Abstract
Twenty four rhizobial strains were isolated from root nodules of Melilotus, Medicago and Trigonella plants growing wild in soils throughout Egypt. The nearly complete 16S rRNA gene sequence from each strain showed that 12 strains (50 %) were closely related to the Ensifer meliloti LMG6133T type strain with identity values higher than 99.0 %, that 9 (37.5 %) strains were more than 99 % identical to the E. medicae WSM419T type strain, and that 3 (12.5 %) strains showed 100 % identity with the type strain of N. huautlense S02T. Accordingly, the diversity of rhizobial strains nodulating wild Melilotus, Medicago and Trigonella species in Egypt is marked by predominance of two genetic types, E. meliloti and E. medicae, although the frequency of isolation was slightly higher in E. meliloti. Sequencing of the symbiotic nodC gene from selected Medicago and Melilotus strains revealed that they were all similar to those of the E. meliloti LMG6133T and E. medicae WSM419T type strains, respectively. Similarly, nodC sequences of strains identified as members of the genus Neorhizobium were more than 99 % identical to that of N. galegae symbiovar officinalis HAMBI 114.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Together with the actinorhizal plants, legumes are best characterized by their ability to establish dinitrogen (N2)-fixing symbiotic associations with soil bacteria collectively referred to as rhizobia. Comprehensive reviews of the associations of bacteria with legumes and the current available knowledge on the phylogenetic diversity of both rhizobia and other endophytic bacteria inhabiting root nodules have been published recently (Rivas et al. 2009; Velázquez et al. 2010; Gyaneshwar et al. 2011; Peix et al. 2015). During the plant-rhizobial interaction process an exchange of molecular signals occurs between the two partners leading to the formation of root nodules, within which symbiotic N2 fixation occurs (Graham 2008).
Among members of the Leguminosae (Fabaceae), the genera Medicago, Melilotus and Trigonella comprise a large number of species of annual herbs, herbaceous perennials and shrubs, mostly native to the Mediterranean region (Lesins and Lesins 1979). These legumes are a biological source of nitrogen that gives them economic significance in cultivation for forage or pasture, as well as environmental value in non-managed ecosystems, and makes them excellent candidates for use in sustainable agricultural systems (Graham 2008; Howieson et al. 2008). The N2-fixing rhizobia that are currently known to nodulate Medicago species are from the genus Ensifer (syn. Sinorhizobium), of which the species E. meliloti (de Lajudie et al. 1994) and E. medicae (Rome et al. 1996) are well characterized microsymbionts (Garau et al. 2005; Peix et al. 2015). In addition to Ensifer, Rhizobium mongolense also nodulates Melilotus ruthenica (van Berkum et al. 1998). Ensifer meliloti has also been isolated from nodules of Melilotus spp. (Yan et al. 2000) and Melilotus officinalis and Medicago monspelliaca (Pandey et al. 2004; del Villar et al. 2008). Information about the symbiosis between Trigonella and rhizobia is relatively scarce even though it is within the Medicago-Melilotus cross inoculation group. However, recent studies from China have shown that the symbionts of T. arcuata are Ensifer (He et al. 2011), and that Rhizobium tibeticum, which was first isolated from root nodules of T. archiducis-nicolai in Tibet (Hou et al. 2009), also nodulated Medicago lupulina, Medicago sativa and Medicago officinalis (Hou et al. 2009). On the other hand, Rajendran et al. (2012) failed to isolate rhizobial strains from nodules of T. foenum-graecum during a study performed to investigate the most common nodule-associated bacteria. Almost nothing is known about the symbionts of Trigonella spp. growing in North African and Mediterranean countries.
Egyptian Medicago, Melilotus and Trigonella species have been studied for years, but they are not well characterized in terms of their symbionts, although there have been several reports from neighbouring North African and Mediterranean countries, such as Tunisia (Zakhia et al. 2004) and Algeria (Sebbane et al. 2006; Arbi et al. 2015). In Egypt, research on their plant-associated microsymbionts has mainly focused on cultivated species, and those of wild legumes have been generally ignored. Medicago, Melilotus and Trigonella grow wild throughout Egypt and are part of the natural vegetation. Because very scarce information about their symbiosis is available, the objective of this study was to explore the diversity of the rhizobia that infect species of the three legume genera via sequencing of their core (16S rRNA) and symbiotic (nodC) genes, and to compare the results obtained with isolates from other legumes within the same inoculation group.
2 Materials and methods
2.1 Isolation of bacteria from nodules and culture conditions
Root fragments (5–10 cm long) containing nodules (5–10/plant) were collected from healthy Melilotus, Medicago, and Trigonella plants growing wild in different locations in Egypt, ranging from the North West Mediterranean coastal region to the Nile Valley (Table 1). Nodules were kept on ice until they were taken to the laboratory. They were surface-sterilized by sequential washing with 96 % ethanol for 10 s, 3 % hydrogen peroxide for 3 min and, finally, rinsed thoroughly in sterile distilled water. Tests to validate surface sterilization of plant tissues were performed by touching the disinfected nodules several times on the surface of solid yeast extract-mannitol (YEM) medium (Vincent 1970) prior to isolation of the interior microbiota. Nodules from the same plant species and location were pooled. Twelve nodules from each pool were placed independently in Petri dishes and crushed in a drop of sterile water with a sterile glass rod. The resulting suspension was streaked onto Petri dishes containing YEM supplemented with 0.025 g L−1 Congo Red and incubated at 30 °C for 10 d. Single colonies were picked and checked for purity by repeated streaking on YEM medium.
2.2 DNA extraction and PCR amplifications
Genomic DNA was isolated from bacterial cells using the RealPure Genomic DNA Extraction kit (Durviz, Spain), according to the manufacturer’s instructions. The quantity of DNA was determined by using a Nanodrop spectrophotometer (NanoDrop ND1000). PCR amplifications of 16S rRNA gene fragments were carried out using the two opposing primers 41f and 1488r as previously reported (Herrera-Cervera et al. 1999). Three forward primers, nodCF, nodCF2 and nodCFn, and the reverse primer nodCI were used for amplification and sequencing of the nodC gene as indicated earlier (Laguerre et al. 2001). Amplification products were purified using the Qiagen PCR product purification system and subjected to cycle sequencing using the same primers as for PCR amplification, with ABI Prism dye chemistry and analyzed with a 3130 xl automatic sequencer at the sequencing facilities of the Estación Experimental del Zaidín, CSIC, Granada, Spain. The 16S rRNA gene sequences were compared to those deposited in EzTaxon-e (Kim et al. 2012) and those of the nodC gene sequences with homologous sequences in GenBank using the Phydit software (Chun 2001). Phylogenetic analyses were performed with the Geneious software package version 7.1.7 (Kearse et al. 2012), inferred using the neighbor-joining algorithm (Saitou and Nei 1987) and visualized with MEGA5 (Tamura et al. 2011).
2.3 Plant nodulation tests
Seeds of Melilotus, Medicago and Trigonella species were surface-sterilized with 2.5 % HgCl2 for 7 min, followed by thorough washing in sterile distilled water. The seeds were then placed in Petri dishes containing 1 % water agar and allowed to germinate at 30 °C in the dark. Seedlings (3 per pot) were planted in autoclaved 1 L Leonard jar assemblies containing sand and vermiculite (1:1, v:v) and inoculated at sowing with 1 mL of a single bacterial strain (~ 108 cells mL−1). The plants were fed with an N-free nutrient solution (Fahraeus 1957), grown in a greenhouse under natural daylight conditions, and harvested at 10 % flowering to check for nodule formation.
2.4 Accession numbers
Accession numbers of the nucleotide sequences of the strains used in this study are shown in the phylogenetic trees.
3 Results
3.1 Phylogenetic analysis of 16S rRNA and nodC genes
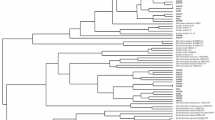
Twenty eight bacterial strains, 12 from Melilotus, 8 from Medicago and 8 from Trigonella species, were isolated from extracts of nodules taken from healthy plants growing wild in different locations in Egypt (Table 1). Partial sequences of the 16S rRNA gene from each strain revealed that 24 strains were members of the family Rhizobiaceae of the Alphaproteobacteria and that the remaining 4 strains belonged to the genera Paenibacillus (2 strains), Variovorax and Brevibacillus, respectively (Table 1). Among the Rhizobiaceae, 21 strains were classified as belonging to the genus Ensifer and 3 belonged to the genus Neorhizobium (Table 1). A neighbor-joining tree (Fig. 1) and EzTaxon-e analysis (Table 1) constructed from the 16S rRNA gene sequences indicated that strains NHBM3B, NHBM5, NHBM10A, NHBM10B, NHBM12 and NHBM13 from Melilotus indicus, NHBM16 from Melilotus messanensis, NHBM18 from Melilotus siculus, NHBM23 from Medicago intertexta and NHBM24 from Medicago polymorpha clustered with E. medicae WSM419T with identity values higher than 99 %. Also, that strains NHBM9, NHBM12 and NHBM14 from Melilotus indicus, strain NHBM17 from Melilotus messanensis, strain NHBM19 from Melilotus siculus, strain NHBM22B from Medicago intertexta, and strains NHBM26 and NHBM27 from Medicago laciniata grouped with E. meliloti LMG6133T with identity values higher than 99.0 %. Strains NHBTR69, NHBTR70, NHBTR72 and NHBTR74 isolated from T. maritima also clustered with E. meliloti LMG6133T with identity values higher than 99.0 %. Together with strains in genus Ensifer, strains NHBM21, NHBM25 and NHBM29 that were isolated from nodules of Medicago sativa, Medicago polymorpha and Medicago laciniata, respectively, showed 100 % identity with the type strain N. huautlense S02T. The 4 non-rhizobial strains from nodules of Melilotus indicus i.e. Paenibacillus strains NHBM4 and NHBM6, Brevibacillus strain NHBM7 and Variovorax strain NHBM15 (Table 1) were not further characterized in this study.
Neighbor-joining phylogenetic tree based on partial 16S rRNA sequences of strains from nodules of Melilotus, Medicago and Trigonella and phylogenetically related species within members of the Rhizobiaceae. Bootstrap values are indicated as percentages derived from 1000 replications. Values lower than 70 are not shown. Bar, 1 nucleotide substitution per 100 nucleotides. The tree is rooted by Azospirillum caulinodans OTS751T
Utilization of different primer combinations resulted in amplification of the nodC gene of strains NHBM3B, NHBM16, NHBM18, NHBM23 and NHBM24 that were chosen as representatives of those previously identified as E. medicae and isolated from different host plants (Table 1). Similar results were obtained when strains NHBM9, NHBM17, NHBM19, NHBM22B and NHBM26, representing those showing identity with E. meliloti (Table 1), were used for PCR amplification of their nodC genes. The primer pair nodCF/nodCI was also useful to amplify the nodC gene of the strains NHBM21, NHBM25 and NHBM29 identified as N. huautlense that were isolated from Medicago sativa, Medicago polymorpha and Medicago laciniata, respectively. Pairwise alignments between globally aligned sequences of nodC from the strains isolated in this study with those of the corresponding Ensifer and Neorhizobium species showed that strains NHBM3B, NHBM16, NHBM18, NHBM23 and NHBM24 varied between 99.42 % and 99.74 % identity to those of the nodC from E. medicae 1037T, that the nodC sequences of strains NHBM9, NHBM17, NHBM19, NHBM22B and NHBM26 were between 99.34 % and 99.74 % identical to those of E. meliloti ATCC 9930T, and that the similarities of the nodC sequences from strains NHBM21, NHBM25 and NHBM29 to those of N. galegae symbiovar (sv.) officinalis HAMBI114 ranged from 98.73 % to 99.45 %. A phylogenetic tree showing the relationship between the nodC genes from the strains isolated from the nodules of Melilotus/Medicago showed that they belonged to the species E. meliloti and E. medicae and were affiliated to the symbiovar meliloti of E. meliloti (Fig. 2), whereas the strains that were related to the species N. huautlense were affiliated with the symbiovar officinalis of N. huautlense (Fig. 3).
Neighbor-joining phylogenetic tree based on partial nodC sequences of strains from nodules of Melilotus and Medicago species and of E. meliloti symbiovars. Bootstrap values are indicated as percentages derived from 1000 replications. Values lower than 70 are not shown. Bar, 2 nucleotide substitution per 100 nucleotides
Neighbor-joining phylogenetic tree based on partial nodC sequences of strains from nodules of Medicago species and of Rhizobium symbiovars. Bootstrap values are indicated as percentages derived from 1000 replications. Values lower than 70 are not shown. Bar, 5 nucleotide substitution per 100 nucleotides
3.2 Plant nodulation tests
Plant nodulation tests under greenhouse conditions showed that the representative strains formed effective nodules on the roots of their corresponding host plants. Indirect indications of effectiveness of the nodules for nitrogen fixation were obtained by visual inspection of the presence of the red leghemoglobin protein in nodule cross-sections and by the dark green intensity of the leaves compared to uninoculated control plants. No nodulation was detected when strains in the genera Paenibacillus, Brevibacillus and Variovorax were used for inoculation.
4 Discussion
In this study, the isolation and identification of rhizobial bacteria from root nodules of Melilotus, Medicago and Trigonella plants growing in the wild is reported. Collectively, out of the 24 strains isolated, 12 were identified as E. meliloti, 9 as E. medicae and 3 as N. huautlense. These results suggest that the diversity of Ensifer strains nodulating species in the Melilotus-Medicago-Trigonella inoculation group in Egypt is marked by the predominance of two genetic types, E. meliloti and E. medicae, although the frequency with which E. meliloti was isolated was slightly higher (50 %) than that of E. medicae (37.5 %). Since the nodules were sampled from wild plants growing in very different locations throughout Egypt, the data suggest that there are no biogeographical differences among the symbionts. A much higher predominance of E. meliloti over E. medicae was reported after isolation of rhizobia from nodules of Melilotus, Medicago and Trigonella growing in a Central Asian soil (Roumiantseva et al. 2002) and from nodules of Medicago sativa, Medicago lupulina and Medicago polymorpha growing in Mexican soils (Silva et al. 2007). However, a predominance of E. medicae has been observed in the nodules of M. lupulina and T. foenum-graecum grown in Spanish soils (Iglesias et al. 2007). With regard to Melilotus, Medicago and Trigonella symbionts from the Mediterranean basin, Zakhia et al. (2004) have shown that, in contrast with our data, Medicago sativa is nodulated by E. meliloti in infra-arid soils from Tunisia, and Sebbane et al. (2006) found that Medicago polymorpha grown in Algeria is not nodulated by E. medicae but by taxonomically unidentified rhizobial strains. In addition, Arbi et al. (2015) have reported that Melilotus indicus growing wild in the Algerian Sahara forms nodules with E. meliloti, which agrees with the present study that both E. meliloti and E. medicae nodulate this legume species. Similar to the Egyptian species, Medicago polymorpha when introduced into Australia is nodulated by E. medicae, and it is particularly associated with annual M. polymorpha that are well adapted to moderately acid soils (Garau et al. 2005). The pH of the soils in which the Egyptian legumes were grown was not determined, so a relationship between pH and the predominant Ensifer species found in the nodules from the present study cannot be determined.
Regardless of the legume species, E. meliloti and E. medicae were isolated from nodules of both Melilotus and Medicago, but E. medicae was not detected within nodules from T. maritima (Table 1). In this study Trigonella were nodulated by E. meliloti, a result coincident with that of He et al. (2011) who showed that T. arcuata is also nodulated by E. meliloti. Indeed, E. meliloti strains CCBAU 83848, CCBAU 83856, CCBAU 83881, CCBAU 83884, and CCBAU 83887 isolated from T. arcuata clustered together with those from T. maritima that were isolated during the present study (Fig. 1). On the other hand, as only four strains were isolated from T. maritima, additional surveys are required to understand the consistency of the association between it and E. meliloti.
With a frequency of 12.5 %, the species N. huautlense was isolated exclusively from nodules of the genus Medicago, including Medicago sativa, Medicago polymorpha and Medicago laciniata (Table 1). To our knowledge this is the first report showing nodulation of Medicago by this rhizobial species. Rhizobium huautlense was first isolated from Sesbania herbacea (Wang et al. 1998) and, together with R. galegae, R. alkalisoli and R. vignae, has been shown to form a separate clade from Rhizobium which represents a new genus, and for which the name Neorhizobium was proposed (Mousavi et al. 2014).
Analysis of the nodC sequences revealed that strains isolated from Melilotus belonged to symbiovar meliloti of E. meliloti (Fig. 2), and that those from nodules of Medicago were affiliated with either E. meliloti symbiovar meliloti or with N. huautlense symbiovar officinalis (Fig. 3). A high degree of similarity (higher than 99 %) was found between the nodC genes of E. medicae and E. meliloti. These results agree with those of Ramírez-Bahena et al. (2015) who suggested that symbiovar meliloti should be recognised within the species E. medicae. Because of the very few divergences found among the sequences within each species, diversity is scarce among the strains isolated in this study. The 12 E. meliloti, 9 E. medicae and the 3 N. huautlense strains identified in this study are true symbionts of their corresponding host legumes as, after nodule isolation, they were able to establish effective symbioses with the Medicago sativa, Medicago polymorpha and T. maritima seedlings that were used for the plant inoculation tests. Cross-inoculation tests were not carried out in this study.
Nodules from Melilotus indicus also harbored species in the genera Paenibacillus, Variovorax and Brevibacillus (Table 1), but the isolation of these endophytes does not necessarily mean that they are restricted to Melilotus indicus. Together with rhizobia, legume nodules are occupied by a variable microbiome composed of very phylogenetically diverse bacteria, mainly species, genera, families and classes within the phyla Proteobacteria, Firmicutes and Actinobacteria (reviewed in Velázquez et al. 2013; Peix et al. 2015). Nodule endophytes have been shown to produce indole acetic acid (IAA) and siderophores, to express both N2-fixation and 1-amino-cyclopropane 1-carboxylate (ACC) deaminase activities, and are also involved in antifungal biocontrol. However, although all the aforementioned traits are related to plant growth promotion ability (Pérez-Montaño et al. 2014), the isolation of endophytic bacteria from nodules is also considered to be a source of potential confusion when trying to identify the actual nodulating symbiont (Gyaneshwar et al. 2011). Moreover, recent work has shown that some endophytes have the capacity to accompany rhizobial cells during the infection process to enter inside the root nodules using it as a niche without any obvious benefit to the plant (Zgadzaj et al. 2015).
References
Arbi SB, Chekireb D, Quatrini P, Catania V, Cheriet D, Ouartsi A (2015) Phenotypic and genotypic characterization of root nodules rhizobia of Medicago littoralis Rhode and Melilotus indicus (L.) All. growing in the Oasis of Touggourt, Oued Righ Valley, in the Algerian Sahara. Symbiosis 66:75–87
Chun J (2001) PHYDIT version 3.1 (available at http://plaza.snu.ac.kr/~jchun/phydit/)
de Lajudie P, Willems A, Pot B, Dewettinck D, Maestrojuan G, Neyra M, Collins MD, Dreyfus B, Kersters K, Gillis M (1994) Polyphasic taxonomy of Rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov., and Sinorhizobium teranga sp. nov. Int J Syst Bacteriol 44:715–733
del Villar M, Rivas R, Peix A, Mateos PF, Martínez-Molina E, van Berkum P, Willems A, Veláquez E (2008) Stable molecular weight RNA profiling showed variations within Sinorhizobium meliloti and Sinorhizobium medicae nodulating different legumes from the alfalfa cross-inoculation group. FEMS Microbiol Lett 282:273–281
Fahraeus A (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16:374–381
Garau G, Reeve WG, Brau L, Deiana P, Yates RJ, James D, Tiwari R, O'Hara GW, Howieson JG (2005) The symbiotic requirements of different Medicago spp. suggest the evolution of Sinorhizobium meliloti and S. medicae with hosts differentially adapted to soil pH. Plant Soil 276:263–277
Graham PH (2008) Ecology of the root-nodule bacteria of legumes. In: Dilworth MJ, James EK, Sprent JI, Newton WE (eds) Nitrogen-fixing leguminous symbioses. Springer, Dordrecht, pp. 23–43
Gyaneshwar P, Hirsch AM, Moulin L, Chen WM, Elliott GN, Bontemps C, Estrada-de Los Santos P, Gross E, Dos Reis FB, Sprent JI, Young JP, James EK (2011) Legume-nodulating betaproteobacteria: diversity, host range and future prospects. Mol Plant-Microbe Interact 24:1276–1288
He YR, Wang JY, Wang ET, Feng G, Chang YL, Sui XH, Chen WX (2011) Trigonella arcuata-associated rhizobia—an Ensifer (Sinorhizobium) meliloti population adapted to a desert environment. Plant Soil 345:89–102
Herrera-Cervera JA, Cabello-Mellado J, Laguerre G, Tichy HV, Requena N, Amarger N, Martínez-Romero E, Olivares J, Sanjuan J (1999) At least five rhizobial species nodulate Phaseolus vulgaris in a Spanish soils. FEMS Microbiol Ecol 30:87–97
Hou BC, Wang ET, Ying LJ, Jia RZ, Chen WF, Gao Y, Don RJ, Chen WX (2009) Rhizobium tibeticum sp. nov., a symbiotic bacterium isolated from Trigonella archiductis-nicolai Vassilez. Int J Syst Evol Microbiol 59:3051–3057
Howieson JG, Yates RJ, Foster JKJ, Real D, Besier RD (2008) Prospects for the future use of legumes. In: Dilworth MJ, James EK, Sprent JI, Newton WE (eds) Nitrogen-fixing leguminous symbioses. Springer, Dordrecht, pp. 363–387
Iglesias O, Rivas R, García-Fraile P, Abril A, Mateos PF, Martinez-Molina E, Velázquez E (2007) Genetic characterization of fast-growing rhizobia able to nodulate Prosopis alba in North Spain. FEMS Microbiol Lett 277:210–216
Kearse M, Moir R, Wilson M, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147:981–993
Lesins KA, Lesins I (1979) Genus Medicago (Leguminosae), a taxonomic study. Junk, The Hague
Mousavi SA, Österman J, Wahlbergb N, Nesmec X, Lavirec C, Vial C, Paulind L, de Lajudie P, Lindström K (2014) Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst Appl Microbiol 37:208–215
Pandey P, Sahgal M, Maheswari DK, Johri BN (2004) Genetic diversity of rhizobia isolated from medicinal legumes growing in the sub-Himalayanm region of Uttaranchai. Curr Sci 86:202–207
Peix A, Ramírez-Bahena MH, Velázquez E, Bedmar EJ (2015) Bacterial associations with legumes. Crit Rev Plant Sci 34:17–42
Pérez-Montaño F, Alias-Villegas C, Bellogin RA, del Cerro P, Espuny MR, Jiménez-Guerrero I, López-Baena FJ, Ollero FJ, Cubo T (2014) Plant growth promotion in cereal and leguminous agricultural important plants: from microorganism capacities to crop production. Microbiol Res 169:325–336
Rajendran R, Patel MH, Joshi SJ (2012) Isolation and characterization of nodule-associated Exiguobacterium sp. from the root nodules of fenugreek (Trigonella foenum-graecum) and their possible role in plant growth promotion. Int J Microbiol 2012:693982
Ramírez-Bahena ME, Vargas M, Martín M, Tejedor C, Velázquez E, Peix A (2015) Alfalfa microsymbionts from different ITS and nodC lineages of Ensifer meliloti and Ensifer medicae symbiovar meliloti established efficient symbiosis with alfalfa in Spanish acid soils. Appl Microbiol Biotechnol 99:4855–4865
Rivas R, García-Fraile P, Velázquez E (2009) Taxonomy of bacteria nodulating legumes. Microbiol Insights 2:251–269
Rome S, Fernández MP, Brunel B, Normand P, Cleyet-Marel JC (1996) Sinorhizobium medicae sp. nov., isolated from annual Medicago spp. Int J Syst Bacteriol 46:972–980
Roumiantseva ML, Andronov EE, Sharypova LA, Dammann-Kalinowski T, Keller M, Young JP, Simarov BV (2002) Diversity of Sinorhizobium meliloti from the Central Asian alfalfa gene Center. Appl Environ Microbiol 68:4694–4697
Saitou N, Nei M (1987) The neigbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sebbane N, Sahnoune M, Zakhia F, Willems A, Benallaoua S, de Lajudie P (2006) Phenotypical and genotypical characteristics of root-nodulating bacteria isolated from annual Medicago spp. in Soummam Valley (Algeria). Lett Appl Microbiol 42:235–241
Silva C, Kan FL, Martinez-Romero E (2007) Population genetic structure of Sinorhizobium meliloti and S. medicae isolated from nodules of Medicago spp. in Mexico. FEMS Microbiol Ecol 60:477–489
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28:2731–2739
van Berkum P, Beyene D, Bao G, Campbell TA, Eardly BD (1998) Rhizobium mongolense sp. nov. is one of three rhizobial genotypes identified which nodulate and form nitrogen-fixing symbioses with Medicago ruthenica [(L.) Ledebour]. Int J Syst Bacteriol 48:13–22
Velázquez E, García-Fraile P, Ramírez-Bahena MH, Peix A, Rivas R (2010) Proteobacteria forming nitrogen fixing symbiosis with higher plants. In: Sezenna ML (ed) Proteobacteria: phylogeny, metabolic diversity and ecological effects. Nova Science Publishers, New York, pp. 37–56
Velázquez E, Martínez-Hidalgo P, Carro L, Alonso P, Peix A, Trujillo ME, Martínez-Molin E (2013) Nodular endophytes: an untapped diversity. In: Rodelas-González MB, González-López J (eds) Beneficial plant-microbial interactions: ecology and applications. CRC Press, Boca Raton, pp. 214–235
Vincent JM (1970) A manual for the practical study of root nodule bacteria. Blackwell Scientific Publications, Oxford
Wang ET, van Berkum P, Beyene D, Sui XH, Dorado O, Chen WX, Martínez-Romero E (1998) Rhizobium huautlense sp. nov., a symbiont of Sesbania herbacea that has a close phylogenetic relationship with Rhizobium galegae. Int J Syst Bacteriol. 3:687–699
Yan AM, Wang ET, Kan FL, Tan ZY, Sui XH, Reinhold-Hurek B, Chen WX (2000) Sinorhizobium meliloti associated with Medicago sativa and Melilotus spp. in arid saline soils in Xinjiang, China. Int J Syst Evol Microbiol 50:1887–1891
Zakhia F, Jeder H, Domergue O, Willems A, Cleyet-Marel JC, Gillis M, Dreyfus B, de Lajudie P (2004) Characterisation of wild legume nodulating bacteria (LNB) in the infra-arid zone of Tunisia. Syst Appl Microbiol 27:380–395
Zgadzaj R, James EK, Kelly S, Kawaharada Y, de Jonge N, Jensen DB, Madsen LH, Radutoiu S (2015) A legume genetic framework controls infection of nodules by symbiotic and endophytic bacteria. PLoS Genet 11:e1005280
Acknowledgments
This study was supported by ERDF-cofinanced grant PE12-AGR1968 from Consejería de Economía, Innovación y Ciencia (Junta de Andalucía, Spain).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
El Batanony, N.H., Castellano-Hinojosa, A., Correa-Galeote, D. et al. The diversity of rhizobia nodulating the Medicago, Melilotus and Trigonella inoculation group in Egypt is marked by the dominance of two genetic types. Symbiosis 67, 3–10 (2015). https://doi.org/10.1007/s13199-015-0365-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-015-0365-8