Abstract
Elucidation of the genetic basis of the control of leaf shape could be of use in the manipulation of crop traits, leading to more stable and increased crop production. To improve our understanding of the process controlling leaf shape, we identified a mutant gene in rice that causes a significant decrease in the width of the leaf blade, termed narrow leaf 7 (nal7). This spontaneous mutation of nal7 occurred during the process of developing advanced backcrossed progeny derived from crosses of rice varieties with wild type leaf phenotype. While the mutation resulted in reduced leaf width, no significant morphological changes at the cellular level in leaves were observed, except in bulliform cells. The NAL7 locus encodes a flavin-containing monooxygenase, which displays sequence homology with YUCCA. Inspection of a structural model of NAL7 suggests that the mutation results in an inactive enzyme. The IAA content in the nal7 mutant was altered compared with that of wild type. The nal7 mutant overexpressing NAL7 cDNA exhibited overgrowth and abnormal morphology of the root, which was likely to be due to auxin overproduction. These results indicate that NAL7 is involved in auxin biosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The morphological traits of leaves, such as size and shape, are major determinants of plant architecture, and strongly affect high-yield performance. However, leaf morphology, as well as other yield-related components, is a highly complex trait. In rice, many quantitative trait loci (QTLs) controlling such traits have been identified and QTLs for the size of leaves were found to be co-located with sink-related traits (Li et al. 1998; Cui et al. 2003). Existing natural variation has proven to be useful for analyzing the genetic basis of leaf morphology in Arabidopsis (Perez-Perez et al. 2002).
In addition, the characterization of isolated mutants has also allowed the identification of genes involved in leaf morphology. Many mutants related to leaf morphology have been identified and classified according to their function in determining leaf morphology (reviewed in Kessler and Sinha 2004; Tsukaya 2006). Research on the genetics of leaf development has used mutagenesis to create loss-of-function mutations that dramatically change leaf shape. These have been categorized into (1) homeobox KNOX genes, (2) auxin distribution in the shoot apical meristem (SAM), and (3) establishment of dorsoventrality (reviewed in Byrne 2005; Hake et al. 2004; Fleming 2005; Castellano and Sablowski 2005). Plant hormones affect diverse developmental processes, e.g., seed germination, seed dormancy, and plant architecture. Auxin has been shown to act as a signal for cell division, cell elongation, and cell differentiation (reviewed in Kepinski 2006; Tax and Durbak 2006). Auxin critically regulates the leaf primordum at the indeterminate SAM and cell growth in leaf development.
Rice is not only a model plant of monocots but also one of the most important crops all over the world. Despite the importance of leaf shape for achieving effective plant architecture for photosynthesis, little is known about the genetic mechanisms that determine leaf morphological characteristics. Several mutants for leaf shape, e.g., narrow-wide mutants, were reported and some of them were mapped on the linkage map based on morphological markers (see Oryzabase; http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp). Recently, by using reverse-genetics of Tos17 and T-DNA insertional rice mutants, the CONSTITUTIVELY WILTED1 (COW1) gene controlling water homeostasis has been identified (Woo et al. 2007). It encodes a member of the YUCCA gene family, which is known to be involved in auxin biosynthesis. The phenotype of the cow1 mutant showed rolled leaf, reduced leaf width, and lower root to shoot ratio.
To improve our understanding of the processes controlling leaf shape, here we identified and cloned a mutant gene for leaf shape. The mutant isolated, now termed narrow leaf 7 (nal7), displays significantly decreased width of the leaf blade. This spontaneous mutation of nal7 occurred while we were developing advanced backcrossed progeny derived from crosses of rice varieties with wild type leaf phenotype. Cloning of the NAL7 locus showed that it encodes a flavin-containing monooxygenase, which shows sequence similarity to the YUCCA gene family, and it is allelic to COW1 (Woo et al. 2007). The YUCCA gene in Arabidopsis catalyzes tryptamine to create N-hydroxytryptamine in auxin biosynthesis (Zhao et al. 2001).
Materials and methods
Plant materials
Hayamasari from Japan and Italica Livorno from Italy, both of which are temperate japonica rice varieties, were used. The mutant exhibiting narrow and curled leaf phenotype was isolated in the advanced backcrossed progeny between Hayamasari and Italica Livorno, who exhibited the normal leaf phenotype. These plants were grown in a paddy field and a greenhouse under standard cultivation conditions and were used for the measurement of leaf shape.
Isolation of nal7 mutant
The mutant exhibiting narrow and curled leaf phenotype was isolated from the advanced backcrossed progeny. BIL116 was crossed with Hayamasari to produce the advanced backcrossed progeny, BC1F2 generation. BIL116 was one line of BILs derived from the cross between Hayamasari and Italica Livorno (Fujino et al. 2004). The mutant appeared and segregated in four of 13 BC1F2 populations derived from the self-pollination of 13 independent BC1F1 plants. One individual with the mutant phenotype was selected as the mutant line, hereafter nal7.
Positional cloning of nal7
To determine the genetic basis of the nal7 mutation, segregation analysis was carried out using the BC1F2 population. In addition, F3 progeny testing derived from 40 randomly selected F2 plants was performed. Less than 20 plants per line were grown and the genotype of the NAL7 locus was determined according to the phenotype of leaf shape. For the initial mapping of nal7, 96 BC1F2 plants from one BC1F1 plant were used. For the high resolution mapping of nal7, the molecular markers listed in Supplementary material Table S1 were used. BC1F2 populations consisting of 414 plants were used for high-resolution mapping. The genotype of each recombinant F2 plant at the NAL7 locus was determined by F3 progeny testing.
Molecular analysis
Genomic DNA was extracted according to the procedure described by Fujino et al. (2004). The genomic DNA was used for PCR analysis to score the co-segregation with the phenotype with PCR-based molecular markers. In addition to one simple sequence repeat (SSR) marker GBR3003 (Fujino et al. 2004), five SSR markers (Table S1) were developed using the Nipponbare genome according to the procedure described by Fujino et al. (2004). For sequencing to detect polymorphisms between the parents in a 180-kb region, PCR products derived from the nal7 mutant were directly sequenced by using a BigDye Terminator v3.1 cycle sequencing kit with a Prism 3700 automated sequencer (Applied Biosystems) (Fujino et al. 2005). DNA sequences were aligned using BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and then aligned visually. All polymorphisms were rechecked from chromatograms with special attention to low-frequency polymorphisms.
Plasmid construction and transformation
To make NAL7 overexpressing plants, a construct with the 35S promoter driving the NAL7 cDNA was developed. A 1,278-bp fragment of the NAL7 cDNA was amplified using the primers: forward primer: GGTCTAGAATGCAGGGGCAGCAGAAGCAGA [an XbaI site (underlined) was added as a linker] and reverse primer: AAGGTACCCTACACCGAGGAGATGTTGGCT [a KpnI site (underlined) was added as a linker]. The PCR product was cloned into the XbaI/KpnI sites of a pBluescript II SK-vector (Stratagene). The insert was subjected to DNA sequencing to confirm the nucleotide sequence. Then the insert was cloned into the pMSH2 Ti-plasmid vector, and Agrobacterium-mediated transformation was performed (Ozawa and Kawahigashi 2006). Plants regenerated from hygromycin-resistant calluses (T0 plants) were grown. T0 transformants were selected on the basis of PCR amplification of the transgene. Self-pollinated plants of each T0 plant (T1) were grown.
Molecular modeling
To understand the molecular basis of the malfunction of the nal7 mutant, models of wild type and mutant NAL7 were prepared. NAL7 displays significant sequence similarity with a bacterial monooxygenase, phenylacetone monooxygenase, for which a crystal structure has been determined (Malito et al. 2004). Using this monooxygenase structure as a template (PDB: 1W4X), a partial structural model of NAL7 could be prepared using the CPH modeling server (http://www.cbs.dtu.dk). Superposition of the modeled structure onto the phenylacetone monooxygenase structure showed that most of the FAD and NADPH binding domains could be modeled (residues 27–232, 26% sequence identity).
Measurement of IAA content
After the fourth leaf was fully expanded, the tip of the fifth leaf emerged 2 months after sowing. The immature fifth leaf was collected for the measurement of indole-3-acetic acid (IAA) content in the leaf. The collected leaf was divided into three regions: tip, middle, and basal. The whole shoots of 4-day-old seedlings were collected as shoots, and the panicle was collected 10 days before heading. Each sample for measurement was collected from 3 to 5 individual plants. Extraction and measurement of free IAA in tissues of the nal7 mutant and wild type Hayamasari was performed according to the GC–MAS method of Nishimura et al. (2006) with a slight modification. In brief, frozen samples were placed in 2.0-ml microtubes with 300 μl of cold 80% methanol containing 2,6-di-tert-butyl-4-methylphenol and 13C6-IAA (Cambridge Isotope Laboratories). The samples were crushed and then centrifuged. The supernatant was dried using a stream of nitrogen gas and IAA was dissolved in 500 μl of distilled water.
RNA isolation and semi-quantitative RT-PCR analysis
Total RNA was extracted from various organs of rice using RNAiso (TAKARA) and was treated with DNase I. Total RNA (0.5 μg) was reverse transcribed using ReverTra Ace (TOYOBO) with an oligo (dT)20 primer. The PCR reaction was performed using KOD-plus (TOYOBO). Each PCR reaction (10 μl) contained 0.5 μl of fivefold-diluted cDNA template. The specificity of each primer for the target gene was confirmed by sequencing of the PCR products. Primers and amplification conditions for RT-PCR analysis are listed in Supplementary material Table S2.
Microscopic analysis
The third leaf of rice plants was collected and fixed using formaldehyde:glacialacetic acid:70% ethanol (1:1:18). Then, the tissue sample was dehydrated in a graded ethanol series to 95%. The samples were embedded in Technovit 7100 resin (Heraeus Kulzer). Microtome sections were applied to glass slides. De-paraffinization of the sections was carried out using xylene. Then, dehydration using a graded ethanol series was performed, and sections were dried overnight before staining with toluidine blue.
Results
Isolation of a narrow leaf mutant
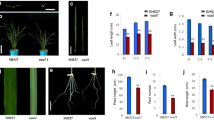
A mutant exhibiting narrow, curled leaves was identified in the advanced backcrossed progeny from the cross between Hayamasari and Italica Livorno, both of which exhibit normal leaf shape. The leaf width in the BC1F2 population in which the mutant appeared exhibited continuous distribution (in Supplementary material Fig. S1). However, the phenotype of the mutant was visually distinguishable (Fig. 1a–c). The segregation ratio in F3 progeny testing (11 wild homozygous:7 mutant homozygous:22 heterozygous) indicated that the mutant is controlled by a single recessive gene. The narrow-leaf phenotype was observed from the third leaf to the flag leaf (Table 1). The mutation only affected the width of leaves. No significant difference in leaf length was detected. Furthermore, no significant difference was observed in the phenotype other than leaf shape.
Characterization of the phenotype of nal7. a, b Representative phenotype of flag leaf in Hayamasari (wild type) and the nal7 mutant. c Leaf transverse section showing the degree of curl in the third leaf in Hayamasari and the nal7 mutant. d Transverse cross section of the leaf blade in Hayamasari and the nal7 mutant. BC and MC indicate bulliform cells and mesophyll cells, respectively
To characterize the phenotype of the narrow leaf mutant in detail, sections of leaf were compared by microscopic examination. A clear difference in number and size was observed only in bulliform cells (Fig. 1d). The bulliform cells in Hayamasari seemed to be fully expanded, while the size of the cells was smaller in the mutant. In addition, number of the bulliform cells was different; two in Haymasari, three in the mutant. Thus, the abnormal morphology of bulliform cells may cause the curled shape of the leaves.
Genetic characterization of nal7
To elucidate the genetic basis of the mutant phenotype, several genetic analyses were performed. Initial mapping of the mutant indicated that the gene for the narrow leaf phenotype was located near the marker GBR3003 on the short arm of chromosome 3 (Fig. 2). The chromosomal locations of six genes for narrow leaf in rice, nal1-6, have been reported (Oryzabase), but none of them corresponded to the region where this mutant was mapped. The gene for the mutant was named narrow leaf 7 (nal7).
Classification of the genotypes of 13 individual BC1F1 plants and of the mutant. The short arm of chromosome 3 was genotyped using five markers. The region of nal7 is indicated by an arrow. White and hatched regions represent the chromosomal segment derived from Hayamasari and heterozygous segments, respectively. The appearance of the nal7 mutant in the BC1F2 populations derived from self-pollination of the BC1F1 plants is shown
As advanced backcrossed progeny, 13 BC1F2 populations were classified into five classes, based on the genotype of the short arm of chromosome 3, where nal7 is located (Fig. 2). Twelve of 13 BC1F2 populations carried the chromosomal region of nal7 from Italica Livorno. However, the nal7 mutant appeared and segregated only in four populations. These phenomena did not match those predicted from the results of genetic mapping. One possibility is that expression of the mutant phenotype is controlled by nal7 and other gene(s) unlinked to nal7. The mutant carried at least two chromosomal regions, chromosomes 3 and 10, from Italica Livorno (data not shown). Another possibility is that the mutation for narrow leaf occurred during the process of developing the advanced backcrossed progeny, for example, spontaneous nucleotide substitution or transposon insertion. Introgression lines of Hoshinoyume (which have wild type phenotype) that carried the chromosomal region corresponding to nal7 from Italica Livorno exhibited normal leaf shape (data not shown). Plants with the mutant phenotype appeared in the F2 population derived from the cross between Hoshinoyume and nal7, exhibiting a ratio of wild and mutant type that was approximately 3:1 (146 wild:30 mutant). These results indicate that the mutation of nal7 occurred during the process of development of the advanced backcrossed progeny. In the backcrossing, BIL116 and its F1 plants were used as the female parent and Hayamasari was used as the pollinator (male parent). The mutant phenotype was linked to the Italica Livorno allele of the marker GBR3003 (Fig. S1), indicating that the mutation occurred in the female plant and is located on the chromosome from Italica Livorno.
Cloning of nal7
The chromosomal region of nal7 was delimited to within 180 kb between the markers AC119797_1 and AC105729_22 (Fig. 3a). Due to the relatively close genetic relationship between Hayamasari and Italica Livorno, there was no polymorphic SSR region. To detect nucleotide polymorphisms between them, 158 of 180 kb in the nal7 mutant was sequenced. A total of six polymorphisms between nal7 and Nipponbare were detected (Supplementary material Table S3). Then, sequence analysis for the regions containing these six polymorphisms was conducted using Hayamasari and Italica Livorno. Among them, SNP5 and SNP98 were polymorphic between Hayamasari and Italica Livorno. Using these markers, the chromosomal region of nal7 was delimited to 150 kb between the markers SNP5 and AC105729_22 and co-segregated with both markers of SNP98 and SNP165. In this region, 13 genes were predicted using RAP-DB (http://rapdb.lab.nig.ac.jp/index.html). One SNP, SNP165 from G to T, in nal7 occurred in the coding region of Os03g0162000, resulting in an amino acid substitution (from glycine to valine) (Fig. 3b, c). Sequence analysis revealed that both Hayamasari and Italica Livorno carry a glycine at the corresponding position. Only the nal7 mutant carries a valine at the respective position. This phenomenon was in good agreement with our findings regarding the genetic basis of the nal7 mutation.
Positional cloning of the nal7 gene. a Initial mapping of the mutant gene revealed that the gene is located near the marker GBR3003 on the short arm of chromosome 3 (upper). High-resolution mapping delimited the nal7 gene to a 180-kb region on chromosome 3 (middle). SNPs in the target region of nal7. The location of nal7 is in the 150-kb region between the markers SNP5 and AC105729_22. The numbers indicate the number of recombinants between the markers. SNP165 is located in Os03g0162000 (AK072466). b The gene structure of nal7. NAL7 encodes a flavin-containing monooxygenase belonging to the YUCCA gene family. c Comparison of nucleotide sequence and amino acid residue between wild type and nal7 mutant
Os03g0162000 is predicted to encode a flavin-containing monooxygenase consisting of 421 amino acids. The protein sequence contains two conserved sequence motifs (GxGxxG) indicative of the presence of two dinucleotide binding domains (Fig. S2), which show sequence homology with a number of FAD-containing, NADPH-dependent monooxygenases (class B flavoprotein monooxygenases) (van Berkel et al. 2006). In the nal7 mutant, the first glycine of the latter sequence motif has been mutated to valine. Modeling of wild type NAL7 revealed that it can nicely accommodate the NADPH coenzyme in a defined binding pocket (Fig. 4a, b). However, inspection of the mutant model indicates that the relatively bulky valine residue causes severe sterical hindrance that prevents effective binding of the adenine moiety of the NADPH coenzyme, resulting in loss of function of NAL7 (Fig. 4c). This mutation may also affect the folding and/or stability of the protein.
Structural modeling. a Superposition of the phenylacetone monooxygenase crystal structure (pdb identifier: 1W4X, in pink) and the modeled partial structure of NAL7 (residues 27–232, in green). b, c Close-up views of the partial structural models of NAL7 (b) and nal7 (c) as shown as a surface representation (green). The FAD (yellow) and NADPH (blue) cofactors are shown as ball and stick representation. The observed mutation in NAL7, Gly → Val, is highlighted (orange) and illustrates the steric clash with the adenine moiety of the NADPH cofactor. The picture was generated using PyMol (pymol.sourceforge.net)
NAL7 shows significant sequence identity with YUCCA from Arabidopsis and FLOOZY from petunia, both of which encode an enzyme required for the biosynthesis of auxin (Zhao et al. 2001; Tobena-Santamaria et al. 2002). The phylogenetic relationships among 14 rice YUCCA genes and 11 Arabidopsis YUCCA genes showed that NAL7 corresponds to OsYUCCA8 and belongs to the same group as OsYUCCA1 and YUCCA1 and YUCCA4 from Arabidopsis (Fig. S3).
IAA content of nal7
Because NAL7 encodes an auxin biosynthesis enzyme, we measured and compared the IAA content in several tissues from Hayamasari and the nal7 mutant. A clear difference in IAA content between them was detected (Fig. 5). In the basal region of leaves, the IAA content was reduced in the nal7 mutant, while the IAA content in the nal7 mutant was increased in the shoots of 4-day-old seedlings and in the panicles. Although the nal7 mutant shows different IAA content compared with WT, no phenotypic difference was observed except for the narrow leaves.
Comparison of IAA content between Hayamasari and the nal7 mutant. Data represent mean ± SD (n = 3). Samples were collected from various tissues: S whole shoot of 4-day-old seedling; LT tip region of leaf; LM middle region of leaf; LB basal region of leaf; P panicle before heading. White columns Hayamasari; black columns the nal7 mutant
Expression patterns of NAL7 and other members of the rice YUCCA gene family
To determine the expression specificity of the rice YUCCA gene family, semiquantitative reverse transcription-PCR analysis was conducted using RNA extracted from various tissues, embryos in seeds after 2 days of incubation, shoots and roots of 6-day-old seedlings, and panicles before heading (Fig. 6). The expression levels of all these genes were quite low, in terms of the signals is detectable in more than 30 PCR cycles. Tissue specificity of the expression of NAL7 mRNA was observed. Relatively strong expression of NAL7 was detected in embryos, while a low expression levels of NAL7 were detected in various tissues from later developmental stages. In embryos, other members were also expressed in lower levels than NAL7. Differences in the tissue specificity of the expression patterns of the rice YUCCA gene family were detected. Strong expression of OsYUCCA7 was detected in roots, and for OsYUCCA5, strong expression was detected in panicles, but no signal was detected in roots. Expression of OsYUCCA6 was detected in all of these tissues, but was weak in embryos. Constitutive low expression of OsYUCCA1 and OsYUCCA3 was observed. In the nal7 mutant, altered expression patterns were detected. However, the difference was only detected in weak expression levels.
Phenotype of plants overexpressing NAL7 cDNA
To investigate the effect of modification of the expression patterns, tissue specificity, and amount, we transformed the nal7 mutant with the NAL7 cDNA fused to the 35S promoter. Due to alteration of the auxin content in the derived transgenic plants, most of the transformed calli exhibited overgrowth of the roots, and the rate of plant regeneration was quite low. In some cases, adventitious roots formed directly from calli, and in other cases, abnormal root morphology, including extensive hairy roots or short, thick roots were observed (Fig. 7a–c). Such phenotypes were nearly identical to those reported for IAA overproduction resulting from, for example, OsYUCCA1 overexpression (Yamamoto et al. 2007), indicating that NAL7 plays a role in IAA biosynthesis or regulation.
Characterization of the phenotype of transgenic nal7 plants overexpressing NAL7. a, b Representative phenotype of transgenic calli after regeneration. c Close-up view of root in b. d Variation of the width of the leaf blade of the flag leaf in four independent lines. e Same leaves as in d, after flattening. Line number of transgenic plants is indicated. Bar 1 cm
Although the overexpression of NAL7 caused abnormal development, ten independent regenerated plants were obtained, which grew normally and were fertile. However, overexpression of the transgene was not detected in these transgenic plants (Fig. S4). Furthermore, altered expression of OsYUCCA genes was observed in these transgenic plants, suggesting that the transgene caused these alterations. The altered phenotype of the transgenic plants might have been the result of very weak expression of the transgene. Nine of these transgenic plants showed wider leaves than the nal7 mutant (Fig. 7d, e). However, variation of the leaf shape, including width and the degree of curl, was observed among the transgenic rice plants. These results suggest that NAL7 might control the development of leaf width.
Discussion
IAA is the main auxin in plants and is known to control a wide range of aspects of plant growth and developmental processes. Due to the multiple auxin synthesis pathways in plants, including several tryptophan-dependent routes and a tryptopan-independent pathway, the molecular details and the physiological roles of each pathway are not well known (Bartel 1997; Cohen et al. 2003; Zazimalova and Napier 2003). In addition, the redundancy of genes involved these pathways make it difficult to dissect the pathways genetically. In this study, we identified a spontaneous mutant, nal7, exhibiting narrow leaves with curl and showing altered IAA content. NAL7 encodes a flavin-containing monooxygenese and is a member of the rice YUCCA gene family.
Based on the sequence, nal7 is allelic to CONSTITUTIVELY WILTED 1 (COW1) (Woo et al. 2007). The cow1 mutant exhibited a phenotype of rolled leaf, reduced leaf width, and lower root to shoot ratio (Woo et al. 2007). These phenotypes were derived from null alleles caused by Tos17 or T-DNA insertion. The nal7 mutant did not exhibit reduced root growth (data not shown), causing the decrease in root to shoot ratio. One possibility is that this nal7 mutant caused by single amino acid substitution might show reduced enzyme activity, it did not show as severe a phenotype as the null allele. The other possibility is the difference of genetical background; nal7 in Hayamasari, cow1 in Hwayoung. Therefore, the nal7 allele is novel for the COW1 locus.
In this study, we directly demonstrated that NAL7 controls or regulates auxin biosynthesis. The IAA content in the nal7 mutant was altered compared with that in wild type Hayamasari (Fig. 5). The phenotype of the overexpression of NAL7 cDNA in the nal7 mutant was likely due to auxin overproduction (Fig. 7). Furthermore, the overexpressing plants exhibited wider leaves than the nal7 mutant. These results indicate that NAL7 is involved in auxin biosynthesis, and the phenotype observed in the nal7 mutant is caused by auxin.
Relationships between leaf morphology and IAA content were demonstrated in several studies. Transgenic petunia plants overexpressing iaaM exhibited much smaller and narrower leaves with extreme curling, and the leaf tissue of the transgenic plants contained very high levels of IAA (Klee et al. 1987). Arabidopsis YUCCA activation-tagging lines exhibited narrow leaves with curl as a result of their elevated IAA content (Zhao et al. 2001). Narrow leaves with curl are also caused by elevated IAA levels in transgenic petunia overexpressing FLOOZY (Tobena-Santamaria et al. 2002), and transgenic Arabidopsis overexpressing UGT84B1 (Jackson et al. 2002), CYP79B2 (Zhao et al. 2002), and YUCCA2 (Zhao et al. 2001). In addition, activation tagging of YUCCA3 (Zhao et al. 2001) and YUCCA5 (Woodward et al. 2005) resulted in the same phenotype and elevated IAA content.
Why does the phenotype of the nal7 mutant with a loss of function of an auxin biosynthesis gene appear similar to those of plants overexpressing auxin biosynthesis genes? The temporal and spatial distribution of IAA during leaf development in Arabidopsis has been reported (Ljung et al. 2001). Actively dividing young leaves contained the highest concentration of IAA, while the expanded leaves had dramatically decreased levels (Ljung et al. 2001). Arabidopsis YUCCA1 is expressed in young leaf primordia (Cheng et al. 2006). The formation of new primordia is preceded by the accumulation of localized relatively high concentrations of auxin (Reinhardt et al. 2003). The nal7 mutant clearly showed altered IAA content in the shoots of seedling and in panicles (Fig. 5). In the SAM, nal7 may cause narrow leaves by affecting the determination of leaf width in the leaf primordia. Further analysis of the relationship between IAA content in the SAM and leaf width in rice will elucidate the determination of leaf width by auxin.
The only difference in the phenotype observed between the nal7 mutant and wild type Hayamasari was in leaf shape. NAL7 may play an important role in the determination of leaf width at the SAM, where other member of OsYUCCA gene family might not have any function. Members of the YUCCA gene family in Arabidopsis are spatially and temporally regulated: e.g., YUCCA1 is expressed at young leaf primordia while YUCCA4 is not (Cheng et al. 2006). Member of rice YUCCA gene family exhibited tissue specificity in gene expression (Fig. 6). Based on the phylogenic relationships within the YUCCA gene family in Arabidopsis and the rice YUCCA gene family, the NAL7 gene is classified into the same clade as YUCCA1 and YUCCA4 (Fig. S3).
Auxin is known to play a critical role in embryogenesis and in leaf development. In Arabidopsis, YUCCA1 and YUCCA4 show distinct and overlapping expression patterns during embryogenesis and seedling development (Cheng et al. 2007). The yuc1 and yuc2 double mutant do not exhibit any obvious defects in embryogenesis and the formation of leaves, while a single nal7 mutant exhibits clear morphological change in the formation of leaves. These indicated that the YUCCA gene family in rice play similar but distinct role in leaf development in Arabidopsis.
References
Bartel B (1997) Auxin biosynthesis. Annu Rev Plant Physiol Plant Mol Biol 48:51–66
Byrne ME (2005) Networks in leaf development. Curr Opin Plant Biol 8:59–66
Castellano MM, Sablowski R (2005) Intercellular signaling in the transition from stem cells to organogenesis. Curr Opin Plant Biol 8:26–31
Cheng Y, Dai X, Zhao Y (2006) Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Gene Dev 20:1790–1799
Cheng Y, Dai X, Zhao Y (2007) Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19:2430–2439
Cohen JD, Slovin JP, Hendrickson AM (2003) Two gentically discrete pathways convert tryptophan to auxin: more redundancy in auxin biosysnthesis. Trends Plant Sci 8:197–199
Cui KH, Peng SB, Xing YZ, Yu SB, Xu CG et al (2003) Molecular dissection of the genetic relationships of source, sink and transport tissue with yield traits in rice. Theor Appl Genet 106:649–658
Fleming AJ (2005) Formation of primordia and phyllotaxy. Curr Opin Plant Biol 8:53–58
Fujino K, Sekiguchi H, Sato T, Kiuchi H, Nonoue Y et al (2004) Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor Appl Genet 108:794–799
Fujino K, Sekiguchi H, Kiguchi T (2005) Identification of an active transposon in intact rice plants. Mol Genet Genomics 273:150–157
Hake S, Smith HMS, Holtan H, Magnani E, Mele G et al (2004) The role of knox genes in plant development. Annu Rev Cell Dev Biol 20:125–51
Jackson RG, Kowalczyk M, Li Y, Higgins G, Ross J et al (2002) Over-expression of an Arabidopsis gene encoding a glucosyltransferase of indole-3-acetic acid: phenotypic characterisation of transgenic lines. Plant J 32:573–583
Kepinski S (2006) Integrating hormone signaling and patterning mechanisms in plant development. Curr Opin Plant Biol 9:28–34
Kessler S, Sinha N (2004) Shaping up: the genetic control of leaf shape. Curr Opin Plant Biol 7:65–72
Klee HJ, Horsch RB, Hinchee MA, Hein MB, Hoffmann NL (1987) The effects of overproduction of two Agrobacterium tumefaciens T-DNA auxin biosynthetic gene products in transgenic petunia plants. Gene Dev 1:86–96
Li Z, Pinson SRM, Stansel JW, Paterson AH (1998) Genetic dissection of the source–sink relationship affecting fecundity and yield in rice (Oryza sativa L.). Mol Breed 4:419–426
Ljung K, Bhalerao RP, Sandberg G (2001) Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28:465–474
Malito E, Alfieri A, Fraaije MW, Mattevi A (2004) Crystal structure of a Baeyer–Villiger monooxygenase. Proc Natl Acad Sci USA 101:13157–13162
Nishimura T, Mori Y, Furukawa T, Kadota A, Koshiba T (2006) Red light causes a reduction in IAA levels at the apical tip by inhibiting de novo biosynthesis from tryptophan in maize coleoptiles. Planta 224:1427–1435
Ozawa K, Kawahigashi H (2006) Positional cloning of the nitrite reductase gene associated with good growth and regeneration ability of calli and establishment of a new selection system for Agrobacterium-mediated transformation in rice (Oryza sativa L.). Plant Sci 170:384–393
Perez-Perez JM, Serrano-Cartagena J, Micol JL (2002) Genetic analysis of natural variations in the architecture of Arabidopsis thaliana vegetative leaves. Genetics 162:893–915
Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K et al (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426:255–260
Tax FE, Durbak A (2006) Meristems in the movies: live imaging as a tool for decoding intercellular signaling in shoot apical meristems. Plant Cell 18:1331–1337
Tobene-Santamaria R, Bliek M, Ljung K, Sandberg G, Mol JNM et al (2002) FLOOZY of petunia is a flavin mono-oxygenase-like protein required for the specification of leaf and flower architecture. Gene Dev 16:753–763
Tsukaya H (2006) Mechanism of leaf-shape determination. Annu Rev Plant Biol 57:477–496
van Berkel WJH, Kamerbeek NM, Fraaije MW (2006) Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol 124:670–689
Woo YM, Park HJ, Suudi M, Yang JI, Park JJ et al (2007) Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol Biol 65:125–136
Woodward C, Bemis SM, Hill EJ, Sawa S, Koshiba T et al (2005) Interaction of auxin and ERECTA in elaborating Arabidopsis inflorescence architecture revealed by the activation tagging of a new member of the YUCCA family putative flavin monooxygenases. Plant Physiol 139:192–203
Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143:1362–1371
Zazimalova E, Napier RM (2003) Points of regulation for auxin action. Plant Cell Rep 21:625–634
Zhao Y, Christensen SK, Fankhauser C, Cashmen JR, Cohen JD et al (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309
Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J et al (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Gene Dev 16:3100–3112
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Yano.
Nucleotide sequence data reported are available in the DDBJ/EMBL/GenBank databases under accession numbers AB354301 and AB354302.
Electronic supplementary material
Below is the link to the electronic supplementary material.
438_2008_328_MOESM1_ESM.tif
Frequency distribution of the width of the leaf blade in the flag leaf in the advanced backcrossed progeny. Arrowhead indicates the mean of Hayamasari (HY). Three classified genotypes, homozygous for the Italica Livorno allele (black), heterozygous (hatched), and homozygous for the Hayamasari allele (white), assessed using the marker GBR3003, are indicated (TIF 46 kb)
438_2008_328_MOESM2_ESM.tif
Sequence alignment of NAL7. The mutation identified in the nal7 mutant allele is indicated by bold type and by an arrowhead above the aligned sequences. The two conserved motifs are boxed (TIF 72 kb)
438_2008_328_MOESM3_ESM.tif
Phylogenetic tree of the YUCCA gene family. The phylogenetic tree of 14 rice and 11 Arabidopsis YUCCA genes was constructed using CLUSTAL W. Bootstrap analysis values are shown at the nodal branches. The indicated scale represents 0.1 amino acid substitution per site. The accession numbers and plant species are shown in green for Arabidopsis, blue for rice from Yamamoto et al. (2007), and red for rice from this study. Gene ID is according to RAP-DB (http://rapdb.lab.nig.ac.jp/index.html (TIF 62 kb)
438_2008_328_MOESM4_ESM.tif
Expression of nal7 in transgenic plants measured by RT-PCR analysis. Ten independent lines (T1), and Hayamasari and the nal7 mutant as non transgenic plants were used. Total RNA was extracted from the whole shoot of the 6-day-old seedling. The numbers on the right indicate the number of PCR cycles. Ubi2 was used as a loading control (TIF 158 kb)
Rights and permissions
About this article
Cite this article
Fujino, K., Matsuda, Y., Ozawa, K. et al. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol Genet Genomics 279, 499–507 (2008). https://doi.org/10.1007/s00438-008-0328-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-008-0328-3