Abstract
The accumulation of carotenoids in higher plants is regulated by the environment, tissue type and developmental stage. In Brassica napus leaves, β-carotene and lutein were the main carotenoids present while petals primarily accumulated lutein and violaxanthin. Carotenoid accumulation in seeds was developmentally regulated with the highest levels detected at 35–40 days post anthesis. The carotenoid biosynthesis pathway branches after the formation of lycopene. One branch forms carotenoids with two β rings such as β-carotene, zeaxanthin and violaxanthin, while the other introduces both β- and ε-rings in lycopene to form α-carotene and lutein. By reducing the expression of lycopene ε-cyclase (ε-CYC) using RNAi, we investigated altering carotenoid accumulation in seeds of B. napus. Transgenic seeds expressing this construct had increased levels of β-carotene, zeaxanthin, violaxanthin and, unexpectedly, lutein. The higher total carotenoid content resulting from reduction of ε-CYC expression in seeds suggests that this gene is a rate-limiting step in the carotenoid biosynthesis pathway. ε-CYC activity and carotenoid production may also be related to fatty acid biosynthesis in seeds as transgenic seeds showed an overall decrease in total fatty acid content and minor changes in the proportions of various fatty acids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Carotenoids comprise a large group of secondary metabolites that are natural pigments present in most higher plants (Demmig-Adams et al. 1996; Vishnevetsky et al.1999; Cuttriss and Pogson 2004; Howitt and Pogson 2006). They are essential components of photosynthetic membranes and provide photoprotection against light damage by channeling excess energy away from chlorophyll (Bassi et al. 1993; Young 1993; Kuhlbrandt et al. 1994; Bartley and Scolnik 1995; Demmig-Adams and Adams 2002). In addition, carotenoids act as membrane stabilizers (Demmig-Adams et al. 1996) and are also precursors in abscisic acid (ABA) biosynthesis (Rock and Zeevaart 1991; Lindgren et al. 2003; Howitt and Pogson 2006). Carotenoids are synthesized and accumulated in the plastids of higher plants (Cunningham and Gantt 1998). Chloroplasts store carotenoids in thylakoid membranes associated with light harvesting (Peter and Thornber 1991; Cunningham and Gantt 1998), while chromoplasts may store high levels of carotenoids in membranes, oil bodies, or other crystalline structures within the stroma (Kirk and Tiliney-Bassett 1978; Cunningham and Gantt 1998; Howitt and Pogson 2006).
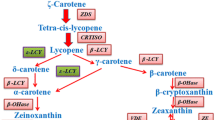
Carotenoids are derived from the isoprenoid pathway with the first committed step being the condensation of two geranylgeranyl diphosphates to form phytoene (Taylor and Ramsay 2005). Phytoene then undergoes four sequential desaturation reactions to form lycopene (Cunningham and Gantt 1998). In higher plants the cyclization of lycopene with lycopene β- and ε-cyclases is the branch-point in carotenoid biosynthesis (Cunningham and Gantt 1998; Fig. 1). On one branch a single enzyme, lycopene β-cyclase (β-CYC), introduces a β-ring at both ends of lycopene to form β-carotene in a two-step reaction. The first dedicated reaction in the other branch of the pathway, leading to lutein, requires both β-CYC and lycopene ε-cyclase (ε-CYC) to introduce one β- and one ε-ring into lycopene to form α-carotene (Pogson et al. 1996; Cunningham and Gantt 1998; Howitt and Pogson 2006). In contrast, a novel ε-CYC from the cyanobacterium Prochlorococcus marinus MED4, was shown to catalyze the simultaneous formation of α-, β- and ε-carotene (Stickforth et al. 2003). Carotenoids with two ε-rings are rare in plants and algae (Goodwin 1980); however an ε-CYC of romaine lettuce uniquely adds two ε-rings to lycopene to form lactucaxanthin (Cunningham and Gantt 2001). A set of reactions in plants, the xanthophyll cycle, rapidly optimizes the concentration of zeaxanthin and violaxanthin in the cell through the action of zeaxanthin epoxidase and violaxanthin de-epoxidase via antheraxanthin (Demmig-Adams and Adams 2002). In Arabidopsis thaliana, nine cis-epoxycarotenoid dioxygenase (NCED) enzymes cleave the cis-isomers of violaxanthin and neoxanthin to form xanthoxin, which is the precursor of ABA (Nambara and Marion-Poll 2005).
Carotenoids are widely used in the food and cosmetics industries (Fraser and Bramley 2004; Taylor and Ramsay 2005; Botella-Pavia and Rodriguez-Concepción 2006), and their importance to human health has been well documented (Bartley and Scolnik 1995; Mayne 1996; Demmig-Adams and Adams 2002; Krinsky and Johnson 2005). For example, β-carotene is the precursor of vitamin A (Lakshman and Okoh 1993) and lutein and zeaxanthin provide protection against macular degeneration (Landrum and Bone 2004). Vitamin A (retinol) deficiency in humans results in symptoms ranging from night blindness to total and irreversible blindness (Ye et al. 2000). The dietary consumption of foods rich in provitamin A (β-carotene) avoids deficiency. Lutein and zeaxanthin also help protect the eye by absorbing potentially harmful blue light radiation (Krinsky and Johnson 2005). Carotenoid levels in many crops used in human and animal diets are inadequate and metabolic engineering provides a promising tool to fortify plants with these essential nutrients. Several examples demonstrating metabolic engineering approaches to increase carotenoid concentrations in plants have been reported (reviewed in Botella-Pavía and Rodríguez-Concepción 2006). Overexpression of a bacterial phytoene synthase (PSY) in a seed-specific manner in Brassica napus resulted in a 50-fold increase in carotenoid concentrations (Shewmaker et al. 1999). Tuber-specific expression of a bacterial PSY in potato enhanced levels of both β-carotene and lutein (Ducreux et al. 2005). Overexpression of the endogenous PSY in the seeds of A. thaliana resulted in a 43-fold average increase in the level of β-carotene (Lindgren et al. 2003) while expression of A. thaliana lycopene β-CYC led to a significant increase in β-carotene content in tomato fruits (Rosati et al. 2000). Expression of the daffodil PSY and a bacterial phytoene desaturase (CrtI) in rice resulted in the production of β-carotene, lutein and zeaxanthin in the endosperm (Ye et al. 2000). Tuber-specific silencing of lycopene epsilon cyclase (LCY-e) or β-carotene hydroxylase (CHY1 and CHY2) increased the levels of β-carotene and total carotenoids in potato tuber (Diretto et al. 2006; Diretto et al. 2007).
In this report, we show differential accumulation of carotenoid compounds in different B. napus organs and in seeds at different developmental stages. In an attempt to enhance the level of β-carotene in the seed of B. napus, we downregulated the expression of ε-CYC using RNAi. This strategy was aimed at reducing the level of lutein present in seeds and diverting lycopene to β-carotene biosynthesis only. Using our RNAi system to repress ε-CYC led to increased levels of β-carotene, lutein, zeaxanthin and violaxanthin in B. napus seeds.
Materials and Methods
Vector construction for RNAi and plant transformation
Two B. napus expressed sequence tags (EST), CL1624 and CL1622, homologous to the 5′- and 3′-ends, respectively, of the A. thaliana ε-CYC (NM_125085) were identified from the B. napus EST collection held at the Saskatoon Research Centre (http://www.brassica.ca). These two ESTs were used to generate RNAi constructs specific to the 5′ and 3′ ends of ε-CYC. The ε-CYC-specific 5′ and 3′ fragments were amplified by the polymerase chain reaction (PCR). Primers P1 and P2 having built-in SpeI and AscI or BamHI and SwaI sites, respectively (Table 1) were used to generate the 5′-end fragment of 352 bp, and a 410 bp from the 3′-end was amplified using primers P3 and P4. Single palindromic repeats of the 5′ and 3′-end PCR products were inserted around a 300 bp spacer of β-glucuronidase in pGSA1285 vector (Fig. 2) (CAMBIA, Canberra, ACT, Australia). The resulting RNAi vectors were designated 710-422 for the 5′-end fragment and 710-423 for the 3′-end fragment.
Growth conditions and plant transformation
Cotyledon explants of B. napus doubled haploid line DH12075 were used for Agrobacterium tumefaciens (GV3101pMP90)-mediated transformation (Moloney et al. 1989). B. napus plants were grown in a controlled environment greenhouse (16 h light/8 h dark, 20°C/17°C). Only those plants shown to be transgenic as determined by PCR to assess the presence of the transgene were subjected to further analysis. The primers used were P2 and P5 for construct 710-422, and P4 and P5 for construct 710-423 (Table 1).
DNA isolation and Southern blot analysis
Total genomic DNA was isolated from leaves of B. napus using the DNeasy Plant Mini Kit (Qiagen, Mississauga, Canada). Approximately 10 μg of genomic DNA was digested with BamHI, EcoRI, EcoRV, SalI, SpeI and SstI, separated on a 0.8% agarose gel and transferred onto Hybond-XL nylon membrane (Amersham Biosciences, Quebec, Canada). The membrane was probed with a 352 bp B. napus ε-CYC-specific fragment amplified by PCR using primers P6 and P7 from the leaf cDNA. Hybridization was performed using Church buffer (Church and Gilbert 1984) at 61°C for 22 h. The filter was washed twice in 2 × SSC, 0.1% SDS for 10 min at 61°C and twice more in 0.2 × SSC, 0.1% SDS for 10 min at 61°C. X-ray film was then exposed to the filter with the aid of intensifying screen.
RNA isolation and semi-quantitative RT-PCR
Total RNA was isolated from leaves, flower petals, roots and seeds at different developmental stages as described by Carpenter and Simon (1998), with some modifications. About 100 mg of ground tissue was extracted with 600 μl of RNA extraction buffer (0.2 M Tris–HCl, pH 9.0, 0.4 M LiCl, 25 mM EDTA, 1% SDS) and an equal volume of Tris–HCl buffered phenol (pH 7.9). Extraction was repeated twice with phenol and followed once with chloroform. Approximately 1/4 volume of 10 M LiCl was added to the decanted aqueous layer, mixed well, stored at 4°C overnight and then centrifuged at 14, 000g for 20 min. The pellet was resuspended in 0.3 ml of DEPC-treated dH2O, to which 30 μl of 3 M sodium acetate (pH 5.3) and 0.7 ml of 95% ethanol were added. The mixture was chilled at −70°C for 10 min and then centrifuged at 14, 000g for 20 min. The pellet was washed and resuspended in 20 μl of DEPC-treated dH2O.
Total RNA was used for one-step semi-quantitative Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) analysis of PSY, PDS, β-CYC and ε-CYC gene expression. This method was chosen considering the low abundance of many carotenoid biosynthetic gene steady state mRNAs (Giuliano et al. 1993). Primers spanning introns were designed for each gene except β-CYC, which does not have an intron, to distinguish between products amplified from cDNA and genomic DNA (Table 1). For β-CYC, a negative control reaction was included in which the RT-PCR reaction lacked reverse transcriptase. Detection of actin mRNA was used as internal standard for RNA levels; two different sized actin amplicons were used to avoid co-migration with the target gene during gel electrophoresis: a 1178 bp actin fragment was co-amplified with PSY, while a 700 bp actin fragment was used with PDS, β-CYC and ε-CYC. For analysis of ε-CYC gene expression in developing seeds of transgenic plants, a 1.8 kb ε-CYC fragment and a 1.178 kb actin fragment were co-amplified. For analysis of ε-CYC gene expression in other tissues, 0.418 kb ε-CYC fragment and a 0.700 kb actin fragment were co-amplified. The primers used in the RT-PCR reactions (Table 1) were P8 and P9 for PSY; P10 and P11 for PDS; P12 and P13 for β-CYC; P14 and P15 for ε-CYC; P16 and P17 for ε-CYC (1.8 Kb); P18 and P19 for actin (0.700 kb); and P20 and P21 for actin (1.178 kb). RT-PCR co-amplification of the internal standard actin gene and test gene fragments was performed using the SuperScript One-Step RT-PCR System (Invitrogen, Burlington, Canada). Approximately 180 ng of DNase I (Amplification Grade, Invitrogen, Burlington, ON, Canada)-treated total RNA was used in a 25 μl RT-PCR reaction mixture. Reverse transcription was performed at 45°C for 30 min, followed by PCR amplification using an initial denaturation at 94°C for 4 min, then 26 cycles at 94°C (30 s), 55°C (30 s), 72°C (extension time varied depending on target gene) and a final extension at 72°C for 5 min. The RT-PCR products were separated on a 1.0% agarose gel and transferred to Hybond-XL membrane (Amersham Biosciences, QC, Canada). The blots were probed with [α-32P]dCTP labelled gene-specific fragment. The ethidium bromide-stained gel photograph was used for the internal control gene, actin.
Extraction of carotenoids from B. napus seeds and HPLC analysis
Approximately 200 mg of seed in 3 ml extraction solvent (hexane/acetone/ethanol, 50/25/25) were pulverised by rapidly shaking for 30 min in a scintillation vial containing a steel rod (adapted from Shewmaker et al. 1999). The sample was centrifuged for 10 min at 1,800g and the supernatant collected. The pellet was washed with another 3 ml extraction solvent and the supernatant collected and pooled. The solvent was removed by evaporation at room temperature under a stream of nitrogen gas. Triacyl glycerides were saponified in the residue by heating at 80°C for 1 h in 5 ml methanolic-KOH (10% w/v KOH in methanol:water [80:20 v/v]). Carotenoids and aqueous compounds were partitioned using 2 ml H2O and 3 ml petroleum ether. The ether phase and two 3 ml ether washes were collected, pooled and the solvent evaporated at room temperature under a nitrogen gas stream. The residue was resuspended in 200 μl of acetonitrile/methylene chloride/methanol (50/40/10 [v/v]) with 0.5% (w/v) butylated hydroxytoluene and filtered through a 0.2 μm pore size nylon syringe filter into an HPLC sample vial. The extract was immediately analysed using HPLC. Aliquots of 20 μl were loaded onto a 4.6 μm × 250 mm reverse-phase C30 YMC “Carotenoid Column” (Waters Ltd, Mississauga, ON, Canada) at 35°C. Mobile phases consisted of methanol (A) and tert-methyl butyl ether (B). A linear gradient starting at 95% A and 5% B, proceeding to 35% A and 65% B over 25 min and a flow rate of 1.2 ml min−1 was used for elution. Compounds in the eluate were monitored at 450 nm using a photodiode array. Peaks were identified by their retention time and absorption spectra compared to those of known standards (CaroteNature, Switzerland). Quantification of carotenoids was conducted using curves constructed with authentic standards.
Fatty acid analysis
The gas chromatography method described by Young et al. (2006) was used to determine fatty acid concentration and profile. Briefly, triplicate samples of approximately 30 mg of seeds were homogenised in hexane containing 0.938 mg ml−1 heptadecanoic acid methyl ester (HAME; Sigma-Aldrich, Oakville, ON, Canada) as an internal standard. Lipids were transesterified in 6.7% sodium methoxide for 30 min and the solution neutralised in 10% citric acid. The hexane layer was filtered through a 0.45 μm PTFE syringe filter and a 1:20 dilution made. One microlitre of diluted methyl ester solution was injected in a DBwax column (10 m long, 0.1 mm ID, 0.2 μm film, Agilent Technologies Canada, Mississauga, ON, Canada) in a Hewlet Packard 6890 gas chroamatograph. Inlet temperature was set at 240°C, with hydrogen carrier gas and a 1/20 split, using nitrogen makeup gas. Column temperatures started at 150°C, ramped to 220°C at 50°C min−1 and were maintained for 7 min. Column pressure started at 50 psi at insertion and dropped to approximately 35 psi after 2 min. Fatty acid methyl esters were detected using a flame ionisation detector.
Microarray analysis
Microarray analysis of gene expression was conducted on developing seeds of wild type DH12075 and ε-CYC-RNAi line BY351. Ambion AminoAllyl MessageAmp II aRNA amplification kit was used for RNA amplification and labelling according to the manufacture’s instructions (Austin, TX, USA). CyDye Post-labelling reactive dye pack was purchased from Amersham (GE healthcare, Baie d’Urfe, QC, Canada). Initial data processing and analysis were performed in BASE database (http://www.base.thep.lu.se). B. napus 15 K oligo arrays were used.
Results
Carotenoid profiles of leaves, petals and developing seeds of B. napus
The carotenoid profiles of B. napus leaves, petals and developing seeds were determined using HPLC analysis. In leaves, lutein, β-carotene, violaxanthin and β-cryptoaxanthin account for 43.30%, 44.16%, 11.46% and 0.84% of total carotenoids, respectively (Table 2). The levels (relative to total carotenoids) of violaxanthin (30.34%) and β-cryptoxanthin (8.85%) in the petals were higher than in the leaves, but the level of β-carotene (13.79%) was lower. We also observed that the profiles of carotenoids accumulating in the seed varied depending on the developmental stage (Table 2). The highest level of violaxanthin was detected in seeds 15–20 days post anthesis (DPA) and it gradually decreased as the seed matured. Seeds 35–40 DPA had the highest levels of lutein and β-carotene, with much lower concentrations observed in fresh mature and dry mature seeds. Trace amounts of zeaxanthin were detected in seeds 15–20 DPA to 35–40 DPA, but it was undetectable in fresh mature and dry mature seeds. β-cryptoxanthin, which is rapidly converted to zeaxanthin, was detectable only in seeds 15–20 DPA.
Expression profiles of genes in the carotenoid biosynthesis pathway
Semi-quantitative RT-PCR analysis was used to determine the transcript levels of some carotenoid biosynthesis genes in the different organs of B. napus (Fig. 3). Primers spanning intron regions were designed for each gene, except for the intron-free β-CYC, to allow PCR products amplified from residual genomic DNA and target cDNA to be distinguished (Fig. 3a).
Expression profiles of carotenoid biosynthesis genes in different organs, and in developing seeds of B. napus. (a) RT-PCR fragment amplified from templates of cDNA (1) and genomic DNA (2). (b) gene expression in different organs of B. napus relative to a co-amplified actin internal control. (c) gene expression in developing B. napus seeds relative to a co-amplified actin internal control. PSY, phytoene synthase; PDS, phytoene desaturase; β-CYC, lycopene, β-cyclase; ε-CYC, lycopene ε-cyclase; DPA, days post-anthesis
RT-PCR analysis revealed that PSY, PDS, and ε-CYC genes were highly expressed in leaves, petals and stems relative to weaker expression in roots (Fig 3b). The expression of β-CYC was highest in petals, and weaker in all other organs studied. Expression of these genes was also analyzed in developing seeds ranging from 20 to 45 DPA. During seed development, the expression of PSY, PDS, β-CYC, and ε-CYC was generally highest in early stages, i.e. up to 30 DPA, but decreased thereafter (Fig. 3c). Although expression of PDS and β-CYC declined during the later stages of seed development, the drop in relative expression levels appeared to be less compared to PSY and ε-CYC.
Silencing of ε-CYC increased levels of β-carotene and lutein
Two RNAi constructs, 710-422 and 710-423, were made to the 5′ and 3′ ends of B. napus ε-CYC and used to transform B. napus DH12075 line. RNA from the 25 DPA developing seeds of two transgenic lines, BY351 (expressing p710-422) and BY371 (expressing p710-423), were subjected to RT-PCR analysis to determine the expression levels of ε-CYC and other carotenoid biosynthesis genes, namely PSY, PDS and β-CYC (Fig. 4). Only the expression of ε-CYC and PSY was reduced in transgenic lines relative to the untransformed wild type control DH12075. Expression of PDS and β-CYC in seeds appeared to be unaltered by the expression of the RNAi construct at 25 DPA.
Visual observation of the color of carotenoid extracts from the mature seeds of ε-CYC silenced lines and the untransformed control DH12075 suggested that significant changes in carotenoid content had occurred (Data not shown). This was confirmed by using HPLC analysis to determine carotenoid profiles in mature seeds of transgenic and untransformed DH12075 plants. Seeds of the 10 transgenic lines tested had 2.6 to 41.7-fold higher concentrations of total carotenoids than those observed in DH12075. In DH12075 seeds, β-carotene and lutein, both of which are derived from lycopene, were the two main carotenoid compounds present (Table 3). However, β-carotene concentrations were at least 5.8-fold higher in the ε-CYC silenced lines than DH12075, with the greatest amount observed in line BY269 (185-fold). Unexpectedly, lutein concentrations were 1.9–22-fold greater in the transgenic lines than in DH12075. The ratio of β-carotene to lutein approximately doubled in the seeds of most transgenic lines, although 4.8, 4.9 and 8-fold in the relative amounts of β-carotene to lutein were observed in lines BY223, BY365 and BY269, respectively. Violaxanthin, zeaxanthin and β-cryptoxanthin were undetectable in DH12075 seeds, but were detected in the 10 transgenic lines with the exception of β-cryptoxanthin in lines BY351, BY58 and BY371. Interestingly, statistically significant differences in carotenoid profiles were not observed in the leaves of transgenic plants compared to untransformed DH12075 even though the CaMV 35S constitutive expression promoter was used (data not shown).
Silencing of ε-CYC has minimal impact on fatty acid profiles
Of the ten transgenic lines tested, eight had lower concentrations of fatty acids than DH12075 (Table 4). For these eight lines the range of fatty acid content ranged from 22.3% to 31.1% of fresh weight (FW). The reductions in fatty acid content were not correlated with increased carotenoid levels, nor were there any patterns associated with the two constructs or generation of seeds used in the analysis.
The amount of palmitic acid in the transgenic seeds increased compared with DH12075, except for BY223 (Table 4). The concentrations of oleic acid and eicosanoic acid decreased compared with DH12075, except for oleic acid in BY371. Overall, the magnitude of the changes to the relative concentrations of fatty acids was small, except for the minor unidentified C18 compound.
Discussion
Our results showed that developing green seeds had higher β-carotene concentrations than mature seeds in wild type DH12075. This last observation was of particular interest since the purpose of this work was to investigate means to produce mature seeds with higher concentrations of β-carotene. Significant increases in carotenoid concentrations could be realised by preventing carotenoid reduction during seed maturation. However, preventing carotenoid breakdown or conversion to other compounds may require that the normal developmental changes that occur to plastids during seed maturation and desiccation be altered. These changes may result in abnormal plastid recovery and behaviour during dormancy and germination, with concomitant changes in seedling vigour (Lindgren et al. 2003). Therefore, rather than blocking catabolism of carotenoids, we tried to divert carotenoid production during development to produce seeds with higher accumulated levels of β-carotene.
Several attempts have been successfully made to engineer higher β-carotene in crop plants. One approach is to upregulate the supply of their precursor-phytoene (Shewmaker et al. 1999; Ducreux et al. 2005). Another strategy to increase β-carotene accumulation was based on upregulating lycopene β-cyclase expression (Rosati et al. 2000; Ravanello et al. 2003). Both β-CYC and ε-CYC were thought to be required for the synthesis of α-carotene and subsequently lutein in higher plants (Pogson et al. 1996). Therefore, we hypothesized that downregulating ε-CYC would divert the substrate, lycopene, from α-carotene (and lutein) production to the other branch of the pathway leading to β-carotene. In addition, as the transcript levels of β-CYC were minimally affected by seed maturation, production of β-carotene should not be inhibited by lack of this enzyme. We used RNAi constructs to repress ε-CYC expression in B. napus.
As expected, reductions in ε-CYC transcript levels in the seeds of the transgenic plants led to increased concentrations of β-carotene compared with the untransformed wild type line, as did the concentrations of other carotenoids derived from β-carotene (Table 3). Unexpectedly, lutein concentrations were also increased dramatically in the transgenic seeds.
Our results suggest that ε-CYC plays a key role in the carotenoid biosynthesis pathway. Of the carotenoid biosynthesis genes examined, only transcription οf ε-CYC and PSY appeared to be affected by expression of the RNAi construct (Fig. 4), yet these plants had major alterations in their carotenoid profiles. This suggests that the accumulation of carotenoids in transgenic seeds was not due to enhanced de novo biosynthesis. ε-CYC was reported to play a key role in controlling the ratio of lutein to β,β-carotenoids (Pogson et al. 1996). Control over the branch point in the pathway could be via substrate competition between ε-CYC and β-CYC. That is ε-CYC could be one of the rate-limiting steps in the pathway and its removal allowed the uninhibited function of β-CYC. The higher concentration of β-carotene and derivatives in the seeds of transgenic plants supports this theory.
The increase in lutein concentrations (Table 3) was unexpected as both β-CYC and ε-CYC are required for the synthesis of α-carotene and subsequently lutein in higher plants (Pogson et al. 1996). This finding appears to be inconsistent with recent reports in the literature, which showed that tuber- specific silencing of ε-CYC in potato resulted in significant increase in β-carotene (up to 14-fold), with only two transgenic lines showing minor increases in lutein levels (1.5–1.8 fold) (Diretto et al. 2006). This suggests regulation of carotenoid biosynthesis varies at the ε-CYC step between potato tubers and B. napus seeds. Three possibilities may explain the production of lutein in the ε-CYC silenced lines of B. napus: (1) The existence of additional ε-CYC gene copies that are not silenced by ε-CYC RNAi, which may have compensated for ε-ring formation. Southern blot analysis using enzymes with either single or no recognition sites within the cDNA sequence showed that the B. napus genome contains at least two homologues of ε-CYC (Fig. 5). These homologues need to be identified and the activity of any gene products determined in RNAi silenced and parental tissues; (2) Formation of the ε-ring by enzymes with a broad substrate range; enzymes with the ability to form ε-rings at the ends of aliphatic compounds may have led to the formation of ε-rings in lycopene; and (3) ε-CYC is not rate-limiting for lutein biosynthesis in seeds, but rather ε-CYC silencing may cause alteration in cellular compartments or sequestration molecules enhancing seed capacity to store lutein.
The increase in total carotenoid synthesis and altered ratio of lutein to β-carotene observed in the seeds of transgenic plants were not seen in the leaves. Differences in capacity to make and store excess carotenoids may explain the differences between the tissues. Our data showed that different tissues have different carotenoid profiles (Table 2), suggesting that regulation of biosynthesis vary from tissue to tissue. Compartmentalisation of the carotenoids may have prevented their catabolism or conversion and led to accumulation in the seeds. In seeds, carotenoids are compartmentalised to elaioplasts (lipid storing plastids), which can store a large amount of carotenoids in specialized lipoprotein-sequestering structures (Kirk and Tiliney-Bassett 1978; Bartley and Scolnik 1995; Vishnevetsky et al. 1999; Howitt and Pogson 2006). However in leaves, carotenoids are synthesized and localized with chlorophyll as a chlorophyll-carotenoid protein complex in chloroplasts (Green and Durnford 1996; Giuliano et al. 2000; Römer et al. 2002). The amounts of β-carotene and lutein are directly proportional to the contents of chlorophyll a and b, respectively. Therefore, carotenoid synthesis may be regulated more stringently to prevent disruption of photosynthesis (Peter and Thornber 1991; Bassi et al. 1993; Pogson et al. 1996). Another possibility is that leaf chloroplasts of transgenic plants may already have reached their limit of carotenoids and no additional storage compartments may be available for excess carotenoid molecules. Alterations in plastid morphology or function have been observed in tissues producing excess carotenoids. A new inclusion body in plastids was observed and assumed to store excess carotenoids when a bacterial PSY was overexpressed in B. napus seeds causing a 50-fold increase in carotenoid accumulation (Shewmaker et al. 1999). In the curds of the cauliflower Or mutant excess β-carotene is stored in chromoplasts that are converted from more non-coloured plastids (Li et al. 2006). When the Or gene was introduced into potato, additional orange bodies were observed in the transgenic tubers (Li and Van Eck 2007). These suggested that enhancing the storage capacity for carotenoid accumulation is an important strategy to alter carotenoid content in plants. Further investigation is needed to determine whether alterations to plastid morphology occurred in the seeds of ε-CYC silenced lines.
The increase in transcript abundance (5-fold) of the late embryogenesis abundant protein M10 (LEA protein M10) in transgenic seeds (Table 5) suggests an increase in ABA production, as LEA proteins are often ABA inducible (Galau et al. 1986; Soeda et al. 2005). Greater concentrations of ABA would have the effect of fore-shortening seed maturation (Fowler and Downey 1970; Bewley 1997). This earlier maturation may be the reason why the transgenic seeds had slightly lower fatty acid contents. Fatty acid accumulation is one of the last metabolic processes to be deactivated by seed desiccation and shorter maturation times lead to lower oil concentrations (Johnson-Flanagan et al. 1992; Si and Walton 2004). Increasing ABA levels would induce earlier maturation and result in the earlier termination of storage molecule accumulation. Reduction in oleic acid concentration in transgenic seeds may be related to a reduction in the expression of stearoyl-ACP desaturase (Table 5), which inserts a cis double bond at the 9 position of C18:0-ACP (Ohlrogge and Jaworski 1997). Another possible reason for a decreased fatty acid could be the diversion of more of the carbon pool to carotenoid biosynthesis, which depleted available carbon precursor for fatty acid biosynthesis. Determination of ABA content and examination of the lipid:protein ratio in the seeds from the RNAi lines would help determine if reducing ε-CYC expression, or increasing carotenoid content, affects the biosynthesis and accumulation of ABA and fatty acids in the seeds.
References
Bartley GE, Scolnik PA (1995) Plant carotenoid: pigments for photoprotection, visual attraction, and human health. Plant Cell 7:1027–1038
Bassi R, Pineau B, Dainese P, Marquardt J (1993) Carotenoid-binding proteins of photosystem II. Eur J Biochem 212:297–303
Bewley JD (1997) Seed germination and dormancy. Plant Cell 9:1055–1066
Botella-Pavía P, Rodríguez-Concepción M (2006) Carotenoid biotechnology in plants for nutritionally improved foods. Physiol Plant 126:369–381
Carpenter CD, Simon AE (1998) Preparation of RNA. In: Martinez-Zapater JM, Salinas J (eds) Methods in molecular biology, vol. 82. Arabidopsis protocols. Humana Press, Totowa, NJ, pp 85–89
Church G, Gilbert W (1984) Genome sequencing. Proc Natl Acad Sci USA 81:1991–1995
Cunningham FX Jr, Gantt E (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49:557–583
Cunningham FX Jr, Gantt E (2001) One ring or two? Determination of ring number in carotenoids by lycopene ε-cyclases. Proc Natl Acad Sci USA 98:2905–2910
Cuttriss AJ, Pogson BJ (2004) Carotenoids. In: Davies KM (ed) Plant pigments and their manipulation. CRC Press, Boca Raton, FL, pp 57–91
Demmig-Adams B, Gilmore AM, Adams WW III (1996) In vivo functions of carotenoids in higher plants. FASEB J 10:403–412
Demmig-Adams B, Adams WW III (2002) Antioxidants in photosynthesis and human nutrition. Science 298:2149–2153
Diretto G, Tavazza R, Welsch R, Pizzichini D, Mourgues F, Papacchioli V, Beyer P, Giuliano G (2006) Metabolic engineering of potato tuber carotenoids through tuber-specific silencing of lycopene epsilon cyclase. BMC Plant Biol 6:13
Diretto G, Welsch R, Tavazza R, Mourgues F, Pizzichini D, Beyer P, Giuliano G (2007) Silencing of beta-carotene hydroxylase increases total carotenoid and beta-carotene levels in potato tubers. BMC Plant Biol 7:11
Ducreux LJM, Morris WL, Hedley PE, Shepherd T, Davies HV, Millam S, Taylor MA (2005) Metabolic engineering of high carotenoid potato tubers containing enhanced levels of β-carotene and lutein. J Exp Bot 56:81–89
Fowler DB, Downey RK (1970) Lipid and morphological changes in developing rapeseed, Brassica napus. Can J Plant Sci 50:233–247
Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43:228–265
Galau GA, Hughes DW, Dure L (1986) Abscisic-acid induction of cloned cotton late embryogenesis-abundant (lea) messenger-RNAs. Plant Mol Biol 7:155–170
Giuliano G, Bartley GE, Scolnik PA (1993) Regulation of carotenoid biosynthesis during tomato development. Plant Cell 5:379–387
Giuliano G, Aquilani R, Dharmapuri S (2000) Metabolic engineering of plant carotenoids. Trends Plant Sci 5:406–409
Goodwin TW (1980) The biochemistry of the carotenoids, 2nd edn., vol 1. Chapman & Hall, London, pp 377
Green BR, Durnford DG (1996) The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 47:685–714
Howitt CA, Pogson BJ (2006) Carotenoid accumulation and function in seeds and non-green tissues. Plant Cell Environ 29:435–445
Johnson-Flanagan AM, Huiwen Z, Geng X-M, Brown DCW, Nykiforuk CL, Singh S (1992) Frost, abscisic acid, and desiccation hasten embryo development in Brassica napus. Plant Physiol 99:700–706
Kirk JT, Tiliney-Bassett RA (1978) Proplastids, etioplasts, amyloplasts, chromoplasts and other plastids. In: Kirck ST, Tiliney-Bassett RA (eds) The plastids:their chemistry, structure, growth and inheritance. Elsevier/North Holland, Biomedical Press, Amsterdam, pp 217–239
Krinsky NI, Johnson EJ (2005) Carotenoid actions and their relation to health and disease. Mol Aspects Med 26:459–516
Kuhlbrandt W, Wang DN, Fujiyoshi Y (1994) Atomic model of plant light-harvesting complex by electron crystallography. Nature 367:614–621
Landrum JT, Bone RA (2004) Dietary lutein and zeaxanthin: reducing the risk of macular degeneration. Agro Food Industry Hi-Tech 15:22–25
Lakshman MR, Okoh C (1993) Enzymatic conversion of all trans-beta-carotene to retinal. Meth Enzymol 214:256–269
Li L, Lu S, Cosman KM, Earle ED, Garvin DF, O’Neill J (2006) β-Carotene accumulation induced by cauliflower Or gene is not due to an increase capacity of biosynthesis. Phytochemistry 67:1177–1184
Li L, Van Eck J (2007) Metabolic engineering of carotenoid accumulation by creating a metabolic sink. Transgenic Res DOI 10.1007/s11248-007-9111-1
Lindgren L, Stahlberg KG, Hoglund AS (2003) Seed-specific overexpression of an endogenous Arabidopsis phytoene synthase gene results in delayed germination and increased levels of carotenoids, chlorophyll, and abscisic acid. Plant Physiol 132:779–785
Mayne ST (1996) Beta-carotene, carotenoids and disease prevention in humans. FASEB J 10:690–701
Moloney MM, Walker JM, Sharma KK (1989) High efficiency transformation of Brassica napus using Agrobacterium vectors. Plant Cell Rep 8:238–242
Nambara E, Marion-Poll A (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56:165–185
Ohlrogge JB, Jaworski JG (1997) Regulation of fatty acid synthesis. Annu Rev Plant Physiol Plant Mol Biol 48:109–136
Peter GF, Thornber JP (1991) Biochemical composition and organization of higher plant photosystem II light-harvesting pigment-protein. J Biol Chem 266:16745–16754
Pogson BJ, McDonald K, Truong M, Britton G, DellaPenna D (1996) Arabidopsis carotenoid mutants demonstrate lutein is not essential for photosynthesis in higher plants. Plant Cell 8:1627–1639
Ravanello MP, Ke D, Alvarez J, Huang B, Shewmaker CK (2003) Coordinate expression of multiple bacterial carotenoid genes in canola leading to altered carotenoid production. Metab Eng 5:255–263
Rock CD, Zeevaart JAD (1991) The aba mutant of Arabidopsis thaliana is impaired in epoxy-carotenoid biosynthesis. Proc Natl Acad Sci USA 88:7496–7499
Römer S, Lübeck J, Kauder F, Steiger S, Adomat C, Sandmann G (2002) Genetic engineering of a zeaxanthin-rich potato by antisense inactivation and co-suppression of carotenoid epoxidation. Metab Eng 4:263–272
Rosati C, Aquilani R, Dharmapuri S, Pallara P, Marusic C, Tavazza R, Bouvier F, Camara B, Giuliano G (2000) Metabolic engineering of beta-carotene and lycopene content in tomato fruit. Plant J 24:413–419
Shewmaker CK, Sheey JA, Daley M, Colburn S, Ke DY (1999) Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J 20:401–412
Si P, Walton GH (2004) Determinants of oil concentration and seed yields in canola and Indian Mustard in the lower rainfall areas of Western Australia. Aust J Agric Res 55:367–377
Soeda Y, Konings MCJM, Vorst O, van Houwelingen AMML, Stoopen GM, Maliepaard CA, Kodde J, Bino RJ, Groot SPC, van der Geest AHM (2005) Gene expression programs during Brassica oleracea seed maturation, osmopriming, and germination are Indicators of progression of the germination process and the stress tolerance level. Plant Physiol 137:354–368
Stickforth P, Steiger S, Hess WR, Sandmann G (2003) A novel type of lycopene ε-cyclase in the marine cyanobacterium Prochlorococcus marinus MED4. Arch Microbiol 179:409–415
Taylor M, Ramsay G (2005) Carotenoid biosynthesis in plant storage organs: recent advances and prospects for improving plant food quality. Physiol Plant 124:143–151
Vishnevetsky M, Ovadis M, Vainstein A (1999) Carotenoid sequestration in plants: the role of carotenoid-associated proteins. Trends Plant Sci 4:232–235
Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid –free) rice endosperm. Science 287:303–305
Young AJ (1993) Factors that affect the carotenoid composition of higher plants and algae. In: Young AJ, Britton G (eds) Carotenoids in photosynthesis. Chapman and Hall, London, pp 161–205
Young LW, Jalink H, Denkert R, Reaney MTJ (2006) Factors affecting the density of Brassica napus seeds. Seed Sci & Technol 34:633–645
Acknowledgements
We are grateful to Mr. Delwin Epp for technical assistance with B. napus tissue culture and Dr. Branimir Gjetvaj for assistance with the microarray analysis. We thank Drs. Kevin Falk, Kevin Rozwadowski and Bhinu V.S. for critical reading of the manuscript, and for helpful suggestions. Funding for this project was provided by the Saskatchewan Agriculture Development Fund.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yu, B., Lydiate, D.J., Young, L.W. et al. Enhancing the carotenoid content of Brassica napus seeds by downregulating lycopene epsilon cyclase. Transgenic Res 17, 573–585 (2008). https://doi.org/10.1007/s11248-007-9131-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-007-9131-x