Abstract
The symbiotic nitrogen-fixing bacteria are found in diverse climatic conditions and are ecologically important. The classification of rhizobia has always been fascinating; with the advent of polyphasic approaches, it is continuously changing by addition of new genera and species and the reclassification and discovery of nontraditional rhizobia. In comparison with crop legumes, the study of symbiotic associations in wild/native legumes has led to the discovery of several genetically diverse rhizobia. In the era of global climate change, increasing desertification, and for food security, the identification and characterization of rhizobia adapted to arid and hot climatic conditions are important. With this aim desert rhizobia associated with several native legumes belonging to different tribes have been broadly studied from less-explored regions of Indian Thar Desert. The diverse legumes in alkaline soils of the Thar Desert are found to be nodulated by traditional rhizobial genera, Ensifer and Bradyrhizobium. On the basis of core gene phylogeny, the Ensifer strains affiliated to mimosoid, cesalpinioid, and papilionoid legumes clustered into novel clades and lineages. Bradyrhizobium strains phylogenetically diversified from the B. yuanmingense type strain are microsymbiont of species of Tephrosia, Alysicarpus, Crotalaria, and Chamaecrista in addition to strains of Ensifer. The tree rhizobia (isolated from Vachellia, Senegalia, Prosopis, Mimosa) have host range restricted to tree species and therefore could be used as an inoculum in forestry practices. The other native rhizobia isolated from wild legumes (Tephrosia and Chamaecrista) are compatible with crop legumes (Vigna, Cyamopsis, Glycine max) and can be useful in preparation of consortia for extension of agricultural practices.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The ability to fix atmospheric nitrogen to ammonia is exclusively confined to prokaryotic organisms (diazotrophs) that contain the nitrogenase enzyme complex and the phenomenon is known as biological nitrogen fixation (BNF). Diazotrophs can be broadly divided into two groups: (i) free-living/nonsymbiotic and (ii) mutualistic/symbiotic. Eukaryotic organisms are incapable to fix nitrogen, and therefore few of them establish a symbiotic relationship with diazotrophic bacteria. In these interactions, eukaryotic organisms supply nutrients and energy to the diazotrophs in exchange for fixed nitrogen. Legume-rhizobia symbiosis is one of such plant-microbe interactions that contribute fixed nitrogen to the biosphere in a cost-effective and eco-friendly manner. The symbiotic diazotrophs invade or colonize the root (and occasionally stem) of the host plant, where they multiply and induce nodule organogenesis. Rhizobia are heterotrophic, aerobic, non-sporulated, gram-negative, and rod-shaped nitrogen-fixing bacteria that have coevolved with host legumes (Sprent 2001). Rhizobia are widely distributed in diverse geographical and ecological niches of the world. Besides diazotrophy, a group of rhizobia have been discovered which show methylotrophy, for example, Methylobacterium strains isolated from species of Crotalaria (Sy et al. 2001), and stem-nodulating photosynthetic bradyrhizobia have been isolated from species of Aeschynomene (Molouba et al. 1999). The rhizobia are among the most extensively studied group of bacteria due to their nitrogen-fixing ability and could replace synthetic nitrogen fertilizers, in nitrogen-poor soils (Zahran 2001). Relatively little is known about the nitrogen-fixing value of most of the wild legume species found in arid conditions (Sprent 2001). Rhizobia associated with wild/native legumes establish effective symbiosis under harsh environmental conditions and are more tolerant to abiotic stresses (salinity, alkalinity, drought, elevated temperatures, etc.) than the crop rhizobia (Zahran 2001). Thus symbiotically efficient rhizobia with increased tolerance to high salt, pH, and temperature could enhance the production of food and forage legumes in semiarid and arid regions of the world. There are many potentially nodulated legumes native to arid and semiarid areas worldwide which need to be explored for their microsymbiont diversity (Sprent and Gehlot 2010; Panwar et al. 2014).
All bacteria isolated from root nodules were initially classified into the genus Rhizobium until the early 1980s. Later two distinct groups of rhizobia were recognized based on growth on culture medium: fast- and slow-growing. The fast-growing rhizobia are acid-producing and associated mainly with temperate legumes, while the slow-growing (bradyrhizobia) are associated with tropical and subtropical legumes. For a long time, the grouping of root nodule bacteria was based on nodulation with certain host plants called as the cross-inoculation group. The Bergey’s manual has played a fundamental role in rhizobial taxonomy. Development in molecular biology techniques and with the advent of numerical taxonomy considering wide range of characteristics led to the definition of new genera and species and also the renaming of some species (Jordan 1982; Graham et al. 1991; Young et al. 2003). Moulin et al. (2001) published in Nature the nodulation of legumes by Burkholderia, a genus that belongs to class beta-proteobacteria. Sawada et al. (2003) reported 44 bacterial species distributed in 12 genera, 10 of which belong to the class “alpha-proteobacteria” (Allorhizobium, Azorhizobium, Blastobacter, Bradyrhizobium, Devosia, Mesorhizobium, Methylobacterium, Ochrobactrum, Rhizobium, and Ensifer) and 2 to the class “beta-proteobacteria” (Burkholderia and Cupriavidus). Peix et al. (2015) reviewed that there are 15 rhizobial genera belonging to proteobacteria under “classical rhizobia” (Azorhizobium, Bradyrhizobium, Mesorhizobium, Rhizobium, and Ensifer) and “new rhizobia” (Aminobacter, Burkholderia, Cupriavidus, Devosia, Herbaspirillum, Methylobacterium, Microvirga, Ochrobactrum, Phyllobacterium, and Shinella) with nodulating and non-nodulating species. Shamseldin et al. (2017) mentioned about 18 genera of rhizobia with more than 250 species belonging to class alpha- and beta-proteobacteria. At the time of writing this book chapter, the following legume-nodulating genera are known: Rhizobium, Neorhizobium, Pararhizobium, Ensifer, Shinella, Mesorhizobium, Phyllobacterium, Aminobacter Bradyrhizobium, Blastobacter, Photorhizobium, Devosia, Azorhizobium, Methylobacterium, Microvirga, and Ochrobactrum in alpha-proteobacteria and Paraburkholderia (Sawana et al. 2014), Cupriavidus, Trinickia, and Herbaspirillum in beta-proteobacteria (List of Prokaryotic Names with Standing in Nomenclature (LPSN); http://www.bacterio.net).
2 Diversity of Classical and Unconventional Rhizobia
Rhizobial systematics is becoming complex and rapidly changing, and recently many new species have been recognized. The term “root nodule bacteria” (RNB) is nowadays used for all groups of bacteria that have been isolated from root nodule; they may be nodulating or non-nodulating. The legume-nodulating bacterial species within class alpha-proteobacteria comprises of six families, namely, Bradyrhizobiaceae, Brucellaceae, Hyphomicrobiaceae, Methylobacteriaceae, Phyllobacteriaceae, and Rhizobiaceae, under the order Rhizobiales and families Burkholderiaceae and Oxalobacteraceae within the order Burkholderiales in class beta-proteobacteria. In addition to classical and new rhizobia, there are few reports from the class gamma-proteobacteria (Benhizia et al. 2004; Shiraishi et al. 2010). Non-rhizobial root nodule endophytes belonging to gamma-proteobacteria (Enterobacter, Klebsiella, Pantoea, Pseudomonas, Stenotrophomonas), Actinobacteria (Arthrobacter, Brevibacterium, Microbacterium, Micromonospora, Mycobacterium, Streptomyces), Firmicutes (Bacillus, Fontibacillus, Paenibacillus), and Sphingobacteria (from Clitoria ternatea) have also been isolated from many indigenous legumes (Hoque et al. 2011; Aserse et al. 2013; de Meyer et al. 2015; Boukhatem et al. 2016). Martínez-Hidalgo and Hirsch (2017) reviewed the existence of many non-rhizobial root nodule endophytes, many of them are nitrogen fixers and few of them were able to induce the formation of nodules. Biogeography of nodulated legumes and associated nitrogen-fixing microsymbionts was reviewed by Sprent et al. (2017) in terms of both longitudinal and latitudinal trends. Our knowledge about rhizobial diversity associated with nodulated legumes is limited due to culture-based approaches used to characterize the strains. The root nodules are house for many conventional and unconventional rhizobia. In this chapter we emphasized mainly the diversity of rhizobial genera belonging to class alpha- and beta-proteobacteria and some recent changes made in classification.
2.1 Alpha-Proteobacteria
2.1.1 Rhizobiaceae
Family Rhizobiaceae comprises of more than 120 sp. that are distributed into the following genera: Agrobacterium, Allorhizobium, Ciceribacter, Ensifer, Neorhizobium, Rhizobium, and Shinella (Mousavi et al. 2014). The largest genus Rhizobium is extremely heterogeneous and has more than 100 species, comprising of both nodulating and few non-nodulating strains. It has gone through major revision based on multilocus sequence phylogenetic analysis of housekeeping genes. The genus Neorhizobium encompasses the former species of Rhizobium (R. alkalisoli, R. huautlense, R. vignae, and R. galegae), and three new species combinations were described, namely, N. galegae, N. huautlense, and N. alkalisoli (Mousavi et al. 2014). The novel genus Pararhizobium recently described by Mousavi et al. (2015) comprised of four new species (P. giardinii, P. capsulatum, P. herbae, and P. sphaerophysae).
The genus Ensifer (formerly Sinorhizobium) (Chen et al. 1988) presently contains 21 species, of which 19 have been validly published based on polyphasic studies. Most of the species have been isolated from root nodules of Glycine max (for details of species, see the review by Shamseldin et al. 2017) and tree species from the African continent (de Lajudie et al. 1994; Nick et al. 1999). Few strains (E. meliloti and E. medicae) nodulate specifically medics, melilots, and spp. of Trigonella (de Lajudie et al. 1994; Rome et al. 1996; El Batanony et al. 2015; Gaur et al. 2018) in different parts of the world. From the New World, species like E. americanus (from Acacia acatlensis) (Toledo et al. 2003) and E. mexicanus (Acacia angustissima) (Lloret et al. 2007) have been reported. Two species, namely, E. abri and E. indiaense, isolated from tropical legumes, Abrus precatorius and Sesbania rostrata, respectively, have been published from India (Ogasawara et al. 2003), but they have not yet been included in the Validation List of the International Journal of Systematic and Evolutionary Microbiology, and no type strains have been submitted so far at international culture collections. Recently new species E. aridi (not validly published) has been reported from hot-arid regions of three continents (Asia, Africa, and America). The reported strains of E. aridi isolated from different legume host were genetically similar but harbored different sym genes (Le Queré et al. 2017). Ensifer sojae has been isolated from root nodules of G. max grown in saline-alkaline soils of China (Li et al. 2011). Similarly, Li et al. (2016) reported genetically diverse rhizobia nodulating Sesbania cannabina in saline-alkaline soils. Cheng et al. (2002) observed that the nodulation response of Medicago sativa and Medicago murex differed with soil acidity.
The genus Allorhizobium described by de Lajudie et al. (1998) contains one species (A. undicola) which effectively nodulates Neptunia natans in Senegal. The merging of the three genera (Agrobacterium, Rhizobium, and Allorhizobium) was done into a single genus, Rhizobium. On the basis of 16S rRNA phylogenetic analyses, four species of Agrobacterium (A. tumefaciens, A. radiobacter, A. rhizogenes, and A. vitis) were transferred to the genus Rhizobium (Young et al. 2003). Bacteria related to Agrobacterium were identified among the root nodules of several tropical leguminous plants from Africa (de Lajudie et al. 1999; Mhamdi et al. 2005). Nodulation and nitrogen-fixing genes were not detected in these strains, and it has been confirmed that these Agrobacterium-like strains enter the nodules by mixed infection with a rhizobia capable of inducing nodule resulting in mixed population within the nodule (Mhamdi et al. 2005).
The genus Shinella has one species (S. kummerowiae), a symbiotic bacterium isolated from root nodules of the herbal legume Kummerowia stipulacea grown in Shandong province of China (Lin et al. 2008). Newly described genus, Ciceribacter (C. lividus), was isolated from rhizosphere soil of Cicer arietinum from Kannivadi, India (Kathiravan et al. 2013).
2.1.2 Phyllobacteriaceae
As per Shamseldin et al. (2017), family Phyllobacteriaceae comprises about 49 nitrogen-fixing species within three genera, namely, Mesorhizobium (40 species), Phyllobacterium (8 species), and Aminobacter (1 species). The genus Mesorhizobium was described by Jarvis et al. (1997) with the aim to reassign some species previously included in the Rhizobium genus. Several Rhizobium species were transferred to this genus (M. loti, M. huakuii, M. ciceri, M. tianshanense, M. mediterraneum). Mesorhizobium is a wide spread rhizobial genus nodulating a wide range of legumes in addition to Cicer arietinum growing mainly in acidic soils (Peix et al. 2015). However, Zhang et al. (2012) suggested that mesorhizobia are the preferred microsymbionts of chickpea growing in alkaline soils of northwest China.
Genus Phyllobacterium was proposed to accommodate bacterial species isolated from leaf nodules of members of Rubiaceae (Pavetta, Psychotria, and Sericanthe) and Myrsinaceae (Ardisia) in tropical continental Africa and Asia (Knösel, 1984). Chromobacterium lividum has also been reported from the leaf-nodulated members of these families. Phyllobacterium trifolii was isolated from root nodules of Trifolium pratense growing in a Spanish soil (Valverde et al. 2005). It harbors symbiotic genes and infectivity tests experiments with this species revealed that it forms nodules on the roots of Trifolium repens and Lupinus albus (Valverde et al. 2005). Recently P. loti was isolated from nodules of Lotus corniculatus (Sánchez et al. 2014).
Maynaud et al. (2012) described Aminobacter anthyllidis, a metal-resistant bacteria nodulating Anthyllis vulneraria a legume host suitable for phytostabilization in mining areas.
2.1.3 Bradyrhizobiaceae
Family Bradyrhizobiaceae contains three nitrogen-fixing genera Bradyrhizobium, Blastobacter, and Photorhizobium. The genus Bradyrhizobium was created to accommodate slow-growing bacteria capable of establishing nitrogen-fixing symbioses with a broad range of plants belonging to three subfamilies of the family Leguminosae and characterized by an alkaline reaction in culture media. The genus Bradyrhizobium presently includes more than 40 species isolated from the nodules of highly divergent legume tribes, including herbaceous and woody species of tropical and temperate origin (Sprent et al. 2017; Shamseldin et al. 2017). Jordan (1982) reported B. japonicum from root nodules of Glycine max, and since then many species (B. daqingense, B. huanghuaihaiense, B. liaoningense, and B. ottawaense) have been isolated which are associated with soybean. Bradyrhizobial species have also been reported as microsymbionts of Arachis hypogaea (B. arachidis, B. guangxiense, B. guangdongense, and B. subterraneum) and Vigna unguiculata (B. kavangense, B. brasilense, B. vignae, and B. manausense). Type strain B. canariense is an acid-tolerant endosymbiont that effectively nodulates shrubs of the tribes Genisteae and Loteae (Vinuesa et al. 2005). Strain B. iriomotense was isolated from a tumor-like root of the legume Entada koshunensis from Japan (Islam et al. 2008). Ramírez-Bahena et al. (2009) described two species (B. pachyrhizi and B. jicamae) that nodulate Pachyrhizus erosus. From Morocco (Africa) B. cytisi was isolated from nodules of Cytisus villosus and B. retamae from Retama monosperma (Chahboune et al. 2011).
The type strains of Bradyrhizobium on the basis of housekeeping genes phylogeny clusters into two mega clades (Ojha et al. 2017). Bradyrhizobium strains belonging to two mega clades do not show a particular geographical pattern as observed for type strains of Ensifer (Sankhla et al. 2017); instead they have been found intermingled in both clades representing different continents from where bradyrhizobia have been reported. Mega clade-I contains several Asian species (B. arachidis, B. daqingense, B. ganzhouense, B. guangxiense, B. guangdongense, B. huanghuaihaiense, B. iriomotense, B. japonicum, B. liaoningense, and B. yuanmingense), African species (B. cytisi, B. kavangense, B. rifense, B. subterraneum, and B. vignae), and few species from Europe (B. betae and B. canariense). From the New World, species such as B. americanum, B. centrosemae, B. centrolobii, B. forestalis, B. ingae, B. manausense, B. neotropicale, B. ottawaense, and B. stylosanthis have been reported. The mega clade-II also contains few Asian (B. erythrophlei, B. ferriligni, and B. lablabi), African (B. namibiense and B. retamae), and European (B. valentinum) species. Mega clade-II is mainly represented by species isolated from the New World such as B. brasilense, B. elkanii, B. embrapense, B. icense, B. jicamae, B. mercantei, B. macuxiense, B. pachyrhizi, B. paxllaeri, B. tropiciagri, and B. viridifuturi.
Symbiotic Bradyrhizobium strains have also been isolated from the nodules of non-legume plants (Trema aspera) growing between rows of tea in the Pangia district of New Guinea (Trinick 1973). The discovery of photosynthetic Bradyrhizobium strains that can induce nitrogen-fixing nodules on stems of the legume Aeschynomene was made (Molouba et al. 1999). The microsymbionts of Aeschynomene indica are unique as they form nodules on plant stems, branches, and roots; and some of them produce the photosynthetic pigments. The genus Photorhizobium contains only a single photosynthetically active species P. thompsonianum efficiently nodulating A. indica (Eaglesham et al. 1990). The aquatic budding bacterium Blastobacter denitrificans also forms nitrogen-fixing symbioses with A. indica (van Berkum and Eardly 2002).
2.1.4 Hyphomicrobiaceae
Family Hyphomicrobiaceae includes two nodulating and nitrogen-fixing genera Devosia and Azorhizobium. The genus Azorhizobium described by Dreyfus et al. (1988) includes nitrogen-fixing root and stem-nodulating microsymbiont species that can also fix nitrogen ex planta under micro-aerobic conditions. Genus Azorhizobium contains three species; two of them effectively fix nitrogen with species of Sesbania. Azorhizobium caulinodans was isolated from the stem nodules of Sesbania rostrata from Africa (Dreyfus et al. 1988), and A. doebereinerae is microsymbiont of Sesbania virgata in Brazil (de Souza Moreira et al. 2006). The unique feature of A. caulinodans is that it fixes nitrogen both in aerobic cultures and in micro-aerobic symbiosis with its legume host S. rostrata. Recently strain A. oxalatiphilum was isolated from macerated petioles of Rumex sp. through enrichment in mineral medium.
The genus Devosia has single nodulating species, D. neptuniae, identified by Rivas et al. (2003) that efficiently nodulates aquatic legume Neptunia natans in India. This strain through horizontal transfer has acquired symbiotic genes from a broad host range strain, Rhizobium tropici (Rivas et al. 2003).
2.1.5 Methylobacteriaceae
Family Methylobacteriaceae comprises of two rhizobial genera Methylobacterium and Microvirga with three and four species, respectively, that can induce nitrogen-fixing nodules on roots of legume plant. The genus Methylobacterium includes pink-pigmented facultative methylotrophic (PPFM) bacteria that are strictly aerobic and able to grow on one-carbon compounds such as formate, formaldehyde, and methanol as sole carbon source. Sy et al. (2001) reported facultative methylotrophic species forming nodules in the Crotalaria species. The type strain M. nodulans was isolated from C. podocarpa from Senegal (Jourand et al. 2004). Methylobacterium strains within the root nodules obtain carbon from photosynthates of host plant as well as from methylotrophy. Madhaiyan et al. (2006) isolated several nodulating and plant-growth promoting Methylobacterium species from tropical legumes.
Ardley et al. (2012) reported three species of Microvirga, namely, M. lupini, isolated from nitrogen-fixing nodules of Lupinus texensis in Texas, and M. lotononidis and M. zambiensis from Listia angolensis collected in Zambia.
2.1.6 Brucellaceae
The family Brucellaceae contains fast-growing, nitrogen-fixing strains in the genus Ochrobactrum, which has two species, namely, O. lupini and O. cytisi, isolated from the nodules of Lupinus honoratus and Cytisus scoparius, respectively (Trujillo et al. 2005; Zurdo-Piñeiro et al. 2007).
2.2 Beta-Proteobacteria
2.2.1 Burkholderiaceae
Family Burkholderiaceae comprises of the two genera Burkholderia sensu lato and Cupriavidus (former Ralstonia). Genus Burkholderia sensu lato is a big and complex group including diverse species with different physiological and ecological properties; pathogenic strains have been isolated from animals and humans; some are phytopathogenic and others are beneficially associated with plant (root nodule symbionts and nonsymbiotic strains) and have also been isolated from a very wide range of environmental habitats (soil and water) [Gyaneshwar et al. 2011]. Taxonomy of Burkholderia sensu lato has undergone major changes several times based on concatenated phylogenetic analysis of housekeeping gene fragments and whole genome sequences. Sawana et al. (2014) based on molecular signatures and phylogenomic analysis proposed for the division of Burkholderia sensu lato containing pathogenic organisms, and a separate new genus Paraburkholderia was proposed for harboring environmental species. Eleven species of the genus Burkholderia were later transferred to genus Paraburkholderia, and a new genus Caballeronia was proposed to accommodate 12 species of the genera Burkholderia and Paraburkholderia (Dobritsa and Samadpour, 2016). Presently this complex group of Burkholderia sensu lato is split into the following genera: Burkholderia sensu stricto (includes the human and animal pathogens), Caballeronia (includes the plant beneficial and environmental strains), and Paraburkholderia (includes the nitrogen-fixing legume microsymbionts) (Sawana et al. 2014; Dobritsa and Samadpour 2016). Based on comparative genomic analysis recently, Estrada-de los Santos et al. (2018) described two novel genera Mycetohabitans and Trinickia which form two distinct and unique clades, including two (M. endofungorum and M. rhizoxinica) and four (T. caryophylli, T. dabaoshanensis, T. soli, and T. symbiotica) new combinations of species.
Initially two legume-nodulating Burkholderia (sensu lato) strains, P. tuberum STM678T and P. phymatum STM815T, were isolated from root nodules of Aspalathus carnosa and Machaerium lunatum, respectively (Moulin et al. 2001). Presently, the following legume-nodulating species of genus Paraburkholderia have been described including P. aspalathi, P. caballeronis, P. caribensis, P. diazotrophica, P. dilworthii, P. kirstenboschensis, P. mimosarum, P. nodosa, P. phenoliruptrix, P. piptadeniae, P. phymatum, P. tuberum, P. rhynchosiae, P. ribeironis, P. sabiae, and P. sprentiae (Estrada-de los Santos et al. 2018). The following species of Paraburkholderia are root nodule symbionts of species of Mimosa (Estrada-de los Santos et al. 2018): P. caribensis (M. pudica and M. diplotricha); P. diazotrophica (Mimosa spp.); P. mimosarum (M. pigra and M. pudica); P. nodosa (M. bimucronata and M. scabrella); P. phenoliruptrix (M. flocculosa); P. phymatum (M. pudica); and P. sabiae (M. caesalpiniifolia). Other beta rhizobia strains (C. taiwanensis and T. symbiotica) belonging to different genera are specifically symbionts of species of Mimosa. Two newly described species P. piptadeniae and P. ribeironis nodulates piptadenia group (Piptadenia gonoacantha, tribe mimoseae) in Brazil (Bournaud et al. 2013, 2017). The following species of Paraburkholderia nodulates papilionoids (Estrada-de los Santos et al. 2018): P. sprentiae (isolated from Lebeckia ambigua root nodules); P. rhynchosiae (from root nodules of Rhynchosia ferulifolia); P. dilworthii (from Lebeckia ambigua root nodules); P. kirstenboschensis (nodulates papilionoid legumes Virgilia oroboides, Hypocalyptus coluteoides, H. oxalidifolius, and H. sophoroides indigenous to South Africa); and P. tuberum (nodulates several Cyclopia species). Paraburkholderia type strains (P. phymatum and P. nodosa) belonging to mimosoid clade also nodulate the promiscuous papilionoid (Phaseolus vulgaris, Vigna unguiculata, and Macroptilium atropurpureum) legumes of tribe Phaseoleae, whereas the T. symbiotica strain has restricted host range (Mimosa spp.) and failed to nodulate these promiscuous legumes. Type strain P. caballeronis originally isolated from tomato has the ability to effectively nodulate P. vulgaris (Martínez-Aguilar et al. 2013). Type strain P. aspalathi was isolated from root nodules of the South African legume Aspalathus abietina (tribe Crotalarieae). The legume-nodulating Paraburkholderia strains of class beta-proteobacteria possess nodulation genes phylogenetically related to those found in legume symbionts of the class alpha-proteobacteria suggesting that the beta rhizobia have evolved through lateral gene transfers (Chen et al. 2003).
Genus Cupriavidus presently contains two rhizobial species, C. taiwanensis nodulating Mimosa sp. which has been repeatedly isolated from root nodules on the pan-tropical weeds M. pudica and M. diplotricha in Taiwan (Chen et al. 2001, 2003) and also from India (Gehlot et al. 2013). Another species is C. necator that was isolated through trap (in soils of Brazil) experiments from root nodules of two promiscuous legume species, P. vulgaris and Leucaena leucocephala (da Silva et al. 2012). This species can effectively nodulate other promiscuous legumes such as M. atropurpureum, V. unguiculata, and M. caesalpiniaefolia (da Silva et al. 2012).
2.2.2 Oxalobacteraceae
The genus Herbaspirillum in family Oxalobacteraceae within the order Burkholderiales was first described to include bacterial strains associated with roots of several cereals. Valverde et al. (2003) reported a nitrogen-fixing bacterium H. lusitanum associated with root nodules of P. vulgaris plants grown in soil from Portugal. Genus Herbaspirillum and another genus Variovorax of the beta-proteobacteria have been isolated from root nodules of species of Acacia (A. salicina and A. stenophylla) from Southeastern Australia (Hoque et al. 2011). It has also been isolated from nodules of Aspalathus linearis, but nodulation has not been confirmed (Hassen et al. 2012).
2.3 Gamma-Proteobacteria
Besides nodulating bacteria from alpha- and beta-proteobacteria, there is a report of gamma-proteobacteria nodulating Hedysarum sp. (Benhizia et al. 2004). Enterobacter cloacae and E. kobei were isolated from root nodules of the three species of Hedysarum (H. carnosum, H. spinosissimum, and H. pallidum) growing in native stands in different habitats in Algeria (Benhizia et al. 2004). Muresu et al. (2008) concluded there are frequent reports of coexistence of symbiotic culturable rhizobia with non-culturable rhizobia and other endophytic bacterial taxa (of the order Enterobacteriales or Pseudomonadales) within root nodules suggesting that diversity of bacterial species nodulating legumes may be broader than expected. Shiraishi et al. (2010) noted the ability of Pseudomonas sp. to nodulate Robinia pseudoacacia. These gamma-proteobacteria strains probably acquired the symbiotic genes from symbiotic-rhizobial species in the soil and rhizosphere through lateral gene transfer.
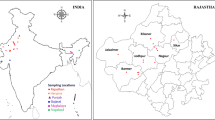
3 Genetic Diversity of Indian Thar Desert Rhizobia
Based on various criterions such as annual precipitation, temperature, soil, geography, and types of plant community, the deserts can be classified to hyper-arid, arid, and semiarid (Sprent and Gehlot, 2010). The Great Indian Desert or the Indian Thar Desert comes under low altitude arid and semiarid region with an area of about 200,000 km2. It is a subtropical desert (https://www.britannica.com/place/Thar-Desert), and most of its part (61%) is present in Western Rajasthan which experiences an arid climatic regime (hot desert climate found under the subtropical ridge). The precipitation is low and varies from hyper-arid areas to semiarid areas, some areas have saline tracts, and soils are alkaline with low fertility. The native legumes of this region are subject to extremely stressful and harsh environmental conditions. In arid regions, heat stress affects both the free-living and symbiotic life of rhizobia, while desertification causes a negative impact on legume-rhizobia symbiosis. Still, a large number of legumes of Thar Desert belonging to different subfamilies were reported to be nodulated (Panwar et al. 2014, Gehlot et al. 2012). As questioned by Sprent and Gehlot (2010) “Nodulated legumes in arid and semiarid environments: are they important?” the answer would be yes, these legumes that have coevolved with their rhizobial partners in the Thar Desert have immense potential for fixing atmospheric nitrogen, reforestation, in controlling soil erosion, and increasing the soil fertility. Sprent and Gehlot (2010) mentioned about some nodulating native, perennial drought-tolerant, and drought-escaping annual legumes from the Thar Desert. The rhizobia of wild legumes have novel genotype and better phenotype/traits than the homologous crop rhizobia. Therefore, isolation of effective and promiscuous rhizobia from wild legumes to inoculate other legume crops is a better strategy to improve the efficiency of the rhizobium-legume symbiosis.
In the era of global climate change and for food security, studies such as exploration, identification, and characterization of indigenous nitrogen-fixing microsymbiont associated with native medicinal and food crop legumes are needed. Such studies are more relevant when working on the microsymbionts specific to legumes growing in a particular soil type and climatic conditions. Earlier research on rhizobia-legume symbiosis was restricted to few agriculturally important food legume crops like soybean, common bean, cowpea, species of Vigna, and associated species. In the last one decade, several studies have been conducted on molecular characterization and analysis of phylogenetic diversity of root nodule bacteria associated with native/wild legumes (such as species of Vachellia, Senegalia, Prosopis, Mimosa, Chamaecrista, Crotalaria, Alysicarpus, Rhynchosia, Tephrosia, Indigofera, Trigonella, and Vigna) of Indian Thar Desert (Gehlot et al. 2012, 2013, 2014, 2016; Tak et al. 2013, 2016a, b; Panwar et al. 2014; Ojha et al. 2015; Sankhla et al. 2015, 2017, 2018; Choudhary et al. 2017, 2018; Rathi et al. 2017, 2018; Gaur et al. 2018). Panwar et al. (2014) reviewed the status of nodulation of more than 30 native legume species belonging to 3 subfamilies and more than 10 legume genera. Gehlot et al. (2012) reported that in the hot-dry and alkaline soils of Indian Thar Desert (Western Rajasthan), native legumes are nodulated by genetically diverse nitrogen-fixing Ensifer, Bradyrhizobium, and Rhizobium strains. The dominant microsymbionts of most legumes were species of Ensifer (Gehlot et al. 2012, 2013; Ojha et al. 2015; Sankhla et al. 2015, 2017, 2018; Tak et al. 2016a, b; Ardley 2017; Choudhary et al. 2017, 2018; Rathi et al. 2017, 2018; Gaur et al. 2018) and few novel Ensifer strains have been characterized at genomic level (Tak et al. 2013; Gehlot et al. 2016; Le Queré et al. 2017). Some native legumes including species of Tephrosia, Chamaecrista, and Alysicarpus in the Thar Desert are effectively nodulated by both Ensifer and Bradyrhizobium strains (Gehlot et al. 2012; Ojha et al. 2015; Tak et al. 2016b; Rathi et al. 2017, 2018).
3.1 Phylogenetic Diversity of Thar Desert Ensifer Strains
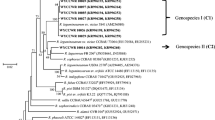
Genetically distinct groups of old-world Ensifer strains have evolved in the Thar Desert and are more adapted to stressed environment and dominating in the alkaline soils. The Thar-Ensifer strains are promiscuous and nodulating papilionoid, mimosoid, and caesalpinioid legumes. In the maximum likelihood phylogenetic tree of the Thar Desert Ensifer strains isolated from root nodules of various wild legumes Vachellia jacquemontii (AJ), Vachellia leucophloea (AL), Senegalia senegal (AS), Crotalaria burhia (CB), Chamaecrista pumila (CP), Mimosa hamata (MH), Mimosa himalayana (MHM), Trigonella foenum-graecum (TFG), Tephrosia falciformis (TF), Tephrosia leptostachya (TL), Tephrosia purpurea (TP), Tephrosia villosa (TV), Tephrosia wallichii (TW), and Vigna trilobata (VT) of Western Rajasthan based on 16S rRNA gene sequences (Fig. 2.1), the strains clustered into novel clades and lineages. Most of the strain isolated from mimosoid trees and shrubs (MH, MHM, AJ, AS, AL), papilionoid shrubs/herbs (CB, TF, TP, TV, TW, VT), and caesalpinioid herb (CP) shared close similarity with E. saheli LMG 7837T (Sesbania cannabina, Senegal) (de Lajudie et al. 1994). Few strains (TV1, CP7, CP40, CP42) shared close similarity with E. terangae LMG 7834T (Senegalia laeta, Senegal) (de Lajudie et al. 1994). Strains CM11 and CP14 shared close affinity with another old-world Ensifer, E. kostiensis HAMBI 1489T (Senegalia senegal, Sudan) (Nick et al. 1999). Strain AS11 clustered with recently reported type strain E. glycinis CCBAU from root nodules of G. max in China. A single strain TL4 clustered with E. adhaerens LMG 20216T and E. sesbaniae CCBAU 65729T (isolated from Sesbania cannabina, China). Microsymbionts of legume, Trigonella foenum-graecum (TFG22 and TFG64) as expected, shared close similarity with type strain E. medicae A321T (Medicago truncatula, France). Few mimosoid strains formed distinct lineages within genus Ensifer (Fig. 2.1).
Maximum likelihood phylogenetic tree of Thar Desert Ensifer strains isolated from root nodules of various wild legumes of Western Rajasthan based on 16S rRNA gene sequences. Bootstrap values more than 50% calculated for 1000 replications are indicated at internodes. The scale bar indicates 1% nucleotide substitution per site. (Abbreviation: superscript T stands for type strain)
The genera in the subfamily Caesalpinioideae found in arid and semiarid regions of India are non-nodulating including species of Cassia, Senna, and Parkinsonia except the genus Chamaecrista. Chamaecrista pumila is a basal legume in which microsymbionts remain trapped in infection thread and are not dropped out, and these infection threads with symbiosomes are called as fixation threads. Novel findings were made related to microsymbionts of C. pumila in the Thar Desert. The genus Chamaecrista is known to be nodulated by slow-growing nitrogen-fixing Bradyrhizobium strains all over the world (Beukes et al. 2016; dos Santos et al. 2017). However, C. pumila growing in arid and semiarid regions of the Thar Desert of India is nodulated by fast-growing Ensifer in addition to slow-growing Bradyrhizobium. This is probably the first report of Ensifer nodulating C. pumila. This suggests that the host legume and naturalized fast-growing Ensifer of Indian Thar Desert have coevolved and both gained the ability to interact and form a symbiotic association (Rathi et al. 2018).
The limitation of 16S rRNA gene phylogeny is that closely related species cannot always be distinguished due to high level of sequence conservation. Therefore to determine the exact taxonomic position of these Ensifer strains, multilocus sequence analysis (MLSA) was performed for few selective strains. The MLSA based on conserved protein-coding housekeeping genes (glnII, atpD, recA, and dnaK) suggests that the Thar Desert Ensifer strains associated with root nodules of members of the papilionoid, mimosoid, and caesalpinioid legumes are novel species of Ensifer as they are significantly divergent from existing type strains of Ensifer (Tak et al. 2016b; Sankhla et al. 2017; Rathi et al. 2018). The Tephrosia-Ensifer (TP6 and TW10) strains were characterized at genomic level and designated as novel species of E. aridi (Le Queré et al. 2017). These Thar Desert Ensifer strains were found to be genetically identical to strains isolated from other parts of the world such as Ensifer strains from root nodules of Acacia raddiana and Acacia gummifera from Merzouga Desert in Morocco (Africa) and from wild species of Phaseolus (Phaseolus filiformis) in the hot desert of Baja California in Mexico. The core genomes of six Ensifer strains from three different continents (Asia, Africa, and America) were compared with Ensifer type strains. Genome-based species delineation tools such as average nucleotide identity (ANI) and in silico-based DNA-DNA hybridization (DDH) demonstrated that they belong to a new species of Ensifer; however these strains were symbiotically distinct (sym genes continent specific). The genomic data suggested several conserved genes specific to genus Ensifer (Le Queré et al. 2017). Previously published work and the finding of predominance of Ensifer in hot-arid alkaline soils of Thar Desert suggest that Ensifer species have been mostly isolated from legumes growing in the arid and alkaline soils of Old World and New World (Tak et al. 2016b; Shamseldin et al. 2017; Sankhla et al. 2017; Rathi et al. 2018). Exception to this is E. medicae that nodulates species of Medicago in acidic soils (Garau et al. 2005). These findings are supported by the comparative genomic studies on Ensifer microsymbionts nodulating soybean growing in alkaline soil of China, suggesting that the strains of Ensifer have specific genes for adaptation to alkalinity, low water potential, salt stress, and high temperature (Tian et al. 2012).
Our studies suggest that the Ensifer strains associated with mimosoid members (Vachellia, Senegalia, and Mimosa) of the Thar Desert on the basis of their symbiotic genes are closer to type strain E. arboris (isolated from Prosopis chilensis, Sudan) [Fig. 2.2]. In species phylogeny these strains shared close similarity with E. saheli; this incongruence with symbiotic gene phylogeny is due to horizontal transfer of the sym genes (Gehlot et al. 2013; Sankhla et al. 2017; Choudhary et al. 2017, 2018). Ensifer strains affiliated with papilionoid and caesalpinioid legumes in the Thar desert showed maximum similarity to symbiotic genes (nodA and nifH) of closely related type strains (E. fredii, E. glycinis, E. shofinae, E. sojae, and E. xinjiangensis) isolated from root nodules of G. max and with broad host range strain (Ensifer sp. NGR234) (Tak et al. 2016a, b; Sankhla et al. 2018; Rathi et al. 2018) (Fig. 2.2). This phylogenetic incongruence observed in species and symbiotic gene phylogeny of Thar Desert Ensifer strains may be attributed to soil alkalinity and hot-arid conditions that play a major role in the evolution of these rhizobial strains. In contrast to this, the symbiotic essential genes of TFG strains had intermediate sequences diversified from closely related E. meliloti and E. medicae , as observed in their species phylogeny based on 16S rRNA gene (Gaur et al. 2018) [Fig. 2.2]. It was surprising to note that no E. aridi type of strains was isolated from root nodules of various mimosoids (Vachellia, Senegalia, and Mimosa) studied from the Thar Desert, although E. aridi (African strains LMR001 and LMR13) have been recovered from the species of Acacia (Vachellia) from Morocco, Africa (Fig. 2.2). The sym gene phylogenies of different E. aridi strains from three continents (American strain LEM457) clearly indicate that these strains are genetically identical but harbor different symbiotic genes specific to local environmental conditions (Fig. 2.2). The Indian E. aridi strains harbor novel sym genes diversified from E. fredii and cross-nodulating papilionoid, mimosoid, and caesalpinioid wild legumes as well as crops (Tak et al. 2016b; Le Queré et al. 2017).
Maximum likelihood concatenated phylogenetic tree of Thar Desert Ensifer strains isolated from root nodules of various wild legumes of Western Rajasthan based on nodA-nifH concatenated gene sequences. Bootstrap values more than 50% calculated for 1000 replications are indicated at internodes. The scale bar indicates 5% nucleotide substitution per site. (Abbreviation: superscript T stands for type strain)
3.2 Phylogenetic Diversity of Thar Desert Bradyrhizobium Strains
Among the various wild legumes of the Thar Desert studied, the slow-growing bradyrhizobial strains were also recovered from root nodules of species of Tephrosia (tribe Millettieae, Papilionoideae) (Gehlot et al. 2012; Ojha et al. 2015), Crotalaria burhia (tribe Crotalarieae, Papilionoideae) (Ojha et al. 2015; Sankhla et al. 2018), Alysicarpus vaginalis (tribe Desmodieae, Papilionoideae) (Rathi et al. 2017), and Chamaecrista pumila (tribe Cassieae, Caesalpinioideae) (Ojha et al. 2015; Rathi et al. 2018). The slow-growing Bradyrhizobium strains isolated from root nodules of these wild herbs and under shrubs on the basis of 16S rRNA and sym (nodA and nifH) gene phylogeny shared close affinity with the type strain B. yuanmingense which was initially isolated from wild legume (Lespedeza cuneata) of tribe Desmodieae in China (Yao et al. 2002) (Figs. 2.3 and 2.4). From India, strains sharing similarities with type strains B. yuanmingense, B. liaoningense, B. elkanii, and B. japonicum have been isolated from G. max growing in Madhya Pradesh, Uttar Pradesh, and Tamil Nadu having hot-arid climate and being mostly neutral to alkaline soils (Appunu et al. 2008, 2009b; Vinuesa et al. 2008). The B. yuanmingense has also been reported to nodulate V. mungo, V. radiata, and V. unguiculata growing in regions of Uttar Pradesh, Andhra Pradesh, and Tamil Nadu (Appunu et al. 2009a) and in another crop Macrotyloma uniflorum growing in two agro-eco-climatic regions of South India (Appunu et al. 2011). As per previous studies from various parts of India, B. yuanmingense is often the preferred symbionts of crops (G. max and Vigna spp.) in the hot-dry tropical climate and alkaline soils. The existing geographical factors and alkaline soil in the Thar Desert are playing a role in the dominance of Ensifer and the sporadic occurrence of B. yuanmingense-like strains in root nodules of species of Tephrosia, Alysicarpus, and Chamaecrista in addition to strains of Ensifer which might be the preferred/primary rhizobia compared to other rhizobial genera.
Maximum likelihood phylogenetic tree of Thar Desert Bradyrhizobium strains isolated from root nodules of various wild legumes of Western Rajasthan based on 16S rRNA gene sequences. Bootstrap values more than 50% calculated for 1000 replications are indicated at internodes. The scale bar indicates 1% nucleotide substitution per site. (Abbreviation B. stands for Bradyrhizobium and superscript T stands for type strain)
Maximum likelihood concatenated phylogenetic tree of Thar Desert Bradyrhizobium strains isolated from root nodules of various wild legumes of Western Rajasthan compared with Bradyrhizobium strains isolated from other sampling sites in India having neutral [Tamil Nadu (TN), Jharkhand (JH), and Uttarakhand (UT)] and acidic [Shillong, Meghalaya] soils, based on nodA-nifH concatenated gene sequences. Bootstrap values more than 50% calculated for 1000 replications are indicated at internodes. The scale bar indicates 5% nucleotide substitution per site. (Abbreviation B. stands for Bradyrhizobium and superscript T stands for type strain)
In contrast to the Thar Desert bradyrhizobial strains (from mega clade-I), novel species of Bradyrhizobium belonging to both mega clade-I and clade-II have been recently isolated from little-studied legumes Eriosema chinense (tribe Phaseoleae, Papilionoideae) and Flemingia vestita (tribe Phaseoleae, Papilionoideae) growing in acidic soil of the sub-Himalayan region of the Indian state of Meghalaya (Ojha et al. 2017). These strains isolated from acidic soils of Shillong harbor novel nodA and nifH genes (Fig. 2.4). This suggests that although selection of rhizobia by the host plants depends upon molecular signaling between the two partners, that too is influenced by ecological factors such as soil alkalinity, soil acidity, precipitation, and soil nutrients’ availability in the region (Rathi et al. 2018). Caesalpinioid legume C. pumila in Thar Desert was nodulated by novel strains of Ensifer and strains divergent from B. yuanmingense, whereas in acidic soils and wet soils of northeastern state Meghalaya, it was found to be exclusively nodulated by diverse species of Bradyrhizobium belonging to both mega clade-I and clade-II. Significant symbiotic (nodA and nifH gene) diversity was observed in C. pumila strains isolated from acidic soils of Shillong (CPS) (Fig. 2.4). In the mimosoid legumes of Thar Desert such as V. leucophloea (Choudhary et al. 2017), P. cineraria, and Dichrostachys cinerea (Unpublished data, HS Gehlot), the primary dominant root nodule microsymbionts were fast-growing species of Ensifer, and occasionally slow-growing Bradyrhizobium strains close to B. yuanmingense were also reported.
4 Conclusions and Future Perspectives
Since the beginning of exploration of native and wild legumes from various geographical regions all over the world, the number of rhizobial genera and species has increased tremendously due to which rhizobial classification is changing. The soil alkalinity, low precipitation, and high temperature in the arid and semiarid regions of the Thar Desert are responsible for evolution and diversification of novel Ensifer strains when compared to slow-growing Bradyrhizobium from tropical and subtropical acidic soil all over the world including temperate regions. Novel Ensifer strains are dominant microsymbionts associated with legumes in the region. Majority of the Ensifer strains adapted to arid conditions of the Thar Desert are promiscuous in nature, showing geographical clustering and mosaic pattern in nucleotide sequences of housekeeping as well as symbiotic genes. Hostile environmental conditions are an important factor underlying horizontal gene transfer among related and diverse genera and species of bacteria. Molecular phylogeny of Thar Desert rhizobia provided information about their evolution and diversification and helped in better understanding the biogeography of rhizobial strains in India and in global context. Genomic characterization of diverse nitrogen-fixing novel strains from different climatic conditions will enrich our knowledge about the unique features of these strains which may open new fields for biotechnologists to improve existing microsymbiont associated with legume crops. Such studies are essential for strengthening the microbial resources in terms of agriculturally important microbes and can reduce the use of chemical fertilizers that are causing environmental pollution and adversely affecting the economy of farmers.
References
Appunu C, N’Zoue A, Laguerre G (2008) Genetic diversity of native bradyrhizobia isolated from soybeans (Glycine max L.) in different agricultural-ecological-climatic regions of India. Appl Environ Microbiol 74:5991–5996
Appunu C, N’Zoue A, Moulin L, Depret G, Laguerre G (2009a) Vigna mungo, V. radiata and V. unguiculata plants sampled in different agronomical-ecological-climatic regions of India are nodulated by Bradyrhizobium yuanmingense. Syst Appl Microbiol 32:460–470
Appunu C, Sasirekha N, Prabavathi VR, Nair S (2009b) A significant proportion of indigenous rhizobia from India associated with soybean (Glycine max L.) distinctly belong to Bradyrhizobium and Ensifer genera. Biol Fertil Soils 46:57–63
Appunu C, Ganesan G, Kalita M, Kaushik R, Saranya B, Prabavathy VR, Sudha N (2011) Phylogenetic diversity of rhizobia associated with Horsegram [Macrotyloma uniflorum (Lam.) Verdc.] grown in South India based on glnII, recA and 16S-23S intergenic sequence analyses. Curr Microbiol 62:1230–1238
Ardley J (2017) Legumes of the Thar Desert and their nitrogen fixing Ensifer symbionts. Plant Soil 410:517–520
Ardley JK, Parker MA, de Meyer SE, Trengove RD, O’Hara GW, Reeve WG, Yates RJ, Dilworth MJ, Willems A, Howieson JG (2012) Microvirga lupini sp. nov., Microvirga lotononidis sp. nov., and Microvirga zambiensis sp. nov. are Alphaproteobacterial root nodule bacteria that specifically nodulate and fix nitrogen with geographically and taxonomically separate legume hosts. Int J Syst Evol Microbiol 62:2579–2588
Aserse AA, Räsänen LA, Aseffa F, Hailemariam A, Lindström K (2013) Diversity of sporadic symbionts and nonsymbiotic endophytic bacteria isolated from nodules of woody, shrub, and food legumes in Ethiopia. Appl Microbiol Biotechnol 97:10117–10134
Benhizia Y, Benhizia H, Benguedouar A, Muresu R, Giacomini A, Squartini A (2004) Gamma proteobacteria can nodulate legumes of the genus Hedysarum. Syst Appl Microbiol 27:462–468
Beukes CW, Stepkowski T, Venter SN, Cłapa T, Phalane FL, le Roux MM, Steenkamp ET (2016) Crotalarieae and Genisteae of the South African great escarpment are nodulated by novel Bradyrhizobium species with unique and diverse symbiotic loci. Mol Phylogenet Evol 100:206–218
Boukhatem ZF, Merabet C, Bekki A, Sekkour S, Domergue O, Dupponois R (2016) Nodular bacterial endophyte diversity associated with native Acacia spp. in desert region of Algeria. Afr J Microbiol Res 10:634–645
Bournaud C, de Faria SM, dos Santos JMF, Tisseyre P, Silva M, Chaintreuil C, Gross E, James EK, Prin Y, Moulin L (2013) Burkholderia species are the most common and preferred nodulating symbionts of the piptadenia group (Tribe mimoseae). PLoS One 8:e63478
Bournaud C, Moulin L, Cnockaert M, de Faria S, Prin Y, Severac D, Vandamme P (2017) Paraburkholderia piptadeniae sp. nov. and Paraburkholderia ribeironis sp. nov., two root-nodulating symbiotic species of Piptadenia gonoacantha in Brazil. Int J Syst Evol Microbiol 67:432–440
Chahboune R, Carro L, Peix A, Barrijal S, Velázquez E, Bedmar EJ (2011) Bradyrhizobium cytisi sp. nov., isolated from effective nodules of Cytisus villosus. Int J Syst Evol Microbiol 61:2922–2927
Chen WX, Yan GH, Li JL (1988) Numerical taxonomic study of fast-growing soybean rhizobia and a proposal that Rhizobium fredii be assigned to Sinorhizobium gen. nov. Int J Syst Bacteriol 38:392–397
Chen WM, Laevens S, Lee TM, Coenye T, de Vos P, Mergeay M, Vandamme P (2001) Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int J Syst Evol Microbiol 51:1729–1735
Chen WM, Moulin L, Bontemps C, Vandamme P, Béna G, Boivin-Masson C (2003) Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J Bacteriol 185:7266–7272
Cheng Y, Watkin ELJ, O’Hara GW, Howieson JG (2002) Medicago sativa and Medicago murex differ in the nodulation response to soil acidity. Plant Soil 238:31–39
Choudhary S, Meghwal RR, Sankhla IS, Tak N, Gehlot HS (2017) Molecular characterization and phylogeny of novel diverse nitrogen fixing microsymbionts associated with Vachellia (Acacia) leucophloea in arid and semi-arid regions of Rajasthan. Indian For 143:266–278
Choudhary S, Tak N, Gehlot HS (2018) Phylogeny and genetic diversity assessment of Ensifer strains nodulating Senegalia (Acacia) senegal (L.) Britton. in arid regions of Western Rajasthan, India. Microbiology 87:127–142
da Silva K, Florentino LA, da Silva KB, de Brandt E, Vandamme P, de Souza Moreira FM (2012) Cupriavidus necator isolates are able to fix nitrogen in symbiosis with different legume species. Syst Appl Microbiol 35:175–182
de Lajudie P, Willems A, Pot B, Dewettinck D, Maestrojuan G, Neyra M, Collins MD, Dreyfus B, Kersters K, Gillis M (1994) Polyphasic taxonomy of Rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int J Syst Bacteriol 44:715–733
de Lajudie P, Laurent-Fulele E, Willems A, Torck U, Coopman R, Collins MD, Kersters K, Dreyfus B, Gillis M (1998) Allorhizobium undicola gen. nov., nitrogen-fixing bacteria that efficiently nodulate Neptunia natans in Senegal. Int J Syst Bacteriol 48:1277–1290
de Lajudie P, Willems A, Nick G, Mohamed TS, Torck U, Filai-Maltouf A, Kersters K, Dreyfus B, Lindström K, Gillis M (1999) Agrobacterium biovar 1 strains isolated from nodules of tropical legumes. Syst Appl Microbiol 22:119–132
de Meyer SE, de Beuf K, Vekeman B, Willems A (2015) A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium). Soil Biol Biochem 83:1–11
de Souza Moreira FM, Cruz L, de Faria SM, Marsh T, Martinez-Romero E, de Pedrosa OF, Pitard RM, Young JPW (2006) Azorhizobium doebereinerae sp. nov. microsymbiont of Sesbania virgata (Caz.) Pers. Syst Appl Microbiol 29:197–206
Dobritsa AP, Samadpour M (2016) Transfer of eleven Burkholderia species to the genus Paraburkholderia and proposal of Caballeronia gen. nov., a new genus to accommodate twelve species of Burkholderia and Paraburkholderia. Int J Syst Evol Microbiol 66:2836–2846
dos Santos JMF, Alves PAC, Silva VC, Rhem MFK, James EK, Gross E (2017) Diverse genotypes of Bradyrhizobium nodulate herbaceous Chamaecrista (Moench) (Fabaceae, Caesalpinioideae) species in Brazil. Syst Appl Microbiol 40:69–79
Dreyfus B, Garcia JL, Gillis M (1988) Characterization of Azorhizobium caulinodans gen. nov., sp. nov., a stem-nodulating nitrogen-fixing bacterium isolated from Sesbania rostrata. Int J Syst Bacteriol 38:89–98
Eaglesham ARJ, Ellis JM, Evans WR, Fleischmann DE, Hungria M, Hardy RWF (1990) The first photosynthetic N2-fixing Rhizobium: characteristics. In: Gresshoff PM, Roth LE, Stacey G, Newton WE (eds) Nitrogen fixation: achievements and objectives. Chapman & Hall, New York, pp 805–811
El Batanony NH, Castellano-Hinojosa A, Correa-Galeote D, Bedmar EJ (2015) The diversity of rhizobia nodulating the Medicago, Melilotus and Trigonella inoculation group in Egypt is marked by the dominance of two genetic types. Symbiosis 67:3–10
Estrada-de los Santos P, Palmer M, Chávez-Ramírez B, Beukes C, Steenkamp ET, Briscoe L, Khan N, Maluk M, Lafos M, Humm E, Arrabit M, Crook M, Gross E, Simon MF, dos Reis Junior FB, Whitman WB, Shapiro N, Poole PS, Hirsch AM, Venter SN, James EK (2018) Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 9:389
Garau G, Reeve WG, Brau L, Deiana P, Yates RJ, James D, Tiwari R, O’Hara GW, Howieson JG (2005) The symbiotic requirements of different Medicago spp. suggest the evolution of Sinorhizobium meliloti and S. medicae with hosts differentially adapted to soil pH. Plant Soil 176:263–277
Gaur S, Tak N, Rathi S, Choudhary S, Gehlot HS (2018) Identification and molecular characterization of root nodule-microsymbiont of Trigonella foenum-graecum L. growing in different soils from Western Rajasthan, India. J Environ Biol 39:684–692
Gehlot HS, Panwar D, Tak N, Tak A, Sankhla IS, Poonar N, Parihar R, Shekhawat NS, Kumar M, Tiwari R, Ardley J, James EK, Sprent JI (2012) Nodulation of legumes from the Thar Desert of India and molecular characterization of their rhizobia. Plant Soil 357:227–243
Gehlot HS, Tak N, Kaushik M, Mitra S, Chen WM, Poweleit N, Panwar D, Poonar N, Parihar R, Tak A, Sankhla IS, Ojha A, Rao SR, Simon MF, dos Reis Junior FB, Perigolo N, Tripathi AK, Sprent JI, Young JPW, James EK, Gyaneshwar P (2013) An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann Bot 112:179–196
Gehlot HS, Tak N, Dagla HR, Davis TD (2014) Indigenous and modern scientific strategies for characterization, conservation and sustainable utilization of bio-resources of the Indian Thar Desert. J Arid Land Stud 24:5–8
Gehlot HS, Ardley J, Tak N, Tian R, Poonar N, Meghwal RR, Rathi S, Tiwari R, Adnawani W, Seshadri R, Reddy TBK (2016) High-quality permanent draft genome sequence of Ensifer sp. PC2, isolated from a nitrogen-fixing root nodule of the legume tree (Khejri) native to the Thar Desert of India. Stand Genomic Sci 11:43
Graham PH, Sadowsky MJ, Keyser HH, Barnet YM, Bradley RS, Cooper JE, de Ley DJ, Jarvis BDW, Roslycky EB, Strijdon BW, Young JPW (1991) Proposed minimal standards for the description of new genera and species of root- and stem- nodulating bacteria. Int J Syst Bacteriol 41:582–587
Gyaneshwar P, Hirsch AM, Moulin L, Chen WM, Elliott GN, Bontemps C, Estrada-de los Santos P, Gross E, dos Reis Junior FB, Sprent JI, JPW Y, James EK (2011) Legume-nodulating beta-proteobacteria: diversity, host range and future prospects. Mol Plant-Microbe Interact 24:1276–1288
Hassen AI, Bopape FL, Habig J, Lamprecht SC (2012) Nodulation of rooibos (Aspalathus linearis Burm. f.), an indigenous South African legume, by members of both the α-proteobacteria and β-proteobacteria. Biol Fertil Soils 48:295–303
Hoque MS, Broadhurst LM, Thrall PH (2011) Genetic characterization of root-nodule bacteria associated with Acacia salicina and A. stenophylla (Mimosaceae) across South-Eastern Australia. Int J Syst Evol Microbiol 61:299–309
Islam MS, Kawasaki H, Muramatsu Y, Nakagawa Y, Seki T (2008) Bradyrhizobium iriomotense sp. nov., isolated from a tumor-like root of the legume Entada koshunensis from Iriomote Island in Japan. Biosci Biotechnol Biochem 72:1416–1429
Jarvis BDW, van Berkum P, Chen WX, Nour SM, Fernandez MP, Cleyet-Marel JC, Gillis M (1997) Transfer of Rhizobium loti, Rhizobium huakuii, Rhizobium ciceri, Rhizobium mediterraneum, and Rhizobium tianshanense to Mesorhizobium gen. nov. Int J Syst Bacteriol 47:895–898
Jordan DC (1982) Transfer of Rhizobium japonicum Buchanan 1980 to Bradyrhizobium gen. nov., a genus of slow-growing, root nodule bacteria from leguminous plants. Int J Syst Bacteriol 32:136–139
Jourand P, Giraud E, Béna G, Sy A, Willems A, Gillis M, Dreyfus B, de Lajudie P (2004) Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root nodule- forming and nitrogen-fixing bacteria. Int J Syst Evol Microbiol 54:2269–2273
Kathiravan R, Jegan S, Ganga V, Prabavathy VR, Tushar L, Sasikala C, Ramana CV (2013) Ciceribacter lividus gen. nov., sp. nov., isolated from rhizosphere soil of chick pea (Cicer arietinum L.). Int J Syst Evol Microbiol 63:4484–4488
Knösel DH (1984) Genus IV Phyllobacterium nom rev. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology. Williams and Wilkins Co, Baltimore, pp 254–256
Le Queré A, Tak N, Gehlot HS, Lavire C, Meyer T, Chapulliot D, Rathi S, Sakrouhi I, Rocha G, Rohmer M, Severac D, Filali-Maltou A (2017) Genomic characterization of Ensifer aridi, a proposed new species of nitrogen-fixing rhizobium recovered from Asian, African and American deserts. BMC Genomics 18:85
Li QQ, Wang ET, Chang YL, Zhang YZ, Zhang YM, Sui XH, Chen WF, Chen WX (2011) Ensifer sojae sp. nov., isolated from root nodules of Glycine max grown in saline-alkaline soils. Int J Syst Evol Microbiol 61:1981–1988
Li Y, Li X, Liu Y, Wang ET, Ren C, Liu W, Xu H, Wu H, Jiang N, Li Y, Zhang X, Xie Z (2016) Genetic diversity and community structure of rhizobia nodulating Sesbania cannabina in saline-alkaline soils. Syst Appl Microbiol 39:195–202
Lin DX, Wang ET, Tang H, Han TX, He YR, Guan SH, Chen WX (2008) Shinella kummerowiae sp. nov., a symbiotic bacterium isolated from root nodules of the herbal legume Kummerowia stipulacea. Int J Syst Evol Microbiol 58:1409–1413
Lloret L, Ormeño-Orrillo E, Rincón R, Martínez-Romero J, Rogel-Hernández MA, Martínez-Romero E (2007) Ensifer mexicanus sp. nov. a new species nodulating Acacia angustissima (Mill.) Kuntze in Mexico. Syst Appl Microbiol 30:280–290
Madhaiyan M, Poonguzhali S, Ryu J, Sa T (2006) Regulation of ethylene levels in canola (Brassica campestris) by 1-aminocyclopropane-1-carboxylate deaminase containing Methylobacterium fujisawaense. Planta 224:268–278
Martínez-Aguilar L, Salazar-Salazar C, Méndez RD, Caballero-Mellado J, Hirsch AM, Vásquez-Murrieta MS, Estrada-de los Santos P (2013) Burkholderia caballeronis sp. nov., a nitrogen fixing species isolated from tomato (Lycopersicon esculentum) with the ability to effectively nodulate Phaseolus vulgaris. Antonie Van Leeuwenhoek 104:1063–1071
Martínez-Hidalgo P, Hirsch AM (2017) The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes 1:70–82
Maynaud G, Willems A, Soussou S, Vidal C, Mauré L, Moulin L, Cleyet-Marel JC, Brunelc B (2012) Molecular and phenotypic characterization of strains nodulating Anthyllis vulneraria in mine tailings, and proposal of Aminobacter anthyllidis sp. nov., the first definition of Aminobacter as legume-nodulating bacteria. Syst Appl Microbiol 35:65–72
Mhamdi R, Mrabet M, Laguerre G, Tiwari R, Aouani ME (2005) Colonisation of Phaseolus vulgaris nodules by Agrobacterium-like strains. Can J Microbiol 51:105–111
Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, de Lajudie P, Masson-Boivin C (1999) Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol 65:3084–3094
Moulin L, Munive A, Dreyfus B, Boivin-Masson C (2001) Nodulation of legumes by members of the β subclass of Proteobacteria. Nature 411:948–950
Mousavi SA, Österman J, Wahlberg N, Nesme X, Lavire C, Vial L, Paulin L, de Lajudie P, Lindström K (2014) Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst Appl Microbiol 37:208–215
Mousavi SA, Willems A, Nesme X, de Lajudie P, Lindström K (2015) Revised phylogeny of Rhizobiaceae: proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst Appl Microbiol 38:84–90
Muresu R, Polone E, Sulas L, Baldan B, Tondello A, Delogu G, Cappuccinelli P, Alberghini S, Benhizia Y, Benhizia H, Benguedouar A, Mori B, Calamassi R, Dazzo FB, Squartini A (2008) Coexistence of predominantly nonculturable rhizobia with diverse, endophytic bacterial taxa within nodules of wild legumes. FEMS Microbiol Ecol 63:383–400
Nick G, de Lajudie P, Eardly BD, Soumalainen S, Paulin L, Zhang X, Gillis M, Lindström K (1999) Sinorhizobium arboris sp. nov. and Sinorhizobium kostiense sp. nov., isolated from leguminous trees in Sudan and Kenya. Int J Syst Bacteriol 49:1359–1368
Ogasawara M, Suzuki T, Mutoh I, Annapurna K, Arora NK, Nishimura Y, Maheshwari DK (2003) Sinorhizobium indiaense sp. nov. and Sinorhizobium abri sp. nov. isolated from tropical legumes, Sesbania rostrata and Abrus precatorius, respectively. Symbiosis 34:53–68
Ojha A, Rao CS, Tak N, Gehlot HS, Rao SR (2015) Genetic diversity analysis of rhizobial symbionts associated with legumes of India for Efficient Biological Nitrogen Fixation (BNF) Technology and Natural Soil Fertility. In: Choudhury H (ed) Biology, biotechnology and sustainable development. Research India Publications, New Delhi, pp 183–196
Ojha A, Tak N, Rathi S, Chouhan B, Rao SR, Barik SK, Joshi SR, Sprent JS, James EK, Gehlot HS (2017) Molecular characterization of novel Bradyrhizobium strains nodulating Eriosema chinense and Flemingia vestita, Important unexplored native legumes of the Sub-Himalayan region (Meghalaya) of India. Syst Appl Microbiol 40:334–344
Panwar D, Tak N, Gehlot HS (2014) Nodulated native legumes in an arid environment of Indian Thar Desert. In: Fulekar MH, Kale RK (eds) Recent trends in plant sciences. IK International Publishing House Pvt. Ltd, New Delhi, pp 284–298
Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ (2015) Bacterial associations with legumes. Crit Rev Plant Sci 34:17–42
Ramírez-Bahena MH, Peix A, Rivas R, Camacho M, Rodriguez-Navarro DN, Mateos PF, Martinez-Molina E, Willems A, Velazquez E (2009) Bradyrhizobium pachyrhizi sp. nov. and Bradyrhizobium jicamae sp. nov., isolated from effective nodules of Pachyrhizus erosus. Int J Syst Evol Microbiol 59:1929–1934
Rathi S, Gaur S, Tak N, Tak A, Gehlot HS (2017) Genetically diverse root nodule bacteria associated with Alysicarpus vaginalis from alkaline soil of Rajasthan, India. Plant Archives 17:495–505
Rathi S, Tak N, Bissa G, Chouhan B, Ojha A, Adhikari D, Barik SK, Satyawada RR, Sprent JI, James EK, Gehlot HS (2018) Selection of Bradyrhizobium or Ensifer symbionts by the native Indian caesalpinioid legume Chamaecrista pumila depends on soil pH and other edaphic and climatic factors. FEMS Microbiol Ecol 94:fiy180. doi.org/10.1093/femsec/fiy180
Rivas R, Willems A, Subba-Rao NS, Mateos PF, Dazzo FB, Kroppenstedt RM, Martínez-Molina E, Gillis M, Velázquez E (2003) Description of Devosia neptuniae sp. nov. that nodulates and fixes nitrogen in symbiosis with Neptunia natans, an aquatic legume from India. Syst Appl Microbiol 26:47–53
Rome S, Fernandez MP, Brunel B, Normand P, Cleyet-Marel JC (1996) Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int J Syst Bacteriol 46:972–980
Sánchez M, Ramírez-Bahena MH, Peix A, Lorite MJ, Sanjuán J, Velázquez E, Monza J (2014) Phyllobacterium loti sp. nov. isolated from nodules of Lotus corniculatus. Int J Syst Evol Microbiol 64:781–786
Sankhla IS, Meghwal RR, Tak N, Tak A, Gehlot HS (2015) Phenotypic and molecular characterization of microsymbionts associated with Crotalaria medicagenia: a native legume of the Indian Thar desert. Plant Archives 15:1003–1010
Sankhla IS, Tak N, Meghwal RR, Choudhary S, Tak A, Rathi S, Sprent JI, James EK, Gehlot HS (2017) Molecular characterization of nitrogen fixing microsymbionts from root nodules of Vachellia (Acacia) jacquemontii, a native legume from the Thar Desert of India. Plant Soil 410:21–40
Sankhla IS, Meghwal RR, Choudhary S, Rathi S, Tak N, Tak A, Gehlot HS (2018) Molecular characterization of microsymbionts associated with root nodules of Crotalaria burhia Buch.-Ham. ex Benth., a native keystone legume species from Thar Desert of India. Indian J Exp Biol 56:373–385
Sawada H, Kuykendall LD, Young JM (2003) Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J Gen Appl Microbiol 49:155–179
Sawana A, Adeolu M, Gupta RS (2014) Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5:429
Shamseldin A, Abdelkhalek A, Sadowsky MJ (2017) Recent changes to the classification of symbiotic, nitrogen-fixing, legume-associating bacteria: a review. Symbiosis 71:91–109
Shiraishi A, Matsushita N, Hougetsu T (2010) Nodulation in black locust by the Gamma-Proteobacteria Pseudomonas sp. and the Beta-Proteobacteria Burkholderia sp. Syst Appl Microbiol 33:269–274
Sprent JI (2001) Nodulation in Legumes. Royal Botanic Gardens. In: Kew
Sprent JI, Gehlot HS (2010) Nodulated legumes in arid and semi-arid environments: are they important? Plant Ecol Divers 3:211–219
Sprent JI, Ardley J, James EK (2017) Biogeography of nodulated legumes and their nitrogen fixing symbionts. New Phytol 215:40–56
Sy A, Giraud E, Jourand P, Garcia N, Willems A, de Lajudie P, Prin Y, Neyra M, Gillis M, Boivin-Masson C, Dreyfus B (2001) Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol 183:214–220
Tak N, Gehlot HS, Kaushik M, Choudhary S, Tiwari R, Tian R, Hill Y, Bräu L, Goodwin L, Han J, Liolios K, Huntemann M, Palaniappan K, Pati A, Mavromatis K, Ivanova N, Markowitz V, Woyke T, Kyrpides N, Reeve W (2013) Genome sequence of Ensifer sp. TW10; a Tephrosia wallichii (Biyani) microsymbiont native to the Indian Thar Desert. Stand Genomic Sci 9:304–314
Tak A, Tak N, Sankhla IS, Meghwal RR, Gehlot HS (2016a) Molecular characterization of nitrogen fixing Ensifer species from Vigna trilobata growing in alkaline soil of Thar Desert. Green Farming 7:300–304
Tak N, Awasthi E, Bissa G, Meghwal RR, James EK, Sprent JS, Gehlot HS (2016b) Multi locus sequence analysis and symbiotic characterization of novel Ensifer strains nodulating Tephrosia spp. in the Indian Thar Desert. Syst Appl Microbiol 39:534–545
Tian CF, Zhou YJ, Zhang YM, Li QQ, Zhang YZ, Li DF, Wang S, Wang J, Gilbert LB, Li YR, Chen WX (2012) Comparative genomics of rhizobia nodulating soybean suggests extensive recruitment of lineage-specific genes in adaptations. Proc Natl Acad Sci U S A 109:8629–8634
Toledo I, Lloret L, Martínez-Romero E (2003) Sinorhizobium americanus sp. nov., a new Sinorhizobium species nodulating native Acacia spp. in Mexico. Syst Appl Microbiol 26:54–64
Trinick MJ (1973) Symbiosis between Rhizobium and the non-legume, Trema aspera. Nature 244:459–460
Trujillo ME, Willems A, Abril A, Planchuelo AM, Rivas R, Ludeña D, Mateos PF, Martínez-Molina E, Velázquez E (2005) Nodulation of Lupinus albus by strains of Ochrobactrum lupini sp. nov. Appl Environ Microbiol 71:1318–1327
Valverde A, Velázquez E, Gutiérrez C, Cervantes E, Ventosa A, Igual JM (2003) Herbaspirillum lusitanum sp. nov., a novel nitrogen-fixing bacterium associated with root nodules of Phaseolus vulgaris. Int J Syst Evol Microbiol 53:1979–1983
Valverde A, Velázquez E, Fernández-Santos F, Vizcaíno N, Rivas R, Mateos PF, Martínez-Molina E, Igual JM, Willems A (2005) Phyllobacterium trifolii sp. nov., nodulating Trifolium and Lupinus in Spanish soils. Int J Syst Evol Microbiol 55:1985–1989
van Berkum P, Eardly BD (2002) The aquatic budding bacterium Blastobacter denitrificans is a nitrogen-fixing symbiont of Aeschynomene indica. Appl Environ Microbiol 68:1132–1136
Vinuesa P, León-Barrios M, Silva C, Willems A, Jarabo-Lorenzo A, Pérez-Galdona R, Werner D, Martínez-Romero E (2005) Bradyrhizobium canariense sp. nov., an acid-tolerant endosymbiont that nodulates endemic genistoid legumes (Papilionoideae: Genisteae) from the Canary Islands, along with Bradyrhizobium japonicum bv. genistearum, Bradyrhizobium genospecies alpha and Bradyrhizobium genospecies beta. Int J Syst Evol Microbiol 55:569–575
Vinuesa P, Rojas-Jiménez K, Contreras-Moreira B, Mahna SK, Prasad BN, Moe H, Selvaraju SB, Thierfelder H, Werner D (2008) Multilocus sequence analysis for assessment of the biogeography and evolutionary genetics of four Bradyrhizobium species that nodulate soybeans on the Asiatic continent. Appl Environ Microbiol 74:6987–6996
Yao ZY, Kan FL, Wang ET, Wei GH, Chen W (2002) Characterization of rhizobia that nodulate legume species of the genus Lespedeza and description of Bradyrhizobium yuanmingense sp. nov. Int J Syst Evol Microbiol 52:2219–2230
Young JM, Kuykendall LD, Martinez-Romero E, Kerr A, Sawada H (2003) Classification and nomenclature of Agrobacterium and Rhizobium-a reply to Farrand et al. (2003). Int J Syst Evol Microbiol 53:1689–1695
Zahran HH (2001) Rhizobia from wild legumes: diversity, taxonomy, ecology, nitrogen fixation and biotechnology. J Biotechnol 91:43–53
Zhang JJ, Lou K, Jin X, Mao PH, Wang ET, Tian CF, Sui XH, Chen WF, Chen WX (2012) Distinctive Mesorhizobium populations associated with Cicer arietinum L. in alkaline soils of Xinjiang, China. Plant Soil 353:123–134
Zurdo-Piñeiro JL, Rivas R, Trujillo ME, Vizcaíno N, Carrasco JA, Chamber M, Palomares A, Mateos PF, Martínez-Molina E, Velázquez E (2007) Ochrobactrum cytisi sp. nov., isolated from nodules of Cytisus scoparius in Spain. Int J Syst Evol Microbiol 57:784–788
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Tak, N., Gehlot, H.S. (2019). Diversity of Nitrogen-Fixing Symbiotic Rhizobia with Special Reference to Indian Thar Desert. In: Satyanarayana, T., Das, S., Johri, B. (eds) Microbial Diversity in Ecosystem Sustainability and Biotechnological Applications. Springer, Singapore. https://doi.org/10.1007/978-981-13-8487-5_2
Download citation
DOI: https://doi.org/10.1007/978-981-13-8487-5_2
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-13-8486-8
Online ISBN: 978-981-13-8487-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)