Abstract
RNA interference (RNAi) is a naturally occurring biological process that regulates plant growth and development, defense against pathogens, and environmental stresses. It is a sequence-specific homology-based silencing mechanism in which the function of a gene is interfered or suppressed. Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are produced inside the plant cell through the activation of RNAi machinery, which downregulates the expression of the target genes at transcriptional and translational levels. RNAi is more specific, precise in its action, and considered as a potential technology for functional genomics studies. In the last 15 years, it has emerged as a scientific breakthrough for crop improvement without affecting other agronomic traits. It has also been employed as a novel method in understanding the basic phenomenon of plant defense and metabolism. Several desirable traits have been improved in the crop varieties through RNAi, which include crop protection against biotic and abiotic stresses, enhancement of nutritional value, alteration in plant architecture for better adaptation to environmental conditions, overexpression or removal of secondary metabolites, enhancement of shelf life of fruits and vegetables, generation of male sterile lines, and development of seedless fruits. In this book chapter, we have discussed RNAi and its applications in crop improvement.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- RNA interference
- siRNAs
- miRNAs
- Gene silencing
- Crop improvement
- Transgenic plants
- Stress tolerance
- Plant architecture
3.1 Introduction
RNA interference is a conserved, naturally occurring gene regulatory mechanism. It is evolved to protect the organisms against the invading foreign nucleic acids. Besides, it is also required for maintaining genomic stability, regulation of transposon movement, epigenetic modification, and control of cellular processes at transcriptional and translational level (Ketting 2011; Castel and Martienssen 2013). Fire et al. (1998) coined the term RNAi for the unknown silencing mechanism observed upon exogenous supply of dsRNAs of sense and antisense transcripts in Caenorhabditis elegans. They have also detected that dsRNAs of target gene induced silencing even in low concentration and mentioned about the existence of amplification process in C. elegans. Earlier to RNAi, similar types of silencing phenomenon were also reported by scientists working on plant and fungal systems (Napoli et al. 1990; Romano and Macino 1992). They found that the introduction of transgene caused downregulation of transgene as well as the endogenous gene. The phenomenon was called as “co-suppression” in plants and “quelling” in fungi. Later, it was demonstrated that protein complexes involved in RNAi and related phenomena were conserved across the kingdoms (Baulcombe 2000; Matzke et al. 2001). So far, RNAi and related mechanisms have been described in prokaryotes such as bacteria (clustered regularly interspaced short palindromic repeats) (Wilson and Doudna 2013) and eukaryotes – algae (Cerutti et al. 2011), fungi (Romano and Macino 1992), moss (Bezanilla et al. 2003), plants (Napoli et al. 1990), nematodes (Fire et al. 1998), Drosophila (Hammond et al. 2000), and mammals (Elbashir et al. 2001).
RNAi involves homology-based sequence-specific degradation of target gene transcripts (Wilson and Doudna 2013). It is triggered by aberrant dsRNAs which can vary in length and origin. These aberrant or foreign dsRNAs are processed into small RNA duplexes of variable sizes ranging from 21 to 28 nucleotides (nt). Small RNA molecules are loaded on protein complex and then directed toward their cognate RNA where they cause cleavage of target gene or suppression of translation. They are also capable of inducing modification at DNA level through methylation or deacetylation (Molnar et al. 2010). RNAi and its executive molecules are, thus, responsible for gene regulation at transcriptional, posttranscriptional, and translational levels (Xie et al. 2003; Brodersen et al. 2008; Molnar et al. 2010; Khraiwesh et al. 2010). The discovery of RNAi gave a new tool in the hand of scientists to manipulate the plants through genetic engineering and to study the functional genomics. Nowadays, RNAi is extensively used for crop improvement through the alteration of desirable traits in plants. Steps involved in the development of effective RNAi-based strategies for crop improvement include the identification of a suitable target, preparation of an efficient RNAi construct, transformation of plants with the RNAi construct, and evaluation of RNAi lines for desirable characteristics (Saurabh et al. 2014). Different bioinformatic tools are used for initial screening of target genes so that the sequence used for preparation of RNAi construct do not bear any off-target effects on plant’s development or nontargeted organisms (Saurabh et al. 2014). This chapter summarizes the various applications of RNAi for crop improvement.
3.2 RNA Interference (RNAi): siRNAs and miRNAs
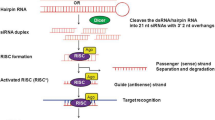
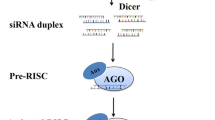
The small noncoding RNAs (ncRNAs) mediate gene silencing at transcriptional and posttranscriptional levels. Both transcriptional gene silencing (TGS) and posttranscriptional gene silencing (PTGS) pathways start with dsRNAs but process through different machineries and mechanisms. PTGS is generally employed for host-induced gene silencing (HIGS), i.e., host plants engineered to produce siRNAs/miRNAs against the target gene. siRNAs and miRNA are the effector molecules for PTGS (Bartel 2004; Zamore and Haley 2005; Vazquez 2006). Both siRNAs and miRNAs are 20–24 nt long and are generated through the processing of long dsRNA. They vary in origin, initial precursor structure, biogenesis pathway, and mode of action (Axtell, 2013). Formation of siRNAs is triggered with the appearance of aberrant dsRNAs from endogenous or exogenous sources (Fire et al. 1998; Tuschl 2001). Plant cell recognizes these aberrant dsRNAs as foreign particles and cleaves them into 21–25 nt small siRNA duplexes with the help of dicer (DCL), an RNAse III endonuclease family member (Hamilton and Baulcombe 1999; Hammond et al. 2000; Bernstein et al. 2001). The generated siRNA duplexes have phosphate group at 5′ end and two nucleotide overhangs at 3′ end (Bernstein et al. 2001; Elbashir et al. 2001). The siRNA duplexes are then loaded onto RNA-induced silencing complex (siRISC) through the recognition at 3′ overhangs. Degradation of passenger/sense strand of siRNAs (strand which has same sequence as the target mRNA) activates the RISC complex and directs the remaining antisense siRNAs toward the cognate mRNA. Argonate (AGO) protein, the main component of RISC complex, then brings out the cleavage of target mRNA based on the sequence-specific homology between antisense siRNA and target mRNA. Plant miRNA biogenesis starts with the endogenous primary miRNA (pri-miRNA) precursor, which has partial double-stranded stem-loop structure and is transcribed by RNA polymerase II in the nucleus (Jones-Rhoades et al. 2006; Zhu 2008). The pri-miRNA is further processed into 70–110-nt-long precursor miRNA (pre-miRNA) by RNase III enzyme DCL1 (dicer-like 1) and other proteins (HYL1, SE, HEN1). The pre-miRNA is cleaved by DCL1 into 22–24-nt-long miRNA duplex, which is then moved to cytoplasm with the help of HASTY protein. The mature miRNA/miRNA* duplex is then recruited into RISC complex, and degradation of sense miRNA by SDN protein takes place leading to the activation of RISC complex. Mature miRNAs bind to the target mRNAs mostly at 3’ UTR (untranslated region) and mediate their cleavage or translational blockage (Bao et al. 2004; Khraiwesh et al. 2010). MicroRNAs are expressed during plant growth and development, synthesis of secondary metabolites, abiotic and biotic stress reactions, etc. Alteration in their expression and biosynthesis could be beneficial for the development of plants with valuable characteristics (Pareek et al. 2015) (Figs. 3.1 and 3.2).
Biogenesis and silencing mechanism of siRNAs (Reproduced from Yogindran and Rajam 2015)
Biogenesis and silencing mechanism of miRNAs in plants (Reproduced from Pareek et al. 2015)
3.3 RNAi for Crop Improvement
Plants are the major source of all kind of food to human being and livestock. Environmental changes, scarcity of land, and depletion of natural resources limit the crop productivity, thereby causing instability in food security and malnutrition across the world. The existing breeding and improvement programs are associated with various physiology, ecological, and biological drawbacks. In recent time, genetic engineering through RNAi has proven its potential for improving crop varieties for different useful agronomic traits.
3.3.1 Biotic Stress Resistance
The pathogens (viruses, bacteria, and fungi), insect pests, and nematode parasites are the biotic factors, which hinder the growth and development of crop plants and affect their quality and yield. Worldwide, biotic factors account for about 40% loss in six major food and cash crops (Oerke 2006). Geometrical elevation in world’s population demands for novel techniques for effective management of biotic factors. RNAi-mediated crop protection against biotic factors opened up a new era in this direction.
Insect pests mostly damage the plants during reproductive stages. Insecticides offer a quick control for eradication of insect pests, but excessive use of insecticides and its persistence in environment and food crop makes it unsuitable for long use. The effectiveness of host-induced RNAi for control of insect pests was first demonstrated by Mao et al. (2007) and Baum et al. (2007). Mao et al. (2007) showed that sensitivity of insect pest toward phytotoxin (gossypol) can be increased by inhibiting the expression of insect P450 monoxygenase gene involved in the detoxification of gossypol through HI-RNAi in cotton. Transgenic maize plants were developed to produce siRNAs against the vital gene of an insect pest, Coleopteran western corn rootworm, V-ATPase for insect resistance. Recently, it has been shown that silencing of chitinase gene in Helicoverpa armigera through HI-RNAi caused downregulation of the target gene transcripts and induced mortality and developmental deformities at larval, pupal, and adult stages (Mamta et al. 2015). Previously, HI-RNAi was used to control different insect pests such as H. armigera (Zhu et al. 2012; Mao et al. 2013; Liu et al. 2015; Jin et al. 2015), Manduca sexta (Kumar et al. 2012), Nilaparvata lugens (Zha et al. 2011; Yu et al. 2014), and Bemisia tabaci (Thakur et al. 2014) through silencing of vital genes of the target pests.
Host genes involved in growth, development, and parasitism are found to be effective targets for control of nematodes through HI-RNAi (Huang et al. 2006; Yadav et al. 2006; Sindhu et al. 2009; Papolu et al. 2013; Tamilarasan and Rajam 2013; Xue et al. 2013; Banerjee et al. 2017). For example, the expression of HgALD dsRNA in soybean gave resistance against nematodes (Youssef et al. 2013). HgALD gene encodes for fructose-1,6-diphosphate aldolase enzyme required during gluconeogenesis. Silencing of FMRF amide-like peptide gene flp-14 and flp-18 through HI-RNAi led to the inhibition of invasion and reproduction pathways and thereby decreases the parasitic responses of root-knot nematode, Meloidogyne incognita (Papolu et al. 2013).
Fungal pathogens not only cause huge crop losses but also produce harmful mycotoxins in crop plants. Ingestion of mycotoxins even in low quantity leads to serious health problems in living beings. Control of fungal infections has been achieved through HI-RNAi in different crop plants (Nowara et al. 2010; Yin et al. 2011; Nunes and dean 2012; Koch et al. 2016). Tinoco et al. (2010) demonstrated HI-RNAi as a proof of concept in β-gluconidase (GUS) expressing necrotrophic fungi, Fusarium verticillioides, through silencing of GUS transgene expression by uptake of GUS-targeted siRNAs generated in transgenic tobacco. A recent study by Chen et al. (2016) showed the development of resistance in barley against Fusarium culmorum through downregulation of β-1, 3-glucan synthase (FcGls1) gene expression. Inhibition of FcGls1 gene induced defects during hyphae growth and development and fungal invasion (Chen et al. 2016). In virus, mostly coat protein (CP) genes are reported to be the potential target for Hi-RNAi (Andika et al. 2005; Kamachi et al. 2007; Kumar et al. 2012; Zhou et al. 2012). For example, production of siRNAs against CP gene of cucumber green mottle mosaic virus (CGMMV) provided resistance against this virus in tobacco (Kamachi et al. 2007). Recently, resistance in tomato (Solanum lycopersicum) against yellow leaf curl virus was achieved through simultaneous silencing of different viral disease-responsive genes by expressing chimeric dsRNA (Chen et al. 2016). Viral resistance can also be achieved by targeting RNAi suppressor proteins. For example, downregulation of viral suppressor proteins (AC2 and AC4) by trans-acting siRNAs generated in tobacco provides high resistance against tomato leaf curl New Delhi virus (ToCNDV) (Singh et al. 2014).
Under biotic stress conditions, expression of various miRNAs gets enhanced or repressed (Khraiwesh et al. 2012; Li et al. 2012; Kumar 2014; Singh et al. 2014). Li et al. (2012) showed that 20 miRNAs were differentially expressed in susceptible soybean variety as compared to the resistant variety against soybean cyst nematodes (SCN – Heterodera glycines). Overexpression of osa-mi7696 provided immunity to rice (Oryza sativa L. ssp. japonica) against blast fungi Magnaporthe oryzae (Campo et al. 2013). This miRNA negatively regulates the expression of natural resistance-associated macrophage protein-6 (OsNramp6) in rice. Thus, RNAi is emerging as a potential alternate approach for gain of resistance under biotic stress conditions.
3.3.2 Abiotic Stress Tolerance
Under natural field conditions, plants are often not able to attain their full growth and development due to the continuous exposure to different abiotic stresses. Drought, salinity, and variation in temperature are the major abiotic conditions, which cause huge crop losses around the world. The changing environment and increasing food demands for growing population exerts great pressure on scientists for development of stress-tolerant crop varieties. Under stress conditions, plants synthesize different noncoding RNAs (ncRNAs) for gene regulation at transcriptional, posttranscriptional, and chromatin level. The ncRNAs and their targets can be utilized for generation of abiotic stress-tolerant variety through RNAi. Downregulation of receptor for activated C-kinase 1(RACK1) through RNAi resulted in development of drought tolerance in rice (Da-Hong et al. 2009). RACK1 is a conserved scaffold protein that regulates expression of antioxidant-related enzymes such as superoxide dismutases (SODs) in plants. Inhibition of RACK1 increases the accumulation of SODs and provides tolerance against drought as well as reactive oxygen species (ROS). Likewise, suppression of farnesyltransferase/squalene synthase (SQS) through siRNA generated from maize squalene synthase enhanced drought tolerance at vegetative and reproductive stages in rice (Manavalan et al. 2012). Increase in endogenous sterol level through silencing of SQS decreases the stomata density and prevents water loss through transpiration, thus preventing the plant from wilting under drought condition. OsTZF1 is a CCCH-type zinc finger protein that gets expressed under drought, salinity, and ROS conditions. Silencing of OsTZF1 gene enhances the tolerance of rice plants to high salt and low water conditions, indicating its role in abiotic stress tolerance (Jan et al. 2013). Low expression of OsTZF1 gene maintains the internal homeostasis of plants through change in hormonal expression at cellular and molecular level under high salt condition. Similarly, suppression of the proline-rich proteins in Poncirus trifoliata through RNAi decreases the tolerance in plants against cold conditions. It was observed that PtrPRP protein gets accumulated at high level in cold condition and its inhibition disrupts the reactive oxidative species homeostasis and membrane permeability in plants (Peng et al. 2015).
Response to abiotic stress is regulated by different miRNAs in economically important crops such as rice, wheat, legumes, sugarcane, radish, etc. (Goswami et al. 2014; Kruszka et al. 2014; Naya et al. 2014; Gentile et al. 2015; Zhang 2015; Sun et al. 2015; Shriram et al. 2016). miRNAs mostly regulate transcription, detoxification, and development processes. Several miRNAs get upregulated under heat stress condition. For example, high expression of mir398 under heat stress condition suppresses copper/zinc super oxide dismutase (CSD) genes. It was observed that the overexpression of miRNA398 decreases the sustainability of Arabidopsis thaliana and common bean (Phaseolus vulgaris) plants under heat stress due to miRNA-mediated degradation of CSD mRNAs (Guan et al. 2013; Naya et al. 2014). Cold-tolerant rice plants can also be generated, without any developmental defect through downregulation of TF by Osa-miRNA319 (Yang et al. 2013). Similarly, a drought-responsive mir168 acts on nuclear factor Y (NF-YA5). Overexpression of soybean NF-YA5 in A. thaliana increases its tolerance against drought condition (Ni et al. 2013). Gao et al. (2011) showed that high expression of Osa-miRNA393 decreases susceptibility of rice toward intolerable salt concentration present in soil. Recently, 22 novel miRNAs were identified in radish (Raphanus sativus) under high salt conditions, which regulate salt-responsive genes such as auxin response factors (ARFs), squamosa promoter-binding-like proteins (SPLs), and nuclear transcription factor Y (NF-Y) (Sun et al. 2015). NAC gene encodes for transcription factor required during plant development and environmental stresses. Thus, miRNAs maintain resistance to different abiotic stresses through up- and downregulation of the target gene transcripts.
3.3.3 Increasing Nutritional Value
Plants are the major source of various biologically active compounds required for overall growth of human. More than 2 billion people are found deficient in one or the other major nutrients and showed “hidden hunger” malnutrition (FAO 2013). Nowadays, various molecular biology and biotechnology techniques are being exploited in order to achieve the required level of nutrition in major staple foods. RNAi offers the new avenue for biofortification of nutrients in crop plants through modification of various physiological and biochemical pathways. Essential fatty acids found in oil are important for smooth functioning of the heart in human. Fatty acid composition in seeds can be easily modified through RNAi technology. The stability and oil quality of soybean oil were improved by downregulation of the expression of alpha-linolenic acid (18:3). Hairpin RNA-mediated tissue-specific suppression of omega-3 fatty acid desaturase enzyme significantly reduced the level of alpha-linolenic acid in transgenic soybean from 1 to 3% as compared to its level in untransformed soybean. Omega-3 fatty acid desaturase enzyme is responsible for the conversion of linoleic acid (18:2) to alpha-linolenic acid (18:3) in seeds (Flores et al. 2008). Opaque 2 gene (O2) encodes basic leucine zipper transcriptional factor, which regulates a storage protein. RNAi-mediated suppression of Opaque 2 gene resulted in increased lysine content in maize seeds without affecting the general function of O2 (Angaji et al. 2010). Wheat expressing high level of amylase can be generated by downregulating the starch-branching enzymes (SBE) through RNAi (Regina et al. 2006). Low glutelin-containing rice is highly recommended for kidney patients due to its easy digestibility property. Kusaba et al. (2003) produced low in glutelin content rice variety, LGC-1 (low gluten content 1), through inhibition of GluB gene expression. When potatoes were grown at low temperature, starch was converted into sucrose and fructose, making the potato sweet, a phenomenon called “cold sweetening.” RNAi-mediated downregulation of sucrose phosphatase gene (SPP) inhibited the cold sweetening phenomena in potato without significantly affecting its other agronomic parameters (Chen et al. 2008). Starch degradation is mediated by phosphorylation and dephosphorylation processes. Inhibition of starch degradation through alteration in the phosphate metabolic genes using RNAi enhanced the starch content in A. thaliana and Zea mays (Weise et al. 2012).
Tomatoes are known for their minerals, fibers, vitamins, and antioxidant property (Rajam et al. 2007). Overexpression of carotenoid or flavonoid synthetic genes or transcription factors increases either carotenoid or flavonoid content. RNAi has been employed in order to improve the level of both carotenoids and flavonoids in tomato fruit. DET1 is a photomorphogenesis regulatory gene, which represses several light-mediated signaling pathways. Expression of dsRNA of DET1 under fruit-specific promoter in tomato suppressed endogenous expression of DET1 and resulted in high levels of flavonoids and carotenoids in tomato fruits (Davuluri et al. 2005). Similarly, downregulation of lycopene epsilon-cyclase (ε-CYC) gene expression through RNAi increased the carotenoid content in rapeseed (Brassica napus). High expression of β-carotene, lutein, zeaxanthin, and violaxanthin was observed in seeds obtained from these RNAi Brassica lines (Yu et al. 2007). Hence, RNAi has tremendous potential to eradicate the malnutrition across the world.
3.3.4 Increase in Shelf Life of Fruits
Fruits and vegetables are rich in various minerals and vitamins. They are harvested, stored, and transported for human consumption. The post-harvest crop losses include losses due to mishandling, spoilage, diseases, and pest infestation during storage and transportation. Delayed in ripening is one of the process through which post-harvest losses can be minimized. Climacteric fruits respond to ripening process according to the concentration of ethylene, where ethylene acts as ripening hormone which initiates, regulates, and coordinates the expression of various ripening-related genes. Blocking of ethylene biosynthesis, ethylene-mediated signaling, and ethylene response elements through RNAi has been shown to delay the ripening process and help in enhancement of shelf life of fruits and vegetables, a trait which is demanded in post-harvesting or transportation industry (Xiong et al. 2005; Gupta et al. 2013; Luo et al. 2013). 1-Aminocyclopropane-1-carboxylate (ACC) oxidase is an enzyme involved in synthesis of ethylene from its precursor ACC. Suppression of ACC oxidase through RNAi decreased the production of ethylene and delayed the ripening process in tomato (Xiong et al. 2005). Synthesis of ethylene precursor ACC is catalyzed by ACC synthase, a critical enzyme in ethylene biosynthetic pathway. Gupta et al. (2013) showed that simultaneous silencing of the three homologs of ACC synthase regulates the ethylene biosynthesis more efficiently. They expressed the chimeric dsRNA resulted from an off-target-free sequences of three tomato ACS homologs under the control of fruit-specific promoter 2A11 and observed a delay in ripening and increase in shelf life for about 45 days in transgenic tomato due to low production of ethylene. They also observed that the expression of ethylene-responsive genes gets affected by low expression of ethylene in RNAi plants. Ripening process also leads to accumulation of carotenoids in fruits. The key carotenoid biosynthesis gene (SlPSY1) is inhibited by STAY-GREEN (SlSGR1) protein. SlSGR1 also coordinates with ripening process through regulation of ethylene signaling and expression of ethylene-responsive genes. RNAi-mediated downregulation of SlSGR1 enhances the shelf life of tomato up to 25–48 days. The low expression of SlSGR1 protein suppressed the production of H2O2 and alters the ethylene-mediated signal transduction in tomato, thus enhances ripening process through ethylene and carotenoid production (Luo et al. 2013).
Fruit development and ripening is a complex process, regulated by a variety of microRNAs (Moxon et al. 2008; Molesini et al. 2012; Karlova et al. 2013). The targets of miRNAs were found to be wide range of transcription factors (TFs), which act as negative or positive regulators for fruit ripening process. For example, tomato mir172 negatively regulates ripening process through downregulation of APETALA2 (SlAP2a) gene (Chung et al. 2010; Karlova et al. 2011). Similarly, mir156 targets squamosa promoter-binding protein (SBP) and negatively regulates ripening process (Manning et al. 2006; Moxon et al. 2008). Inhibition of SBP through mir156 induced colorless never ripe (CNR) phenotype in tomato (Moxon et al. 2008). Karlova et al. (2013) proposed that a correlation exists between CNR and AP2 during ripening but these TFs are negatively regulated by miRNA 156 and 172, respectively. A recent study showed that the ripening inhibitor (RIN) transcription factor binds directly to the promoter sequence of mir172a and positively regulate its expression. Thus, fruit ripening process is coordinated by ripening inhibitors, miRNAs, and ethylene response elements (Gao et al. 2015)
3.3.5 Production of Seedless Fruits
Seedless fruits and vegetables are highly desirable in the market for fresh consumption as well as for production of processed food (Molesini et al. 2012). Various fruit characteristics also get improved through seedlessness, e.g., the absence of seed formation in watermelon and cucumber increases yield and shelf life (Pandolfini 2009). Seedless fruits are generally produced through parthenocarpy, a naturally occurring process which involves direct development of fruit from ovary without pollination or fertilization (Gorguet et al. 2005). It can also be artificially induced by disrupting the genes involved in the process of seed set and seed formation. The complex process of seed formation is temporally and spatially mediated by phytohormones. Generally, seedless fruits obtained through mutation and phytohormone alteration methods are generally found to bear pleiotropic effects such as reduced fruit size, effect on its taste, etc. (Varoquaux et al. 2000; Wang et al. 2005). Therefore, more efficient methods are now employed for generation of parthenocarpic fruits. RNAi-mediated downregulation of chalcone synthase, first gene in flavonoid biosynthesis, leads to development of parthenocarpy in tomato (Schijlen et al. 2007). Recent study showed that suppression of flavonol synthase through RNAi induced seedlessness in tobacco (Nicotiana tabacum cv Xanthi) (Mahajan et al. 2011). Flavonol synthase involved in formation of flavonols in flavonoid biosynthesis pathway is required for seed formation. Similarly, silencing of auxin-responsive element (ARF7) in tomato through RNAi causes seedlessness (De Jong et al. 2009). Parthenocarpy can also be induced through miRNA-mediated regulation of target genes. For example, miRNA167 regulates expression of auxin-responsive element (ARF8) and alteration in the expression of ARF8 through aberrant expression of miRNA-induced parthenocarpy in both Arabidopsis and tomato (Molesini et al. 2009). Besides phytohormones, protein synthesis genes have also been utilized for generation of parthenocarpic fruits in tomato. Silencing of protein synthesis gene AUCSIA caused seedlessness in tomato through uncoupling of fruit formation with fertilization (Molesini et al. 2009). Thus, RNAi approach provides a good alternative to achieve parthenocarpy or seedlessness in fruits.
3.3.6 Modification of Flower Color
Various attributes of flower contribute to million dollar ornamental industry worldwide. Flower color is one of them, which is governed by combination of different pigments such as flavonoids, carotenoids, and betalains (He et al. 2013). These pigments also act as attractant to pollinators and protect the plants from harmful UV rays (He et al. 2013). RNAi-mediated genetic manipulation of pigment biosynthetic pathways offers a new platform for development of commercially valuable color pattern in flowers. Temporal and spatial regulation of gene expression in a highly specific manner through RNAi can induce desirable variation in flower color. Anthocyanins are the most prominent flavonoids, responsible for orange to red and purple to blue color in flowers. Anthocyanins are derived from a branch of flavonoid biosynthesis pathway. The first step in anthocyanin is mediated by chalcone synthase enzyme (CHS). Silencing of anthocyanin biosynthetic genes can be manifested into diverse flower colors. For example, downregulation of three anthocyanin biosynthetic genes – chalcone synthase (CHS), anthocyanidin synthase (ANS), and flavonoid 3′ 5′-hydroxylase (F 3′ 5′ H) – through RNAi induced variable color patterns in Gentiana spp. (Nakatsuka et al. 2008). RNAi-CHS plants showed pure white to pale-blue color petals, and RNAi-ANS plants exhibited only pale-blue color, whereas RNAi plants expressing dsRNA of F 3′ 5′ H gene developed magenta flowers. The involvement of these genes at different steps in anthocyanin biosynthetic pathway is found to be responsible for observed variation in color patterns in three different gentian RNAi plants. Later, the same group demonstrated that more variation in flower color can be introduced by suppressing the target genes together. Silencing of anthocyanin biosynthesis gene 3′-aromatic acyltransferase (5/3′ AT) and F 3′ 5′ H through chimeric RNAi produced variable colors in gentian flower ranging from liliac to pale blue (Nakatsuka et al. 2010). Downregulation of these enzymes caused accumulation of blue color dolphin dine pigment in flowers. Similarly, silencing of CHS gene in Tricyrtis sp., a monocotyledon plant by RNAi-induced alternation in flower color in sepals (Kamiishi et al. 2012). RNAi can also be used in combination of other techniques for improvement and change in color. For example, He et al. (2013) produced blue-colored Chrysanthemum flowers through RNAi-mediated silencing of F3’H and overexpression of the exogenous Senecio cruentus F 3′ 5′ H gene. Thus, commercial value of ornamental plant can be increased through production of desirable color variation in flowers using RNAi approach.
3.3.7 Development of Male Sterile Lines
Hybrids are contributing significantly to meet the future demand for increasing food worldwide. Hybridization leads to production of offsprings with superior characteristics in comparison to their parents, the process known as hybrid vigor or heterosis. Male sterility in female and its restoration in future generations is a prerequisite to produce hybrids in self-pollination plants. Male sterility is a naturally occurring phenomenon in some of the cross-pollinating plants such as grasses and safflower (Duvick 1999). To widen the scope for production of hybrid seeds in self-pollinating plants, male sterility can be artificially induced through various conventional and genetic engineering methods. RNAi-mediated suppression of genes involved in tapetum and pollen development was found to be more effective in producing male sterility (Nawaz-ul-Rehman et al. 2007; Tehseen et al. 2010; Sinha and Rajam 2013). Tapetum is a layer in the microsporangium which provides nutrition to the developing pollen grains. RNAi-mediated silencing of TA29, an anther-specific gene involved in pollen development in tobacco, resulted in male sterility (Nawaz-ul-Rehman et al. 2007). Similarly downregulation of another anther-specific gene Bcp1 arrested the pollen development and induced male sterility in A. thaliana. The male sterile RNAi lines were found to be phenotypically normal and produced viable seeds when restored through cross-fertilization with male-fertile plants. Bcp 1 expressed during diploid tapetum and haploid microspore development and, thus, inhibition of its expression affected the pollen development adversely (Tehseen et al. 2010). S-adenosyl methionine decarboxylase (SAMDC) is a key enzyme in polyamine biosynthesis, required during pollen maturation and germination (Sinha and Rajam 2013). Thus, expression of chimeric (SAMDC) dsRNA under the control of tapetum-specific A9 promoter caused simultaneous silencing of three SAMDC isoforms in tapetum tissue, which resulted in formation of male sterile SAMDC-RNAi lines without affecting their female fertility.
MicroRNA-mediated regulation of male sterility has been reported in various plant species such as cotton (Yang et al. 2013; Wei et al. 2013; Yang et al. 2016), citrus (Fang et al. 2014), and radish (Zhang et al. 2016). For example, rsa-miR159a regulates the expression of transcription factor required during anther and pollen development. High expression of ras-mir159a decreases the expression of MYB101 TF and, thus, induces male sterility through inhibition of normal pollen development in radish plants (Zhang et al. 2016). Yang et al. (2016) observed differential expression of 49 conserved and 51 novel miRNAs during male sterility in cotton. Hence, RNAi-mediated silencing of various genes involved in pollen development opens a new door for production of hybrid seeds in various plant systems.
3.3.8 Production of Secondary Metabolites
Plant secondary metabolites are major sources of pigments, fragrances, drugs, food additives, and pesticides. It is estimated that 70–80% of worldwide population fulfill their primary health requirements from the herbal medicines obtained from the plant secondary metabolites (Canter et al. 2005). Complex array of genes is responsible for synthesis of secondary metabolites. RNAi is recognized as an effective strategy for manipulation of secondary metabolites (Borgio 2009). Replacement of morphine with non-narcotic alkaloid reticuline in opium poppy (Papaver somniferum) presented a best example of metabolic engineering through RNAi. Allen et al. (2004) were the first to report RNAi-mediated silencing of multiple genes involved in different steps of a complex biochemical pathway. They designed hpRNA construct which caused simultaneous downregulation of all members of codeine reductase (COR) gene family. Silencing of COR gene family caused accumulation of (S)-reticuline, a non-narcotic alkaloid precursor in transgenic plants at the expense of morphine, codeine, and opium. Cassava (Manihot esculenta) is the third largest source of carbohydrates and major staple food in tropical countries. Presence of cyanogenic glucosides compound in cassava makes it unsuitable for food consumption at large scale. Silencing of cytochrome P450 through RNAi reduced the cyanogenic glucoside to a significant level in leaves as well as in tubers (Jørgensen et al. 2005). Potato (Solanum tuberosum L.) tubers have emerged as efficient bioreactors for production of recombinant human therapeutic glycoproteins. RNAi technology has been employed in inhibition of endogenous patatin expression at transcriptional and translational level. The developed potato tubers showed high accumulation of heterologous patatin glycoprotein, which has fastened the purification of recombinant protein (Kim et al. 2008).
Caffeine is a stimulant for the central nervous, respiratory, and circulatory system. It also gives protection against type 2 diabetes, Parkinson’s disease, and liver disorders. However, high consumption of caffeine causes insomnia, restlessness, and palpitations. Decaffeinated coffee (DECAF) occupy only 10% of the world coffee market. Suppression of CaMXMT1 (7-N-methylxanthine methyltransferase or theobromine synthase) by the RNAi resulted in reduction of caffeine content up to 70% in the silenced transgenic plant (Ogita et al. 2003). Similarly, low caffeine producing tea was generated through downregulation of caffeine synthase gene (CS) without affecting its stimulating property (Mohanpuria et al. 2011).
3.3.9 Removal of Allergens from Food Crops
Consumption of allergens containing food causes various health problems in humans, which even cannot be cured with the use of existing therapies. Allergens are naturally occurring compounds found in various food crops, capable of producing allergic response even if consumed in small quantities. Besides, they also cause hindrance in extraction of pure desirable products. Elimination of these unwanted compounds from plants is a costly and cumbersome process, requiring various chemical reactions and engineering processes, which reduces the nutritional value of food materials. RNAi has emerged as a powerful technology for removal of allergens through the alteration in their biosynthetic pathway or biochemical responses. It helps to enhance the edibility and food quality of crop plants without affecting their physiological processes. Major apple (Malus domestica) contains a pathogen-related protein PR10 allergens Mal d 1 which induce IgE-mediated hypersensitive response in organisms. Expression of Mal d1 dsRNA sequence reduces the expression of endogenous gene in developed RNAi apple plants and lowered the allergic response upon consumption (Gilisen et al. 2005). Le et al. (2006) provided a promising design for development of allergen-free tomato plants through RNAi. They identified a novel allergen Lyce3 in tomato which encodes a hydrophilic, non-specific lipid transfer protein (ns-LTP). Specific downregulation of Lyce3 gene through RNAi resulted in suppression of Lyce 3 accumulation in tomato. Further, potential of allergens was tested with histamine test for the developed RNAi tomato plants. RNAi tomato plants showed reduced allergenicity and, thus, increased the edibility of tomato to allergen-sensitive population (Le et al. 2006). Consumption of heavy metals even in low concentration produces irreversible damaging effect on various physiological processes in human. Rice can accumulate cadmium (Cd) to a significant level in its seeds due to the presence of phytochelatin synthase (PCS) genes. RNAi-mediated suppression of phytochelatin synthase (OsPCS1) gene reduced the accumulation of Cd in rice (Li et al. 2007). Thus, accumulation of heavy metals in rice seeds can be regulated through RNAi even when plants are grown in heavy metal-polluted soil. Tearless onion can be generated through RNAi-mediated suppression of tear-inducing lachrymatory factor synthases gene (LFS). LFS is responsible for production of tear-inducing lachrymatory factor, Propanethial S-oxidase (LF), from 1-propenyle sulfenic acid. Inhibition of LFS resulted in generation of tearless onion due to low production of LF (Eady et al. 2008). RNAi can also be employed for removal of neurotoxic and carcinogenic compounds from food crops. Consumption of neurotoxin found in chickpeas causes lathyrism, a severe paralytic neurotoxic disease. RNAi-mediated downregulation of BOAA (β-N-oxalylamino-L-alanine) neurotoxin lowered its concentration in crop plants to a level, which is found safe for consumption. The production of carcinogenic compound in tobacco can also be minimized through RNAi-mediated silencing of nicotine demethylase gene (DM). DM is responsible for production of carcinogenic precursor from nicotine (Lewis et al. 2008). Cotton is a major cash crop, known for its fibers and oil worldwide. The cotton seeds are rich in proteins and calories, but they largely remain unutilized due to the presence of high amount of gossypol terpenoid. Gossypol is found in all parts of cotton plant and provides protection against herbivores. Downregulation of gossypol synthesis gene (δ-Cadinene synthase) in tissue-specific manner resulted in development of gossypol-free transgenic seeds, without affecting its expression in other parts of the plant (Sunilkumar et al. 2006). Even, ultralow gossypol-containing cotton seeds (ULGCS) can be produced by tight regulation of gossypol biosynthetic δ-Cadinene synthase gene through RNAi (Rathore et al. 2012).
3.3.10 Change in Plant Architecture
Plant architecture controls several important agronomic traits in plants. For example, plant height, pattern of shoot branching, plant morphology, inflorescence, crop yield, and resistance to environmental stresses (Khush 2001; Camp 2005; Wang and Li 2006). Plant architecture can also be modified in order to minimize the negative effects of climate change on crop productivity. For example, plants are grown in drought and nutrient-deficient soil by manipulating its root architecture for maximum absorption of water and nutrients (de Dorlodot et al. 2007). Understanding of molecular basis of plant architecture has served as platforms for RNAi-mediated alternation in plant architecture (Wang and Li 2008). Shorter plants with erect leaf architecture were produced through RNAi-mediated silencing of OsDWARF4 gene in rice (Feldmann 2006). RNAi-mediated downregulation of ornithine decarboxylase (ODC) gene, which is a key gene involved in polyamine biosynthesis resulted in significant physiological and morphological changes including reduced leaf size, decreased abiotic stress tolerance, delayed flowering, and early onset of senescence in tobacco (Choubey and Rajam 2017). Increase in biomass was observed in RNAi plants due to increase in rate of photosynthesis in lower erect leaves. Biofuel production can be enhanced through low lignin content in plant material. Lignin makes the plant material recalcitrant for conversion to ethanol. Low lignin-containing plants can be produced through downregulation of lignin biosynthetic gens by RNAi. For instance, RNAi-mediated downregulation of lignin-associated genes such as Cinnamate-4-hydroxylase, shikimate hydroxycinnamoyl transferase, and 4-coumarate-CoA ligase reduced the lignin content and increased its accessibility to cellulose to degradation (Hisano et al. 2009). RNAi technology is used to enhance the crop yield by manipulating the plant architecture. Taller rice variety QX1 was converted to semi-dwarf variety through RNAi-mediated suppression of GA 20-oxidase (OsGA20ox2) gene. The developed transgenic rice exhibited high yield due to the significant increase in panicle length, number of seeds per panicle, and weight of individual seeds (Qiao et al. 2007).
Plant architecture has been found to be regulated by miRNAs. The manipulation of miRNA expression directly or indirectly affected the plant architecture, biomass accumulation, and yield (Chuck et al. 2011; Wang et al. 2012; Rubinelli et al. 2013). Corngrass1 (Cg1) miRNA that belongs to the mir156 family regulates vegetative growth and flowering in plants. Overexpression of Cg1 miRNA caused prolongation of vegetative phase and delay in flowering time in maize (Chuck et al. 2011). Similarly, phenotype was also observed when Cg1 was overexpressed in other plant species, for example, overexpressing Cg1miRNA in Populus plants showed significant shortening of internode length, increase in the growth of axillary meristem, and ~30% reduction in stem lignin content as compared to the untransformed control (Rubinelli et al. 2013). The miRNA-mediated manipulation of plant architecture enhanced the grain yield in rice (Jiao et al. 2010; Miura et al. 2010; Wang et al. 2012). Osa-miR156 regulates the expression of OsSPL14 (squamosa promoter-binding protein-like 14) and is found to have positive effects on plant architecture and yield in rice (Jiao et al. 2010; Miura et al. 2010). Overexpression of Osa-miR397 elevated grain production up to 25% in RNAi rice plants due to increase in panicle branching and grain size. Osa-miR397 downregulates a brassinosteroid-sensitive gene OsLAC (coding for a laccase-like protein) and, thus, directs the energy toward the growth of plants. Since miR397 is found to be highly conserved across different plant species, similar strategy can be used in other crops for increasing grain yield (Zhang et al. 2013). Thus, RNAi technology has a wide utility in manipulating the plant architecture for high yield, increase in biomass, flowering, and removal of undesirable phenotypes. Rose plant can be easily modified for its thorn characteristic or the plant architecture in mulberry, and tea plants can be manipulated for easy plucking of leaves.
3.4 Conclusions and Future Prospects
The main challenge for agriculture in the twenty-first century is to provide food security for existing and expanding population. Besides, malnutrition is also a major problem faced by people in developing countries. To ensure supply of balanced food to the world, it is necessary to develop biofortified staple food, vegetables, and fruits, enriched in essential compounds and elements such as fatty acids, vitamins, amino acids, and micro- or macronutrients. Increase in resistance toward the present technology, changing environment, increasing population, and pollution have put a high pressure on the existing natural resources for high crop productivity. The development of crop varieties resistant to pathogens and pests and tolerant to changing environmental conditions such as high temperature, drought, flood, oxidative stresses, high salt concentration, and heavy metal-polluted soil can be a blow for world food security, malnutrition, and famine problems. RNAi-based technology has proven its potential in development of crop varieties resistant to biotic and abiotic stresses. Besides, RNAi is creating a milestone in genetic manipulation of crop varieties for highly recommended agronomic traits. Crops have been engineered through RNAi for novel and commercially important agronomic traits including decaffeinated tea, coffee, nicotine-free tobacco, allergen-free cereals, low glutelin-containing wheat, healthy fatty acid-enriched oil crops, blue rose, browning-free apple, tear-free onion, easily packageable tomato, etc. Additionally, RNAi has been employed to remove carcinogenic, neurotoxin, and mycotoxin compounds from food crops. Most of the RNAi-based studies involve a single-gene silencing for the improvement of useful traits in crops. However, silencing of more than one gene can be achieved through chimeric RNAi constructs, which will be useful for improvement of several traits in crop plants simultaneously (Gupta et al. 2013; Sinha and Rajam 2013; Yogindran and Rajam 2015). RNAi-mediated crop improvement strategies hold a tremendous potential for enhancement of desirable traits and eradication of undesirable traits in crop plants (Fig. 3.3).
RNAi-mediated strategies are the most preferred and powerful alternative for crop improvement as compared to the existing approaches. It uses the existing conserved machinery for sequence-specific silencing at posttranscriptional level. So, there is no extra load on plant for production of effective proteins, and even, there are fewer chances of allergic responses. RNAi effector molecules can tolerate mutation without affecting the silencing efficiency; thus, it is very unlikely that the target pest or pathogen would gain resistance. However, RNA-based technology can result in off-target effects in the same or different plants and development of undesirable pleiotropic effects because of sequence homology. Therefore, RNAi-based transgenic approach should be planned in such a way that no or minimal off-target and pleiotropic effects will arise. For example, an in-depth in silico analysis should be performed before selecting the target gene sequence for preparation of hairpin RNAi construct. The applications of RNAi technology should move from lab to field and from model plant to crop plants (Jagtap et al. 2011; Katoch and Thakur 2013; Koch and Kogel 2014; Saurabh et al. 2014; Kamthan et al. 2015). In the near future, RNAi-mediated crop improvement programs in combination with other technologies will change the food security parameter across the world and improve the way of life.
References
Allen RS, Millgate AG, Chitty JA, Thisleton J, Miller JA, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi mediated replacement of morphine with the non-narcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22:1559–1566
Andika IB, Kondo H, Tamada T (2005) Evidence that RNA silencing mediated resistance to beet necrotic yellow vein virus is less effective in roots than in leaves. Mol Plant-Microbe Interact 18:194–204
Angaji SA, Hedayati SS, Poor RH, Poor SS, Shiravi S, Madani S (2010) Application of RNA interference in plants. Plant Omics J 3:77–84
Axtell MJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64:137–159
Banerjee S, Banerjee A, Gill SS, Gupta OP, Dahuja A, Jain PK, Sirohi A (2017) RNA interference: a novel source of resistance to combat plant parasitic nematodes. Front Plant Sci 8:834. https://doi.org/10.3389/fpls.2017.00834
Bao N, Lye KW, Barton MK (2004) MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7:653–662
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Baulcombe D (2000) Unwinding RNA silencing. Science 290:1108–1109. https://doi.org/10.1126/science.290.5494.1108
Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326
Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366
Bezanilla M, Pan A, Quatrano RS (2003) RNA interference in the moss Physcomitrella patens. Plant Physiol 133:470–474
Borgio JF (2009) RNA interference (RNAi) technology: a promising tool for medicinal plant research. J Med Plant Res 3:1176–1183
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190. https://doi.org/10.1126/science.1159151
Camp WV (2005) Yield enhancement genes: seeds for growth. Curr Opin Biotechnol 16:147–153
Campo S, Peris-Peris C, Sire C, Moreno AB, Donaire L, Zytnicki M, Notredame C, Llave C, San Segundo B (2013) Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol 199:212–227
Canter PH, Thomas H, Ernst E (2005) Bringing medicinal plants into cultivation: opportunities and challenges for biotechnology. Trends Biotechnol 23:180–185
Castel SE, Martienssen RA (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14:100–112
Cerutti H, Ma X, Msanne J, Repas T (2011) RNA-mediated silencing in algae: biological roles and tools for analysis of gene function. Eukaryot Cell 10:1164–1172. https://doi.org/10.1128/EC.05106-11
Chen S, Hajirezaei MR, Zanor MI, Hornyik C, Debast S, Lacomme C, Fernie AR, Sonnewald U, Bornke F (2008) RNA interference-mediated repression of sucrose-phosphatase in transgenic potato tubers (Solanum tuberosum) strongly affects the hexose-to-sucrose ratio upon cold storage only minor effects on total soluble carbohydrate accumulation. Plant Cell Environ 31:165–176
Chen W, Kastner C, Nowara D, Oliveira-Garcia E, Rutten T, Zhao Y, Deising HB, Kumlehn J, Schweizer P (2016) Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J Exp Bot 67:4979–4991. https://doi.org/10.1093/jxb/erw263
Choubey A, Rajam MV (2017) Transcriptome response and developmental implications of RNAi-mediated ODC knockdown in tobacco. Funct Integr Genomics DOI. https://doi.org/10.1007/s10142-016-0539-3
Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D, Arora R, Bragg JN, Vogel JP, Singh S, Simmons BA, Pauly M, Hake S (2011) Overexpression of the maize corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc Natl Acad Sci U S A 108:17550–17555. https://doi.org/10.1073/pnas.1113971108
Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J (2010) A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J 64:936–947
Da-Hong L, Hui L, Yan-li Y, Ping-ping Z, Jian-sheng L (2009) Down-regulated expression of RACK1 gene by RNA interference enhances drought tolerance in rice. Rice Sci 16:14–20
Davuluri GR, Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, Bramley PM, Pennings HMJ, Bowle C (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23:890–895
de Dorlodot S, Forster B, Pages L, Price A, Tuberosa R, Draye X (2007) Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 12:474–481
De Jong M, Wolters-Arts M, Feron R, Mariani C, Vriezen WH (2009) The Solanum lycopersicum 7 (auxin response factorSlARF7) regulates auxin signaling during tomato fruit set and development. Plant J 57:160–117
Duvick DN (1999) Heterosis: feeding people and protecting natural resources. In: Coors JG, Pandey S (eds) Genetics and exploitation of heterosis in crops. American Society of Agronomy/Crop Science Society of America, Madison, pp 19–29
Eady CC, Kamoi T, Kato M, Porter NG, Davis S, Shaw M, Kamoi A, Imai S (2008) Silencing onion lachrymatory factor synthase causes a significant change in the sulfur secondary metabolite profile. Plant Physiol 147:2096–2106. https://doi.org/10.1104/pp.108.123273
Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–508
Fang YN, Qiu WM, Wang Y, Wu XM, Xu Q, Guo WW (2014) Identification of differentially expressed microRNAs from a male sterile Ponkan mandarin (Citrus reticulata Blanco) and its fertile wild type by small RNA and degradome sequencing. Tree Genet Genome 10:1567–1581. https://doi.org/10.1007/s11295-014-0780-7
FAO (2013) The state of food insecurity in the world, executive summary. Rome, Italy, FAO
Feldmann KA (2006) Steroid regulation improves crop yield. Nat Biotechnol 24:46–47
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Flores T, Karpova O, Su X, Zeng P, Bilyeu K, Sleper DA, Nguyen HT, Zhang ZJ (2008) Silencing of GmFAD3 gene by siRNA leads to low alpha-linolenic acids (18:3) of fad 3-mutant phenotype in soybean [Glycine max (Merr.)]. Transgenic Res 17:839–850
Gao P, Bai X, Yang L, Lv D, Pan X, Li Y, Cai H, Ji W, Chen Q, Zhu Y (2011) osa-MIR393: a salinity and alkaline stress-related microRNA gene. Mol Biol Rep 38:237–242. https://doi.org/10.1007/s11033-010-0100-8
Gao C, Ju Z, Cao D, Zhai B, Qin G, Zhu H, Fu D, Luo Y, Zhu B (2015) MicroRNA profiling analysis throughout tomato fruit development and ripening reveals potential regulatory role of RIN on microRNAs accumulation. Plant Biotechnol J 13:370–382
Gentile A, Dias LI, Mattos RS, Ferreira TH, Menossi M (2015) MicroRNAs and drought responses in sugarcane. Front Plant Sci 6:58. https://doi.org/10.3389/fpls.2015.00058
Gilisen LJ, Bolhaar ST, Matos CI, Rouwendal GJ, Boone MJ, Krens FA et al (2005) Silencing of major apple allergen Mal d 1 by using the RNA interference approach. J Allergy Clin Immunol 115:364–369. https://doi.org/10.1016/j.jaci.2004.10.014
Gorguet B, van Heusden AW, Lindhout P (2005) Parthenocarpic fruit development in tomato. Plant Biol 7:131–139
Goswami S, Kumar RR, Rai RD (2014) Heat-responsive microRNAs regulate the transcription factors and heat shock proteins in modulating thermo-stability of starch biosynthesis enzymes in wheat (Triticum aestivum L.) under the heat stress. Aust J Crop Sci 8:697–705
Guan Q, Lu X, Zeng H, Zhang Y, Zhu J (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermo tolerance in Arabidopsis. Plant J 74:840–851. https://doi.org/10.1111/tpj.12169
Gupta A, Pal RK, Rajam MV (2013) Delayed ripening and improved fruit processing quality in tomato by RNAi-mediated silencing of three homologs of 1-aminopropane-1-carboxylate synthase gene. J Plant Physiol 170:987–995
Hamilton AJ, Baulcombe DC (1999) A novel species of small antisense RNA in posttranscriptional gene silencing. Science 286:950–952
Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296
He H, Ke H, Keting H, Qiaoyan X, Silan D (2013) Flower colour modification of Chrysanthemum by suppression of F3'H and overexpression of the exogenous Senecio cruentus F3' 5' H gene. PLoS One 8:e74395
Hisano H, Nandakumar R, Wang ZY (2009) Genetic modification of lignin biosynthesis for improved biofuel production. In Vitro Cell Dev Biol Plant 45:306–313. https://doi.org/10.1007/s11627-009-9219-5
Huang G, Allen R, Davis E, Baum T, Hussey R (2006) Engineering broad rootknot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc Natl Acad Sci U S A 103:14302–14306
Jagtap UB, Gurav RG, Bapat VA (2011) Role of RNA interference in plant improvement. Naturwissenschaften 98:473–492
Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, Shinozaki K, Nakashima K, Yamaguchi-Shinozaki K (2013) OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol 161:1202–1216
Jiao Y, Wang Y, Xue D, Wang J, Yan M, Liu G, Dong G, Zeng D, Lu Z, Zhu X, Qian Q, Li J (2010) Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet 42:541–544
Jin S, Singh ND, Li L, Zhang X, Daniell H (2015) Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant Biotech J 13:435–446
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Jørgensen K, Bak S, Busk PK, Sørensen C, Olsen CE, Puonti-Kaerlas J, Moller BL (2005) Cassava plants with a depleted cyanogenic glucoside content in leaves and tubers. Plant Physiol 139:363–374
Kamachi S, Mochizuki A, Nishiguchi M, Tabei Y (2007) Transgenic Nicotiana benthamiana plants resistant to cucumber green mottle mosaic virus based on RNA silencing. Plant Cell Rep 26:1283–1288. https://doi.org/10.1007/s00299-007-0358-z
Kamiishi Y, Otani M, Takagi H, Han DS, Mori S, Tatsuzawa F, Okuhara H, Kobayashi H, Nakano M (2012) Flower color alteration in the liliaceous ornamental Tricyrtis sp. by RNA interference-mediated suppression of the chalcone synthase gene. Mol Breed 30:671–680
Kamthan A, Chaudhuri A, Kamthan M, Datta A (2015) Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci 6:208. https://doi.org/10.3389/fpls.2015.00208
Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23:923–941
Karlova R, van Haarst JC, Maliepaard C, van de Geest H, Bovy AG, Lammers M, Angenent GC, de Maagd RA (2013) Identification of microRNA targets in tomato fruit development using high-throughput sequencing and degradome analysis. J Exp Bot 64:1863–1878
Katoch R, Thakur N (2013) RNA interference: a promising technique for the improvement of traditional crops. Int J Food Sci Nutr 64:248–259. https://doi.org/10.3109/09637486.2012.713918
Ketting RF (2011) The many faces of RNAi. Dev Cell 20:148–161
Khraiwesh B, Arif MA, Seumel GI, Ossowski S, Weigel D, Reski R, Frank W (2010) Transcriptional control of gene expression by microRNAs. Cell 140:111–122. https://doi.org/10.1016/j.cell.2009.12.023
Khraiwesh B, Zhu JK, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819:137–148
Khush GS (2001) Green revolution: the way forward. Nat Rev Genet 2:815–822
Kim YS, Lee YH, Kim HS, Kim MS, Hahn KW, Ko JH, Joung H, Jeon JH (2008) Development of patatin knockdown potato tubers using RNA interference (RNAi) technology, for the production of human-therapeutic glycoproteins. BMC Biotechnol 8:36. https://doi.org/10.1186/1472-6750-8-36
Koch A, Kogel KH (2014) New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol J 12:821–831. https://doi.org/10.1111/pbi.12226
Koch A, Biedenkopf D, Furch A, Weber L, Rossbach O, Abdellatef E, Linicus L, Johannsmeier J, Jelonek L, Goesmann A, Cardoza V, McMillan J, Mentzel T, Kogel KH (2016) An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog 12:e1005901
Kruszka K, Pacak A, Swida-Barteczka A, Nuc P, Alaba S, Wroblewska Z, Karlowski W, Jarmolowski A, Szweykowska-Kulinska Z (2014) Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J Exp Bot 65:6123–6135. https://doi.org/10.1093/jxb/eru353
Kumar R (2014) Role of microRNAs in biotic and abiotic stress responses in crop plants. Appl Biochem Biotechnol 174:93–115
Kumar P, Pandit SS, Baldwin IT (2012) Tobacco rattle virus vector: a rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS One 7:e31347
Kusaba M, Miyahara K, Iida S, Fukuoka H, Takano T, Sassa H, Nishimura M, Nishio T (2003) Low glutelin content 1: a dominant mutation that suppresses the glutelin multigene family via RNA silencing in rice. Plant Cell 15:1455–1467
Le LQ, Lorenz Y, Scheurer S, Fotisch K, Enrique E, Bartra J, Biemelt S, Vieths S, Sonnewald U (2006) Design of tomato fruits with reduced allergenicity by dsRNAi-mediated inhibition of ns-LTP (Lyc e 3) expression. Plant Biotechnol J 4:231–242. https://doi.org/10.1111/j.1467-7652.2005.00175.x
Lewis RS, Jack AM, Morris JW, Robert VJ, Gavilano LB, Siminszky B, Bush LP, Hayes AJ, Dewey RE (2008) RNA interference (RNAi)-induced suppression of nicotine demethylase activity reduces levels of a key carcinogen in cured tobacco leaves. Plant Biotechnol J 6:346–354. https://doi.org/10.1111/j.1467-7652.2008.00324.x
Li JC, Guo JB, Xu WZ, Ma M (2007) RNA interference-mediated silencing of phytochelatin synthase gene reduces cadmium accumulation in rice seeds. J Integr Plant Biol 49:1032–1037. https://doi.org/10.1111/j.1672-9072.2007.00473.x
Li X, Wang X, Zhang S, Liu D, Duan Y, Dong W (2012) Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS One 7:e39650. https://doi.org/10.1371/journal.pone.0039650
Liu F, Wang XD, Zhao YY, Li YJ, Liu YC, Sun J (2015) Silencing the HaAK gene by transgenic plant-mediated RNAi impairs larval growth of Helicoverpa armigera. Int J Biol Sci 11:67–74. https://doi.org/10.7150/ijbs.10468
Luo Z, Zhang J, Li J, Yang C, Wang T, Ouyang B, Li H, Giovannoni J, Ye Z (2013) A STAY-GREEN protein SlSGR1 regulates lycopene and β-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytol 198:442–452. https://doi.org/10.1111/nph.12175
Mahajan M, Ahuja PS, Yadav SK (2011) Post-transcriptional silencing of flavonol synthase mRNA in tobacco leads to fruits with arrested seed set. PLoS One 6:e28315. https://doi.org/10.1371/journal.pone.0028315
Mamta, Reddy KR, Rajam MV (2015) Targeting chitinase gene of Helicoverpa armigera by host-induced RNA interference confers insect resistance in tobacco and tomato. Plant Mol Biol 90:281–292
Manavalan LP, Chen X, Clarke J, Salmeron J, Nguyen HT (2012) RNAi-mediated disruption of squalene synthase improves drought tolerance and yield in rice. J Exp Bot 63:163–175
Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni J, Seymour GB (2006) A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet 38:948–952
Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen X (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25:1307–1313
Mao YB, Xue XY, Tao XY, Yang CQ, Wang LJ, Chen XY (2013) Cysteine protease enhances plant-mediated bollworm RNA interference. Plant Mol Biol 83:119–129
Matzke M, Matzke A, Pruss G, Vance V (2001) RNA-based silencing strategies in plants. Curr Opin Genet Dev 11:221–227. https://doi.org/10.1016/S0959-437X(00)00183-0
Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M (2010) OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet 42:545–549
Mohanpuria P, Kumar V, Ahuja PS, Yadav SK (2011) Producing low-caffeine tea through post-transcriptional silencing of caffeine synthase mRNA. Plant Mol Biol 76:523–234. https://doi.org/10.1007/s11103-011-9785-x
Molesini B, Pandolfini T, Rotino GL, Dani V, Spena A (2009) Aucsia gene silencing causes parthenocarpic fruit development in tomato[C][W]. Plant Physiol 149:534–548
Molesini B, Pii Y, Pandolfini T (2012) Fruit improvement using intragenesis and artificial microRNA. Trends Biotechnol 30:80–88. https://doi.org/10.1016/j.tibtech.2011.07.005
Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC (2010) Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328:872–875. https://doi.org/10.1126/science.1187959
Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V, Dalmay T (2008) Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res 18:1602–1609
Nakatsuka T, Mishiba K, Abe Y, Kubota A, Kakizaki Y, Yamamura S, Nishihara M (2008) Flower color modification of gentian plants by RNAi-mediated gene silencing. Plant Biotechnol TOKYO 25:61–68
Nakatsuka T, Mishiba KI, Kubota A, Abe Y, Yamamura S, Nakamura N, Tanaka Y, Nishihara M (2010) Genetic engineering of novel flower colour by suppression of anthocyanin modification genes in gentian. J Plant Physiol 167:231–237
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of chimeric chalcone synthase gene into Petunia results in reversible cosuppression of homologous genes in trans. Plant Cell 2:279–289. https://doi.org/10.1105/tpc.2.4.279
Nawaz-ul-Rehman MS, Mansoor S, Khan AA, Zafar Y, Briddon RW (2007) RNAi-mediated male sterility of tobacco by silencing TA29. Mol Biotechnol 36:159–165
Naya L, Paul S, Valdés-López O, Mendoza-Soto AB, Nova-Franco B, Sosa-Valencia G, Reyes JL, Hernández G (2014) Regulation of copper homeostasis and biotic interactions by microRNA 398b in common bean. PLoS One 9:e84416. https://doi.org/10.1371/journal.pone.0084416
Ni Z, Hu Z, Jiang Q, Zhang H (2013) GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol 82:113–129
Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, Hensel G, Kumlehn J, Schweizer P (2010) HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22:3130–3141
Nunes CC, Dean RA (2012) Host – induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol Plant Pathol 13:519–529. https://doi.org/10.1111/j.1364-3703.2011.00766.x
Oerke EC (2006) Crop losses to pests. J Agric Sci 144:31–43
Ogita S, Usefuji H, Yamaguchi Y, Koizumi N, Sano H (2003) Producing decaffeinated coffee plants. Nature 423:823. https://doi.org/10.1038/423823a
Pandolfini T (2009) Seedless fruit production by hormonal regulation of fruit set. Nutrients 1:168–177
Papolu PK, Gantasala NP, Kamaraju D, Banakar P, Sreevathsa R, Rao U (2013) Utility of host delivered RNAi of two FMRF amide like peptides, flp-14 and flp-18, for the management of root knot nematode, Meloidogyne incognita. PLoS One 8:e80603
Pareek M, Yogindran S, Mukherjee SK, Rajam MV (2015) Plant micro RNAs: biogenesis, functions and applications. In: Bahadur B, Rajam MV, Sahijram L, Krishnamurthy KV (eds) Plant biology and biotechnology, vol II.: Plant Genomics and Biotechnology. Springer, India, pp 639–661
Peng T, Jia MM, Liu JH (2015) RNAi-based functional elucidation of PtrPRP, a gene encoding a hybrid proline rich protein, in cold tolerance of Poncirus trifoliata. Front Plant Sci 29(6):808. https://doi.org/10.3389/fpls.2015.00808
Rajam MV, Madhulatha P, Pandey R, Hazarika PJ, Razdan MK (2007) Applications of genetic engineering in tomato. In: Razdan MK, Mattoo AK (eds) Genetic improvement of Solanaceae Crops: tomato, vol 2. Science Publishers, Enfield, pp 285–311
Rathore KS, Sundaram S, Sunilkumar G, Campbell LM, Puckhaber L, Marcel S, Palle SR, Stipanovic RD, Wedegaertner TC (2012) Ultra-low gossypol cottonseed: generational stability of the seed-specific, RNAi-mediated phenotype and resumption of terpenoid profile following seed germination. Plant Biotechnol J 10:174–183. https://doi.org/10.1111/j.1467-7652.2011.00652.x
Regina A, Bird A, Topping D, Bowden S, Freeman J, Barsby T, Kosar-Hashemi B, Li Z, Rahman S, Morell M (2006) High amylose wheat generated by RNA-Interference improves indices of large bowel health in rats. Proc Natl Acad Sci U S A 103:3546–3551
Romano N, Macino G (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol 6:3343–3353
Rubinelli PM, Chuck G, Li X, Meilan R (2013) Constitutive expression of the corn-grass1 microRNA in poplar affects plant architecture and stem lignin content and composition. Biomass Bioenergy 54:312–321
Saurabh S, Vidyarthi AS, Prasad D (2014) RNA interference: concept to reality in crop improvement. Planta 239:543–564. https://doi.org/10.1007/s00425-013-2019-5
Schijlen EGWM, de Vos RCH, Martens S, Jonker HH, Rosin FM, Molthoff JW, Tikunov YM, Angenent GC, van Tunen AJ, Bovy AG (2007) RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol 144:1520–1530
Shriram V, Kumar V, Devarumath RM, Khare TS, Wani SH (2016) MicroRNAs as potential targets for abiotic stress tolerance in plants. Front Plant Sci 7:817. https://doi.org/10.3389/fpls.2016.00817
Sindhu AS, Maier TR, Mitchum MG, Hussey RS, Davis EL, Baum TJ (2009) Effective and specific in planta RNAi in cyst nematodes: expression interference of four parasitism genes reduces parasitic success. J Exp Bot 60:315–324
Singh A, Taneja J, Dasgupta I, Mukherjee SK (2014) Development of plants resistant to tomato Gemini viruses using artificial trans-acting small interfering RNA. Mol Plant Pathol 16:725–734
Sinha R, Rajam MV (2013) RNAi silencing of three homologues of S-adenosylmethionine decarboxylase gene in tapetal tissue of tomato results in male sterility. Plant Mol Biol 82:169–180
Sun X, Xu L, Wang Y, Yu R, Zhu X, Luo X, Gong Y, Wang R, Limera C, Zhang K, Liu L (2015) Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genomics 16:197. https://doi.org/10.1186/s12864-015-1416-5
Sunilkumar G, Campbell LM, Puckhaber L, Stipanovic RD, Rathore KS (2006) Engineering cottonseed for use in human nutrition by tissue-specifi c reduction of toxic gossypol. Proc Natl Acad Sci U S A 103:18054–18059
Tamilarasan S, Rajam MV (2013) Engineering crop plants for nematode resistance through host-derived RNA interference. Cell Dev Biol 2:114. https://doi.org/10.4172/2168-9296.1000114
Tehseen M, Imran M, Hussain M, Irum S, Ali S, Mansoor S, Zafar Y (2010) Development of male sterility by silencing Bcp1 gene of Arabidopsis through RNA interference. Afr J Biotechnol 9:2736–2741
Thakur N, Upadhyay SK, Verma PC, Chandrashekar K, Tuli R, Singh PK (2014) Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase A gene. PLoS One 9:e87235
Tinoco ML, Dias BB, Dall’Astta RC, Pamphile JA, Aragao FJ (2010) In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol 31:27
Tuschl T (2001) RNA interference and small interfering RNAs. Chembiochem 2:239–245
Varoquaux F, Blanvillain R, Delseny M, Gallois P (2000) Less is better: new approaches for seedless fruit production. Trends Biotechnol 18:233–242
Vazquez F (2006) Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci 11:460–468. https://doi.org/10.1016/j.tplants.2006.07.006
Wang Y, Li J (2006) Genes controlling plant architecture. Curr Opin Biotechnol 17:123–129
Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech J-C, Bouzayen M (2005) The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17:2676–2692
Wang S, Wu K, Yuan Q, Liu X, Liu Z, Lin X, Zeng R, Zhu H, Dong G, Qian Q, Zhang G, Fu X (2012) Control of grain size, shape and quality by OsSPL16 in rice. Nat Genet 44:950–954. https://doi.org/10.1038/ng.2327
Wei MM, Wei HL, Wu M, Song MZ, Zhang JF, Yu JW, Fan S, Yu S (2013) Comparative expression profiling of miRNA during anther development in genetic male sterile and wild type cotton. BMC Plant Biol 13:66. https://doi.org/10.1186/1471-2229-13-66
Weise SE, Aung K, Jarou ZJ, Mehrshahi P, Li Z, Hardy AC, Carr DJ, Sharkey TD (2012) Engineering starch accumulation by manipulation of phosphate metabolism of starch. Plant Biotechnol J 10:545–554. https://doi.org/10.1111/j.1467-7652.2012.00684.x
Wilson RC, Doudna JA (2013) Molecular mechanisms of RNA interference. Annu Rev Biophys 42:217–239
Xie Z, Kasschau KD, Carrington JC (2003) Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 13:784–789. https://doi.org/10.1016/S0960-9822(03)00281-1
Xiong A, Yao Q, Peng R, Li X, Han P, Fan H (2005) Different effects on ACC oxidase gene silencing triggered by RNA interference in transgenic tomato. Plant Cell Rep 23:639–646
Xue B, Hamamouch N, Li C, Huang G, Hussey RS (2013) The 8D05 parasitism gene of Meloidogyne incognita is required for successful infection of host roots. Phytopathology 103:175–181
Yadav BC, Veluthambi K, Subramaniam K (2006) Host-generated double stranded RNA induces RNAi in plant-parasitic nematodes and protects the host from infection. Mol Biochem Parasitol 148:219–222
Yang C, Li D, Mao D, Liu X, Ji C, Li X, Zhao X, Cheng Z, Chen C, Zhu L (2013) Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ 36:2207–2218. https://doi.org/10.1111/pce.12130
Yang X, Zhao Y, Xie D, Sun Y, Zhu X, Esmaeili N, Yang Z, Wang Y, Yin G, Lv S, Nie L, Tang Z, Zhao F, Li W, Mishra N, Sun L, Zhu W, Fang W (2016) Identification and functional analysis of microRNAs involved in the anther development in cotton genic male sterile line Yu98-8A. Int J Mol Sci 17:1677. https://doi.org/10.3390/ijms17101677
Yin C, Jurgenson JE, Hulbert SH (2011) Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol Plant-Microbe Interact 24:554–561
Yogindran S, Rajam MV (2015) RNAi for crop improvement. In: Bahadur B, Rajam MV, Sahijram L, Krishnamurthy KV (eds) Plant biology and biotechnology: Volume II: Plant genomics and biotechnology. Springer, India, pp 623–637
Youssef RM, Kim KH, Haroon SA, Matthews BF (2013) Post-transcriptional gene silencing of the gene encoding aldolase from soybean cyst nematode by transformed soybean roots. Exp Parasitol 134:266–274
Yu B, Lydiate DJ, Young LW, Schafer UA, Hannoufa A (2007) Enhancing the carotenoid content of Brassica napus seeds by down regulating lycopene epsilon cyclase. Transgenic Res 17:573–585
Yu R, Xu X, Liang Y, Tian H, Pan Z, Jin S, Wang N, Zhang W (2014) The insect ecdysone receptor is a good potential target for RNAi-based pest control. Int J Biol Sci 10:1171–1180
Zamore PD, Haley B (2005) Ribo-genome: the big world of small RNAs. Science 309:1519–1524
Zha WJ, Peng XX, Chen RZ, Du B, Zhu LL, He GC (2011) Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the Hemipteran insect Nilaparvata lugens. PLoS One 6:e20504
Zhang B (2015) MicroRNA: a new target for improving plant tolerance to abiotic stress. J Exp Bot 66:1749–1761. https://doi.org/10.1093/jxb/erv013
Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, Liao JY, Wang XJ, Qu LH, Chen F, Xin P, Yan C, Chu J, Li HQ, Chen YQ (2013) Over expression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol 31:848–852
Zhang W, Xie Y, Xu L, Wang Y, Zhu X, Wang R, Zhang Y, Muleke EM, Liu L (2016) Identification of microRNAs and their target genes explores miRNA-mediated regulatory network of cytoplasmic male sterility occurrence during anther development in radish (Raphanus sativus L.). Front Plant Sci 7:1054. https://doi.org/10.3389/fpls.2016.01054
Zhou Y, Yuan Y, Yuan F, Wang M, Zhong H, Gu M, Liang G (2012) RNAi-directed down-regulation of RSV results in increased resistance in rice (Oryza sativa L.). Biotechnol Lett 34:965–972. https://doi.org/10.1007/s10529-012-0848-0
Zhu JK (2008) Reconstituting plant miRNA biogenesis. Proc Natl Acad Sci U S A 105:9851–9852
Zhu JQ, Liu S, Ma Y, Zhang JQ, Qi HS, Wei ZJ, Yao Q, Zhang WQ, Li S (2012) Improvement of pest resistance in transgenic tobacco plants expressing dsRNA of an insect associated gene EcR. PLoS One 7:e3857
Acknowledgments
The financial assistance from the Department of Biotechnology (DBT) and Department of Science and Technology (DST), New Delhi, for RNAi work in the lab is acknowledged. We also thank the University Grants Commission (UGC) for Special Assistance Programme (DRS-III), DST for FIST (Level 2) program, and DU-DST PURSE (Phase II) grant.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Mamta, B., Rajam, M.V. (2018). RNA Interference: A Promising Approach for Crop Improvement. In: Gosal, S., Wani, S. (eds) Biotechnologies of Crop Improvement, Volume 2. Springer, Cham. https://doi.org/10.1007/978-3-319-90650-8_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-90650-8_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-90649-2
Online ISBN: 978-3-319-90650-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)