and degradation. The identification of a complete network of auxin metabolic pathways would allow us to effectively modulate auxin levels in plants with temporal and spatial control and thus greatly facilitate the dissection of the molecular mechanisms by which auxin regulates various aspects of plant growth and development.
References
Abu-Zaitoon YM, Bennett K, Normanly J, Nonhebel HM (2012) A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol Plant 146:487–499
Bak S, Feyereisen R (2001) The involvement of two p450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol 127:108–118
Bartel B, Fink GR (1994) Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc Natl Acad Sci U S A 91:6649–6653
Bartel B, Fink GR (1995) ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268:1745–1748
Bentley JA, Farrar KR, Housley S, Smith GF, Taylor WC (1956) Some chemical and physiological properties of 3-indolylpyruvic acid. Biochem J 64:44–49
Bialek K, Michalczuk L, Cohen JD (1992) Auxin biosynthesis during seed germination in Phaseolus vulgaris. Plant Physiol 100:509–517
Boerjan W, Cervera MT, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inze D (1995) Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7:1405–1419
Carreno-Lopez R, Campos-Reales N, Elmerich C, Baca BE (2000) Physiological evidence for differently regulated tryptophan-dependent pathways for indole-3-acetic acid synthesis in Azospirillum brasilense. Mol Gen Genet 264:521–530
Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20:1790–1799
Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19:2430–2439
Cheol Park H, Cha JY, Yun DJ (2013) Roles of YUCCAs in auxin biosynthesis and drought stress responses in plants. Plant Signal Behav 8(6):e24495
Dai X, Mashiguchi K, Chen Q, Kasahara H, Kamiya Y, Ojha S, DuBois J, Ballou D, Zhao Y (2013) The biochemical mechanism of auxin biosynthesis by an arabidopsis YUCCA flavin-containing monooxygenase. J Biol Chem 288:1448–1457
Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B (1999) IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11:365–376
de Vos M, Kriksunov KL, Jander G (2008) Indole-3-acetonitrile production from indole glucosinolates deters oviposition by Pieris rapae. Plant Physiol 146:916–926
Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA (2011) Gene structure and spatiotemporal expression profile of tomato genes encoding YUCCA-like flavin monooxygenases: the ToFZY gene family. Plant Physiol Biochem 49:782–791
Fujino K, Matsuda Y, Ozawa K, Nishimura T, Koshiba T, Fraaije MW, Sekiguchi H (2008) NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics 279:499–507
Gallavotti A, Barazesh S, Malcomber S, Hall D, Jackson D, Schmidt RJ, McSteen P (2008) sparse inflorescence1 encodes a monocot-specific YUCCA-like gene required for vegetative and reproductive development in maize. Proc Natl Acad Sci U S A 105:15196–15201
Hagen G, Martin G, Li Y, Guilfoyle TJ (1991) Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol Biol 17:567–579
Hoffmann M, Lehmann T, Neu D, Hentrich M, Pollmann S (2010) Expression of AMIDASE1 (AMI1) is suppressed during the first two days after germination. Plant Signal Behav 5:1642–1644
Hull AK, Vij R, Celenza JL (2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci U S A 97:2379–2384
Kai K, Horita J, Wakasa K, Miyagawa H (2007) Three oxidative metabolites of indole-3-acetic acid from Arabidopsis thaliana. Phytochemistry 68:1651–1663
Kim JI, Sharkhuu A, Jin JB, Li P, Jeong JC, Baek D, Lee SY, Blakeslee JJ, Murphy AS, Bohnert HJ, Hasegawa PM, Yun DJ, Bressan RA (2007) yucca6, a dominant mutation in Arabidopsis, affects auxin accumulation and auxin-related phenotypes. Plant Physiol 145:722–735
Kim JI, Baek D, Park HC, Chun HJ, Oh DH, Lee MK, Cha JY, Kim WY, Kim MC, Chung WS, Bohnert HJ, Lee SY, Bressan RA, Lee SW, Yun DJ (2012) Overexpression of Arabidopsis YUCCA6 in potato results in high-auxin developmental phenotypes and enhanced resistance to water deficit. Mol Plant 6:337–349
Lai WG, Farah N, Moniz GA, Wong YN (2010) A Baeyer-Villiger oxidation specifically catalyzed by human flavin-containing monooxygenase 5. Drug Metab Dispos 39:61–70
Lasswell J, Rogg LE, Nelson DC, Rongey C, Bartel B (2000) Cloning and characterization of IAR1, a gene required for auxin conjugate sensitivity in Arabidopsis. Plant Cell 12:2395–2408
LeClere S, Tellez R, Rampey RA, Matsuda SP, Bartel B (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem 277:20446–20452
Leznicki AJ, Bandurski RS (1988a) Enzymic synthesis of indole-3-acetyl-1-O-beta-d-glucose. II. Metabolic characteristics of the enzyme. Plant Physiol 88:1481–1485
Leznicki AJ, Bandurski RS (1988b) Enzymic synthesis of indole-3-acetyl-1-O-beta-d-glucose. I. Partial purification and characterization of the enzyme from Zea mays. Plant Physiol 88:1474–1480
Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6:645–657
Liu H, Ying YY, Zhang L, Gao QH, Li J, Zhang Z, Fang JG, Duan K (2012) Isolation and characterization of two YUCCA flavin monooxygenase genes from cultivated strawberry (Fragaria x ananassa Duch.). Plant Cell Rep 31:1425–1435
Mashiguchi K, Tanaka K, Sakai T, Sugawara S, Kawaide H, Natsume M, Hanada A, Yaeno T, Shirasu K, Yao H, McSteen P, Zhao Y, Hayashi K, Kamiya Y, Kasahara H (2011) The main auxin biosynthesis pathway in Arabidopsis. Proc Natl Acad Sci U S A 108:18512–18517
Michalczuk L, Bandurski RS (1982) Enzymic synthesis of 1-O-indol-3-ylacetyl-beta-D-glucose and indol-3-ylacetyl-myo-inositol. Biochem J 207:273–281
Normanly J (2010) Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb Perspect Biol 2:a001594
Normanly J, Cohen JD, Fink GR (1993) Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc Natl Acad Sci U S A 90:10355–10359
Normanly J, Grisafi P, Fink GR, Bartel B (1997) Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9:1781–1790
Ostin A, Kowalyczk M, Bhalerao RP, Sandberg G (1998) Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol 118:285–296
Ouyang J, Chen M, Li J (1999) Measurement of soluble tryptophan and total indole-3-acetic acid in Arabidopsis by capillary electrophoresis. Anal Biochem 271:100–102
Peer WA, Cheng Y, Murphy AS (2013) Evidence of oxidative attenuation of auxin signalling. J Exp Bot 64:2629–2639
Pencik A, Simonovik B, Petersson SV, Henykova E, Simon S, Greenham K, Zhang Y, Kowalczyk M, Estelle M, Zazimalova E, Novak O, Sandberg G, Ljung K (2013) Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell 25:3858–3870
Phillips KA, Skirpan AL, Liu X, Christensen A, Slewinski TL, Hudson C, Barazesh S, Cohen JD, Malcomber S, McSteen P (2011) vanishing tassel2 encodes a grass-specific tryptophan aminotransferase required for vegetative and reproductive development in maize. Plant Cell 23:550–566
Pollmann S, Neu D, Lehmann T, Berkowitz O, Schafer T, Weiler EW (2006) Subcellular localization and tissue specific expression of amidase 1 from Arabidopsis thaliana. Planta 224:1241–1253
Qin G, Gu H, Zhao Y, Ma Z, Shi G, Yang Y, Pichersky E, Chen H, Liu M, Chen Z, Qu LJ (2005) An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17:2693–2704
Rampey RA, LeClere S, Kowalczyk M, Ljung K, Sandberg G, Bartel B (2004) A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol 135:978–988
Reinecke DM, Bandurski RS (1983) Oxindole-3-acetic acid, an indole-3-acetic acid catabolite in Zea mays. Plant Physiol 71:211–213
Romano CP, Robson PR, Smith H, Estelle M, Klee H (1995) Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol Biol 27:1071–1083
Sakurai A, Fujioka S (1993) The current status of physiology and biochemistry of brassinosteroids. Plant Growth Regul 13:147–159
Simon S, Kubes M, Baster P, Robert S, Dobrev PI, Friml J, Petrasek J, Zazimalova E (2013) Defining the selectivity of processes along the auxin response chain: a study using auxin analogues. New Phytol 200:1034–1048
Staswick P (2009a) Plant hormone conjugation: a signal decision. Plant Signal Behav 4:757–759
Staswick PE (2009b) The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol 150:1310–1321
Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17:616–627
Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM (2008) TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133:177–191
Stepanova AN, Yun J, Robles LM, Novak O, He W, Guo H, Ljung K, Alonso JM (2011) The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23:3961–3973
Strader LC, Culler AH, Cohen JD, Bartel B (2010) Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol 153:1577–1586
Strader LC, Wheeler DL, Christensen SE, Berens JC, Cohen JD, Rampey RA, Bartel B (2011) Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23:984–999
Sugawara S, Hishiyama S, Jikumaru Y, Hanada A, Nishimura T, Koshiba T, Zhao Y, Kamiya Y, Kasahara H (2009) Biochemical analyses of indole-3-acetaldoxime-dependent auxin biosynthesis in Arabidopsis. Proc Natl Acad Sci U S A 106:5430–5435
Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, Cheng Y, Lim J, Zhao Y, Ballare CL, Sandberg G, Noel JP, Chory J (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176
Tobena-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JN, Souer E, Koes R (2002) FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Genes Dev 16:753–763
Walz A, Park S, Slovin JP, Ludwig-Muller J, Momonoki YS, Cohen JD (2002) A gene encoding a protein modified by the phytohormone indoleacetic acid. Proc Natl Acad Sci U S A 99:1718–1723
Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y (2011) Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci U S A 108:18518–18523
Woo YM, Park HJ, Su’udi M, Yang JI, Park JJ, Back K, Park YM, An G (2007) Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 65:125–136
Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot 95:707–735
Wright AD, Sampson MB, Neuffer MG, Michalczuk L, Slovin JP, Cohen JD (1991) Indole-3-acetic acid biosynthesis in the mutant maize orange pericarp, a tryptophan auxotroph. Science 254:998–1000
Yamada T, Palm CJ, Brooks B, Kosuge T (1985) Nucleotide sequences of the Pseudomonas savastanoi indoleacetic acid genes show homology with Agrobacterium tumefaciens T-DNA. Proc Natl Acad Sci U S A 82:6522–6526
Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M (2009) The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151:168–179
Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143:1362–1371
Yang SF, Hoffmann NE (1984) Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Biol 35:155–189
Zhao Y (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61:49–64
Zhao Y (2013) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5:334–338
Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309
Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16:3100–3112
Zhao Z, Zhang Y, Liu X, Zhang X, Liu S, Yu X, Ren Y, Zheng X, Zhou K, Jiang L, Guo X, Gai Y, Wu C, Zhai H, Wang H, Wan J (2013) A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell 27:113–122
Ziegler DM (1988) Flavin-containing monooxygenases: catalytic mechanism and substrate specificities. Drug Metab Rev 19:1–32
Ziegler DM (2002) An overview of the mechanism, substrate specificities, and structure of FMOs. Drug Metab Rev 34:503–511
Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156:1323–1337
Acknowledgment
Our research was supported by the NSF Plant Genome grant (DBI-0820729 to YZ) and the NIH (R01GM68631 to YZ).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Wien
About this chapter
Cite this chapter
Gao, Y., Zhao, Y. (2014). Auxin Biosynthesis and Catabolism. In: Zažímalová, E., Petrášek, J., Benková, E. (eds) Auxin and Its Role in Plant Development. Springer, Vienna. https://doi.org/10.1007/978-3-7091-1526-8_2
Download citation
DOI: https://doi.org/10.1007/978-3-7091-1526-8_2
Published:
Publisher Name: Springer, Vienna
Print ISBN: 978-3-7091-1525-1
Online ISBN: 978-3-7091-1526-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)