Abstract
Aims
Rhizobia associated with white clover (Trifolium repens) grown in alkaline soils of China have never been investigated. This is the first survey to report of their genetic and biogeographical diversity.
Methods
Nodule bacteria were isolated from white clover grown in alkaline soils (pH 8.18–8.99) in North and East China and were characterized by multilocus sequence analysis (MLSA) of the housekeeping genes (atpD, recA, and glnII), and phylogenies of 16S rRNA gene and symbiotic genes nodC and nifH. The biogeographic distribution of rhizobial species was analyzed in relation to the soil factors.
Results
A total of 83 new strains could be affiliated to Rhizobium that shared 100 % sequence similarity of 16S rRNA gene with R. leguminosarum, R. acidisoli, R. anhuiense, R. indigoferae, R. sophorae, and R. laguerreae. Three genospecies were further distinguished based on the housekeeping gene analysis among these new strains: R. anhuiense, R. leguminosarum, and a hypothetical novel Rhizobium genospecies. Highly conserved symbiotic genes corresponding to those of symbiovar trifolii in R. leguminosarum were observed among all the new strains. Unique rhizobial communities associated with white clover were detected in the tested alkaline soils, and soil characteristics such as pH and nutrient levels were estimated as the determinant factors.
Conclusions
White clover established symbiosis with three Rhizobium genospecies harboring similar symbiotic genes in alkaline soils in China. Biogeographic pattern exists in clover rhizobia that was determined by the soil pH and nutrient levels.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Clover (Trifolium) species have been documented as hosts for symbiotic nitrogen-fixing bacteria, so called rhizobia. Despite their wide geographic distribution, Rhizobium strains mainly corresponding to R. leguminosarum sv. trifolii (Jordan 1984) have been isolated from root nodules of the clover plants (Duodu et al. 2007; Kumar et al. 2014; Marek-Kozaczuk et al. 2013; Ramirez-Bahena et al. 2009; Seguin et al. 2001; Shamseldin et al. 2014; Tesfaye and Holl 1999). Therefore, clover has been considered as a highly specific host towards the rhizobial symbionts. Here, we studied white clover, not considered by the works cited above.
White clover (Trifolium repens L.) is a legume widely distributed all over the world, cultured as ornamental or foliage plant or wild plant in the grasslands. In China, white clover has been widely planted as an ornamental plant in cities or as foliage in pastoral areas based upon its adaptation to diverse environments. Grown in the tropical and subtropical regions of South China, where soils are usually acidic and both iron- and aluminum-rich, white clover was reported to be nodulated by seven genospecies belonging to Bradyrhizobium, Rhizobium, and Sinorhizobium, with R. leguminosarum as the majority (Liu et al. 2007). These results suggested that the clover rhizobia might have diversified in Chinese soils to adapt to the local conditions, as it was described for rhizobia associated with soybean (Han et al. 2009; Zhang et al. 2011). However, this hypothesis needed to be verified with further studies on the clover rhizobia isolated from different zones or soil types. Unfortunately, soil physicalchemical characteristics have not been considered in most of the previous studies on clover rhizobia, although these characters are the most important structuring factor for distribution and diversification of rhizobia.
Unlike acidic soils, alkaline soils, rich in calcium, are common in the northern region and occasionally prevalent in the eastern (subtropical) region of China. So far, no report is available on systematic characterization of clover rhizobia in the alkaline soils of China. Therefore, the present study was undertaken to increase knowledge on the clover-rhizobial symbiont interactions in regard to soil conditions, and to understand the diversity and geographic distribution of clover rhizobia. Specifically, rhizobia nodulating white clover grown in alkaline soils of the subtropical region (Henan Province and Shanghai City) were isolated and characterized to estimate their phylogenetic diversity and geographic distribution.
Materials and methods
Isolation of rhizobia
Root nodules of clover plants were collected in December 2014 from six gardens in three cities: four gardens in Zhengzhou City (Xiliuhu at E 113° 34ʹ 56.78ʺ-N 34° 46ʹ 44ʺ, Zhengzhou University at E 113° 31ʹ 49.17ʺ-N 34° 49ʹ 4ʺ, Jinshui at E 113° 39ʹ 5ʺ-N 34° 48ʹ 23ʺ, and Ruizhi at E 113° 33ʹ 4ʺ-N 34° 47ʹ 44ʺ), one in Ruzhou City (Meishan Park at E 112° 49ʹ 33ʺ-N 34° 10ʹ 36ʺ), and one in Shanghai City (Zhongshan Park at E 121° 24ʹ 53ʺ-N 31° 13ʹ 25ʺ). Five plants were sampled at each site by a cross-strategy, and two to three pink and healthy nodules were selected randomly from each plant to isolate the rhizobia with standard procedures on yeast extract mannitol agar (YMA) (Vincent 1970). The rhizobial isolates were purified by successive repeated streaking on the same medium. In all cases, the isolates were incubated at 28 °C and stored in YMA slants at 4 °C for further use and in YM broth supplied with glycerol (25 % w/v) at −80 °C for long-term storage.
Nodulation tests
All the obtained isolates were tested for their ability to nodulate with white clover grown in sterilized vermiculite using the standard procedures (Wei et al. 2009). Briefly, clover seeds of white clover bought from the market in Zhengzhou were surface sterilized by NaClO solution (2.5 %, w/v) for 5 min, pre-germinated on 0.8 % water-agar at 28 °C in dark for 3 days, transplanted into a sterile glass tube (40 × 300 mm) containing 25 ml of nitrogen-free solution and sterilized vermiculates (one seedling per tube) (Wei et al. 2009), and inoculated with 0.1 ml of log phase culture of each isolate (109 cells ml−1). The inoculation test was carried out in triplicates, and the inoculated plants were placed in a growth cabinet with the cycle of 16 h with illumination at 28 °C and 8 h in dark at 18 °C, at about 50 % relative humidity. Uninoculated seedlings were included as control. Nodulation was checked after 45 days of growth (Wei et al. 2009).
Soil sampling and characterization
Soil (about 50 g) was sampled from the clover fields (0 to 20 cm in depth) in parallel to the nodule collection. Air-dried soil samples were grinded and passed through 2-mm mesh screens for physical and chemical analysis. The pH, electrical conductivity (EC), organic matter (OM), and contents of available phosphorus (P), potassium (K), and total nitrogen (N) of soil samples were analyzed at the Department of Chemical and Material Engineering (ZZULI), using standard methods (AFNOR 2005; Appunu et al. 2009; Han et al. 2009; Olsen 1954; Simonis 1996).
PCR-based restriction fragment length polymorphism of 16S rDNA and 16S–23S rRNA intergenic spacer
Genomic DNA of each rhizobial isolate was extracted according to Terefework et al. (2001) and was used as template for 16S rRNA gene and intergenic spacer (IGS) amplifications using primers P1 and P6 (Zhang et al. 2012) and FGPS1490 (forward) and FGPS132′ (reverse) (Laguerre et al. 1996), respectively. PCR products (about 1449 bp for 16S rRNA gene and 900 bp for IGS) were digested separately with each of the following restriction endonucleases: MspI and HaeIII for both restriction processes, AluI and HinfI only for ARDRA, and HhaI only for IGS restriction fragment length polymorphism (RFLP). Separation and visualization of genomic DNA, PCR products, and restriction fragments were done by agarose gel electrophoresis in 0.8, 1.5, and 2.5 % (w/v), respectively (Wang et al. 1999) containing GoldView type I nucleotide dye (Solarbio, Lot.No.20140820). The results were analyzed by DNR Bio-Imaging System (MiniBIS Pro, made in Israel). Then, the rRNA gene type of each strain was designated according to its restriction pattern, and a grouping analysis was performed (Wang et al. 1999) with the UPGMA method of Nei and Li (1979).
Phylogeny of 16S rRNA gene and multilocus sequence analysis of housekeeping genes
16S rRNA gene amplicons from representative strains for different IGS types were further sequenced according to Zhang et al. (2012). All the new nucleotide sequences were Blasted for similarity search and deposited into GenBank database (NCBI) (accession numbers are available in figures and supplementary materials). Sequences were aligned with the related sequences, including the sequences of white clover symbionts from South China, obtained from the NCBI database using MEGA version 6.0 software (Tamura et al. 2013). Phylogenetic reconstructions were performed using the NJ method (Neighbor-Joining) with 1000 bootstrap replications of each sequence (Zhang et al. 2012).
Multilocus sequence analysis (MLSA) of the housekeeping genes atpD (encoding for ATP synthase beta chain), recA (recombinase A), and glnII (glutamine synthetase II) has been widely used to differentiate rhizobial species (Martens et al. 2008; Vinuesa et al. 2005a, b). These three genes were independently amplified by using primer pairs atpD255F/atpD782R, recA41F/recA640R, and glnII12F/glnII689R, respectively (Vinuesa et al. 2005b). The purified PCR products were sequenced directly using the same primers. The acquired sequences (465 bp for atpD, 479 bp for recA, and 600 bp for glnII) were concatenated (Vinuesa et al. 2005a, b) and aligned using ClustalW with the manually concatenated sequences of the same housekeeping genes from type strains of the defined Rhizobium species obtained from NCBI database. Distance calculation and construction of the concatenated gene tree were performed using the NJ method and bootstrapping algorithms contained in the MEGA 6.0 (Tamura et al. 2013).
Phylogenetic analysis of symbiotic genes nodC and nifH
The symbiotic genes nodC (N-acetyglucosaminyl-trasferase) and nifH (nitrogenase dehydrogenase gene) have been described as good molecular markers to estimate the host specificity of rhizobia (Laguerre et al. 2001). Both nodC and nifH genes of the representative strains were amplified by using the primer pairs nodCF/nodCR and nifHF/nifHR, respectively (Laguerre et al. 2001). The acquired sequences were deposited in the NCBI database and were used for alignment and reconstruction of the phylogenetic tree with the same methods described above for 16S rRNA genes.
Estimation of genetic diversity in different soils
To estimate the genetic diversity of white clover rhizobia in the sampling sites, genospecies defined by MLSA and IGS types were used to calculate the Shannon-Wiener index (Hill et al. 2003) using the vegan package integrated in the R statistical language (version 2.12.0; http://www.r-project.org/). Redundancy analysis (RDA) and the canonical version of principal component analysis (PCA) (Braak and Smilauer 2002) were used to examine the multiple relationships between soil factors (N, P, K content, EC values, OM and soil pH) and genospecies of white clover rhizobia from different ecological sites.
Results
Isolation of rhizobia from root nodules and soil characterization
A total of 83 isolates, exhibiting convex colonies of 2–4 mm in diameter after 3–4 days of incubation (YMA, 28 °C), were obtained from the clover nodules. All isolates could effectively nodulate white clover, as testified by the red color inside the root nodules and dark green leaves of the plants. The uninoculated controls grew poorly with no root nodule and yellowish-green leaves.
The soils from the sampling sites were alkaline with pH 8.18–8.99. Soil EC values ranged between 9.1 and 19.8 ms/m. The soil from Ruzhou (Henan) had the highest values of available phosphorus (38.3 mg kg−1), organic matter (68.75 g kg−1), total nitrogen (2.71 g kg−1), and available potassium (161 mg kg−1) (Supplementary Table S1).
PCR-based typing and phylogenies of 16S rRNA gene and housekeeping genes
In the RFLP analyses, the 83 isolates shared a single 16S rRNA type, and they were further divided into 14 IGS types. Seventeen representative strains standing for the IGS types and the geographic origins (Ruzhou, Zhengzhou, and Shanghai) were chosen for further sequencing and phylogenetic analysis, as shown in Table 1. The 16S rRNA gene-based phylogeny showed that all the isolates formed a single clade in the genus Rhizobium, presenting sequence similarities ranging from 99.8 to 100 % among themselves and from 99.3 to 100 % with the type strains for 15 defined species, including R. leguminosarum USDA 2370T and R. lentis BLR 27T and 99.9–100 % with reference strains R. leguminosarum CCBAU 43201 and CCBAU 61381 from acid soils (detail available as Supplementary Fig. S1).
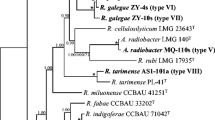
In the phylogenetic analysis based on the concatenated sequences of atpD and recA, the 17 representative strains formed three clusters (genospecies) within the genus Rhizobium (Fig. 1), that were similar with the phylogeny in the individual housekeeping gene trees (Supplementary Fig. S2) and in the tree of concatenated sequences of atpD, recA, and glnII (Supplementary Fig. S3). Cluster 1 (C1) includes the strains exhibiting IGS types 3, 8, 9, and 13 (44 isolates); cluster 2 (C2) includes only one strain WYCCWR 10014 belonging to IGS type 10; cluster 3 (C3) includes the strains representing IGS types 1, 2, 4–7, 11, 12, and 14 (38 isolates). The MLSA similarities were 98.8–100 % between strains inside a cluster, and 93.7–96.5 % between clusters. The C3 members showed similarities of 98.5–100 % with R. anhuiense CCBAU 23525T and of less than 95.0 % with other references. The C1 members showed 99.9–100 and 96.4–96.6 % similarities with R. leguminosarum sv. viciae 3841 and CCBAU 73064, respectively; 96.1–96.2 % with R. sophorae CCBAU 03386T and 95.8–95.9 % with R. laguerreae FB 206T. The single strain in C2, WYCCWR 10014, showed 97.8 % similarity with R. leguminosarum sv. viciae CCBAU 73064, 97.1 % with both of R. sophorae CCBAU 03386T and R. laguerreae FB 206T, and 96.5 % with R. leguminosarum sv. viciae 3841 (Table 1).
Neighbor-joining phylogenetic tree based on concatenated atpD-recA gene sequences, showing the relationships of the bacteria isolated from nodules of white clover. The tree was constructed by using a distance matrix (Kimura 2-parameter model). Bootstrap confidence levels >50 % are indicated at the internodes. Bar = 2 % nucleotide divergence. All the acquired sequences were used for phylogenetic tree reconstruction
Phylogenies of nodC and nifH genes
In the nodC phylogenetic tree (Fig. 2), the reference strains for symbiovars trifolii, viciae, and phaseoli formed separate branches, while all the 17 representative strains were grouped with sv. trifolii in the species of R. leguminosarum, R. pisi, and R. etli, with similarities of 95.1–99.8 % among them, and less than 70.3 % with the reference strains for other symbiovars. The nifH gene phylogeny (Supplementary Fig. S4) showed similar topology to that of the nodC gene, although only 12 nifH genes from the 17 representatives were obtained.
Rhizobial distribution
The diversity (Shannon index) of clover-rhizobial community considering both MLSA clustering and IGS type affiliation presented the following order in the sampling soils: overall of the three areas (2.28, 5.28) > Shanghai (0.94, 2.90) > Ruzhou (0.79, 1.86) > Zhengzhou (0.55, 0.52) (Table 1). The MLSA clusters C1 and C3 were found in all the three sampling areas, but C1 was the dominant group in Zhengzhou (87.5 %), while C3 was dominant in Ruzhou (76.2 %) and Shanghai (77.3 %) (Table 1). C2 contained only one strain from Shanghai. At the level of IGS type, types 1 and 2 (C3) were found in all the three areas, while type 1 was dominant in Ruzhou (47.6 %) and Shanghai (36.4 %). IGS types 3 (C1) and 4 (C3) were found in Ruzhou (23.8, 9.5 %) and Shanghai (4.5, 18.2 %), while type 13 was shared by Zhengzhou and Shanghai, at the relative abundance of 87.5 and 4.5 %, respectively.
According to the RDA results (Fig. 3), soil pH, available phosphorus (P), total nitrogen (N), available potassium (K), organic matter (OM), and EC had all strong effects on the distribution of the clover-rhizobial groups (MLSA cluster and IGS types). The higher EC value strongly selected the IGS types 13 (C1) and 14 (C3) and negatively selected the other 11 types except type 2. The higher pH value was correlated with IGS types 7, 8, 9, 11, 12 (C3), and 10 (C2). In addition, IGS types 3 (C1), 1, 4, 5, and 6 (C3) were comprehensively selected by higher values of pH, P, OM, N, and K; and type 2 isolates were selected by both the high EC and nutrient contents (P, N, K, and organic matter). The pH, P, N, K, and OM were negatively correlated to the IGS types 13 (C1) and 14 (C3).
Biplot of the RDA on the 14 IGS types and soil characteristics in sampling sites by CANOCO. P available phosphate, EC electrical conductivity, OM organic matter, N total nitrogen, K available potassium. Canonical correspondence analyses (RDA) were used to evaluate influence. The longer the arrow is, the greater the influence; the smaller the angle between two arrows, the closer their relationship. The arrows representing P, EC, OM, N, and K are in black color and in gray color for IGS types
Discussion
Since the time when Jordan (1984) designated the clover-nodulating rhizobia as R. leguminosarum sv. trifolii, the taxonomy of rhizobia has been greatly developed (Ormeño-Orrillo et al. 2015). During the last three decades, the diversity and taxonomy of clover-nodulating rhizobia have been studied in several cases (Duodu et al. 2007; Kumar et al. 2014; Liu et al. 2007; Marek-Kozaczuk et al. 2013; Ramirez-Bahena et al. 2009; Seguin et al. 2001; Shamseldin et al. 2014; Tesfaye and Holl 1999). Although diverse genomic groups within the genera Bradyrhizobium, Mesorhizobium, Rhizobium, and Ensifer (Sinorhizobium) were detected in the nodules of Trifolium species, only Rhizobium pisi has been identified for the clover microsymbionts (Marek-Kozaczuk et al. 2013) in addition to R. leguminosarum. Comparing with the previous studies, the present study offers some important information about the clover rhizobia.
In this study, white clover rhizobia isolated from alkaline soils were identified as three genomic species, R. anhuiense, R. leguminosarum, and a novel genospecies in Rhizobium, based on the results of MLSA (Fig. 1 and Supplementary Figs. S1 and S3), which were also supported by PCR-based RFLP of 16S rDNA and IGS, as well as 16S rRNA gene sequence phylogeny. R. leguminosarum sv. trifolii strains were isolated as a dominant group from clover in this study, consistent to previous reports (Duodu et al. 2007; Mauchline et al. 2014; Ramirez-Bahena et al. 2009; Seguin et al. 2001; Liu et al. 2007). The inclusion of reference strains R. leguminosarum CCBAU 43201, R. leguminosarum CCBAU 61381, and Rhizobium sp. CCBAU 33107 originated from white clover in acidic soil of South China (Liu et al. 2007) in this group (Fig. 1) evidenced that R. leguminosarum sv. trifolii has a universal distribution in both the acid and alkaline soils. R. anhuiense was originally isolated from Vicia faba and Pisum sativum in acidic soils from Southern China (Zhang et al. 2015). In this study, R. anhuiense was defined as a dominant microsymbiont of Trifolium repens in alkaline soils in North and East China (Table 1 and Fig. 1), and a novel symbiovar trifolii was found among the isolates (Fig. 2 and Supplementary Fig. S4). Both the detection of WYCCWR 10014 as a putative Rhizobium novel genospecies and identification of R. anhuiense in this study enlarged the diversity of clover rhizobia. Our results are consistent with the conclusion of Liu et al. (2007) that diverse rhizobial species associated with Trifolium species exist in different types of soils in China.
Since the main rhizobial group associated with white clover plants in South China were R. leguminosarum sv. trifolii (Liu et al. 2007), the identification of R. anhuiense as the main rhizobia in this study implied that the rhizobial communities associated with white clover in the North-East regions and in the South regions of China might be different. Both the study of Liu et al. (2007) and our present study revealed the biogeographic patterns of clover rhizobia in China. Furthermore, although R. leguminosarum sv. trifolii was detected in both South (Liu et al. 2007) and North-East regions of China (Fig. 1), its relative abundance varied dramatically in different sampling sites. It seems that this bacterium is more adapted to acidic soils in China. Even in the three sampling areas involved in this study, the rhizobial communities varied as shown by both the presence/absence of some minor IGS types and differences in the relative abundances of the major/common IGS types and genomic species (Table 1).
Consistent with previous studies (Han et al. 2009; Man et al. 2008; Zhang et al. 2011), the biogeographic pattern could be related to the soil characteristics, like EC and pH value, and P, OM, N, and K contents (Fig. 3). In addition to the biogeography of rhizobia, the soil features also altered the diversity of rhizobia, since variation in Shannon index among the sampling areas was detected in this study (Table 1 and Supplementary Table S1). Low contents of phosphorus and organic matter and high EC (salinity) are accompanied with rhizobial diversity decrease in the soils of Zhengzhou (Table 1). The biogeographic pattern (Table 1) and the RDA results (Fig. 3) imply that soil directs the symbiosis between white clover and rhizobia in China, and that rhizobia have to undergo the selection pressures from both host legume and soil conditions, as reported earlier (Han et al. 2009; Man et al. 2008; Zhang et al. 2011).
Lateral transfer of the symbiotic genes is an important way for rhizobial evolution and emerging of new symbiotic bacteria. The highly similar nodC and nifH genes among new and reference strains (Fig. 2 and Supplementary Fig. S4) evidenced that lateral transfer of the symbiotic genes might have occurred among the clover-nodulating rhizobia in China. It was clear that the symbiotic genes corresponding to symbiovar trifolii were shared across the clover-nodulating strains in R. leguminosarum, R. pisi, R. etli, and R. anhuiense (Fig. 2) (Marek-Kozaczuk et al. 2013; Rogel et al. 2011; Shamseldin et al. 2014). This phenomenon indicates that white clover stringently selects the symbiotic gene background of its microsymbionts (Laguerre et al. 2001; Mauchline et al. 2014; Ramirez-Bahena et al. 2009) which might have driven the lateral gene transfer from the introduced clover rhizobia to the more adaptive native bacteria. These results suggested that horizontal gene transfer between closely related species possibly directed the diversification and evolution of clover-nodulating rhizobia to help the clover plants colonizing diverse environments.
R. anhuiense sv. trifolii and the putative Rhizobium novel genospecies sv. trifolii found here may be regarded as specific to alkaline soils since they were not reported in other regions of China. The symbiotic genes might have been transferred from R. leguminosarum sv. trifolii present in the soils of the three sampling sites of this study (Table 1) when the plant was introduced, similar to what proposed for chickpea rhizobia in Europe (Rivas et al. 2007) and in China (Zhang et al. 2012) and for Lotus nodulating Mesorhizobium in New Zealand (Sullivan and Ronson 1998). Alternatively, white clover-nodulating rhizobia may have been introduced together with the plants; then, the symbiotic genes were transferred from the introduced R. leguminosarum sv. trifolii strains to native bacteria adapted to local conditions. Lateral gene transfer of symbiotic genes between different species in soils has been claimed to explain the incongruence between phylogenies of symbiotic and core genes (Kumar et al. 2014; Laguerre et al. 2001; Lan and Reeves 2000; Rogel et al. 2011; Sullivan and Ronson 1998; Wernegreen and Riley 1999).
In conclusion, eighty-three strains of white clover rhizobia isolated from alkaline soils in China were identified as a suspicious novel Rhizobium genospecies, R. anhuiense and R. leguminosarum. Biogeographic patterns of clover rhizobia were detected in China. All the clover rhizobia harbored symbiotic genes similar to R. leguminosarum sv. trifolii reference strains, suggesting lateral transfer of symbiotic genes among different rhizobial genospecies and the existence of sv. trifolii in R. anhuiense. This is the first systematic survey on white clover rhizobia in alkaline soils in North and East China.
References
AFNOR (2005) Soil quality, determination of pH. NF ISO 10390. AFNOR, Paris
Appunu C, Sasirekha N, Prabavathy VR, Nair S (2009) A significant proportion of indigenous rhizobia from India associated with soybean (Glycine max L.) distinctly belong to Bradyrhizobium and Ensifer genera. Biol Fert Soils 46:57–63
Braak CT, Smilauer P (2002) CANOCO reference manual and CanoDraw for Windows user’s guide: sofware for canonical community ordination (version 4.5). Section on Permutation Methods Microcomputer Power, Ithaca, Newyork.
Duodu S, Carlsson G, Huss-Danell K, Svenning MM (2007) Large genotypic variation but small variation in N2 fixation among rhizobia nodulating red clover in soils of northern Scandinavia. J Appl Microbiol 102:1625–1635
Han L, Wang E, Han T, Liu J, Sui X, Chen W, Chen W (2009) Unique community structure and biogeography of soybean rhizobia in the saline-alkaline soils of Xinjiang, China. Plant Soil 324:291–305
Hill TC, Walsh KA, Harris JA, Moffett BF (2003) Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol 43:1–11
Jordan DC (1984) Family III. Rhizobiaceae Conn 1938, 321AL. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology. Williams & Wilkins, Baltimore
Kumar N, Lad G, Giuntini E, Kaye M, Udomwong P, Shamsani N, Young J, Bailly X (2014) Bacterial genospecies that are not ecologically coherent: population genomics of Rhizobium leguminosarum. Open Biol 5:140133
Laguerre G, Mavingui P, Allard MR, Charnay MP, Louvrier P, Mazurier SI, Rigottier-Gois L, Amarger N (1996) Typing of rhizobia by PCR DNA fingerprinting and PCR-restriction fragment length polymorphism analysis of chromosomal and symbiotic gene regions: application to Rhizobium leguminosarum and its different biovars. Appl Environ Microbiol 62:2029–2036
Laguerre G, Nour SM, Macheret V, Sanjuan J, Drouin P, Amarger N (2001) Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiol (England) 147:981–993
Lan R, Reeves PR (2000) Intraspecies variation in bacterial genomes: the need for a species genome concept. Trends Microbiol 8:396–401
Liu XY, Wang ET, Li Y, Chen WX (2007) Diverse bacteria isolated from root nodules of Trifolium, Crotalaria and Mimosa grown in the subtropical regions of China. Arch Microbiol 188:1–14
Man CX, Wang H, Chen WF, Sui XH, Wang ET, Chen WX (2008) Diverse rhizobia associated with soybean grown in the subtropical and tropical regions of China. Plant Soil 310:77–87
Marek-Kozaczuk M, Leszcz A, Wielbo J, Wdowiak-Wróbel S, Skorupska A (2013) Rhizobium pisi sv. trifolii K3.22 harboring nod genes of the Rhizobium leguminosarum sv. trifolii cluster. Syst Appl Microbiol 36:252–258
Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A (2008) Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214
Mauchline TH, Hayat R, Roberts R, Powers SJ, Hirsch PR (2014) Assessment of core and accessory genetic variation in Rhizobium leguminosarum symbiovar trifolii strains from diverse locations and host plants using PCR-based methods. Lett Appl Microbiol 59:238–246
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A 76:5269–5273
Olsen SR (1954) Estimation of available phosphorus in soils by extraction with sodium bicarbonate. USDA, Washington, DC
Ormeño-Orrillo E, Servín-Garcidueñas LE, Rogel MA, González V, Peralta H, Mora J, Julio Martínez-Romero J, Martínez-Romero E (2015) Taxonomy of rhizobia and agrobacteria from the Rhizobiaceae family in light of genomics. Syst Appl Microbiol 38:287–291
Ramirez-Bahena MH, Velazquez E, Fernandez-Santos F, Peix A, Martinez-Molina E, Mateos PF (2009) Phenotypic, genotypic, and symbiotic diversities in strains nodulating clover in different soils in Spain. Can J Microbiol 55:1207–1216
Rivas R, Laranjo M, Mateos PF, Oliveira S, Martinez-Molina E, Velazquez E (2007) Strains of Mesorhizobium amorphae and Mesorhizobium tianshanense, carrying symbiotic genes of common chickpea endosymbiotic species, constitute a novel biovar (ciceri) capable of nodulating Cicer arietinum. Lett Appl Microbiol 44:412–418
Rogel MA, Ormeno-Orrillo E, Martinez Romero E (2011) Symbiovars in rhizobia reflect bacterial adaptation to legumes. Syst Appl Microbiol 34:96–104
Seguin P, Graham PH, Sheaffer CC, Ehlke NJ, Russelle MP (2001) Genetic diversity of rhizobia nodulating Trifolium ambiguum in North America. Can J Microbiol 47:81–85
Shamseldin A, Moawad H, Abd El-Rahim WM, Sadowsky MJ (2014) Near-full length sequencing of 16S rDNA and RFLP indicates that Rhizobium etli is the dominant species nodulating Egyptian winter Berseem clover (Trifolium alexandrinum L.). Syst Appl Microbiol 37:121–128
Simonis AD (1996) Effect of temperature on extraction of phosphorus and potassium from soils by various extracting solutions. Commun Soil Sci Plant Anal 27:665–684
Sullivan JT, Ronson CW (1998) Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci U S A 95:5145–5149
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Terefework Z, Kaijalainen S, Lindstrom K (2001) AFLP fingerprinting as a tool to study the genetic diversity of Rhizobium galegae isolated from Galega orientalis and Galega officinalis. J Biotechnol 91:169–180
Tesfaye M, Holl FB (1999) Rhizobium strains that nodulate Trifolium semipilosum Fres. are phylogenetically distinct. Plant Soil 207:147–154
Vincent JM (1970) A manual for the practical study of root nodule bacteria. International Biological Programme (By) Blackwell Scientific, Oxford
Vinuesa P, Silva C, Lorite MJ, Izaguirre-Mayoral ML, Bedmar EJ, Martinez-Romero E (2005a) Molecular systematics of rhizobia based on maximum likelihood and Bayesian phylogenies inferred from rrs, atpD, recA and nifH sequences, and their use in the classification of Sesbania microsymbionts from Venezuelan wetlands. Syst Appl Microbiol 28:702–716
Vinuesa P, Silva C, Werner D, Martínez-Romero E (2005b) Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation. Mol Phylogenet Evol 34:29–54
Wang E, Van Berkum P, Sui X, Beyene D, Chen W, Martínez-Romero E (1999) Diversity of rhizobia associated with Amorpha fruticosa isolated from Chinese soils and description of Mesorhizobium amorphae sp. nov. Int J Syst Bacteriol 49:51–65
Wei G, Chen W, Zhu W, Chen C, Young JPW, Bontemps C (2009) Invasive Robinia pseudoacacia in China is nodulated by Mesorhizobium and Sinorhizobium species that share similar nodulation genes with native American symbionts. FEMS Microbiol Ecol 68:320–328
Wernegreen JJ, Riley MA (1999) Comparison of the evolutionary dynamics of symbiotic and housekeeping loci: a case for the genetic coherence of rhizobial lineages. Mol Biol Evol 16:98–113
Zhang YM, Li Y Jr, Chen WF, Wang ET, Tian CF, Li QQ, Zhang YZ, Sui XH, Chen WX (2011) Biodiversity and biogeography of rhizobia associated with soybean plants grown in the North China Plain. Appl Environ Microbiol 77:6331–6342
Zhang J, Lou K, Jin X, Mao P, Wang E, Tian C, Sui X, Chen W, Chen W (2012) Distinctive Mesorhizobium populations associated with Cicer arietinum L. in alkaline soils of Xinjiang, China. Plant Soil 353:123–134
Zhang YJ, Zheng WT, Everall I, Young JP, Zhang XX, Tian CF, Sui XH, Wang ET, Chen WX (2015) Rhizobium anhuiense sp. nov. isolated from effective nodules of Vicia faba and Pisum sativum. Int J Syst Evol Microbiol 65:2960–2967
Acknowledgments
This work was financed by the National Natural Science Foundation of China (Project No. 31400008) and the Scientific Research Funds of Zhengzhou University of Light Industry (Project No. 2014bsjj006). ETW is financially supported by grants SIP20150597 authorized by Instituto Politécnico Nacional, Mexico. Thanks to JunWei Zhang from Ruzhou and Xianchao Yang from Shanghai for assistance in the soil sampling.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Katharina Pawlowski.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PPTX 260 kb)
Rights and permissions
About this article
Cite this article
Zhang, J.J., Jing, X.Y., de Lajudie, P. et al. Association of white clover (Trifolium repens L.) with rhizobia of sv. trifolii belonging to three genomic species in alkaline soils in North and East China. Plant Soil 407, 417–427 (2016). https://doi.org/10.1007/s11104-016-2899-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-016-2899-9