Abstract
Glucosinolates are a group of secondary metabolites involved in plant defense and found mainly in the Brassicales order. While the breakdown products of some glucosinolates are beneficial to human health, many glucosinolates are toxic. The recently sequenced genomes of Brassica napus and its parental species Brassica rapa and Brassica oleracea provided the Brassica scientific community with a valuable tool for systematically investigating glucosinolate biosynthesis, transport, and breakdown genes, elucidating the relationship between variation of glucosinolate profiles and the evolution of glucosinolate-related genes in Brassica crops. In this chapter, we summarized the variation in glucosinolate composition and content in Brassica crops and identified 166, 167, 191, 333 genes in B. rapa, B. oleracea var. capitata, B. oleracea var. italica, and B. napus, respectively, as orthologs of 78 glucosinolate biosynthetic, transport, and breakdown genes in Arabidopsis thaliana. Among these glucosinolate-related genes, transcription factor, side-chain modification, and breakdown genes experienced significant expansion in the four Brassica crops. Moreover, phylogenetic and expression pattern analyses of the glucosinolate-related genes HAG1, MAM, AOP, and GTR correspond with the glucosinolate profiles and total seed glucosinolate contents in B. napus and its parental species. These results, together with those published previously, provide a valuable resource for understanding the genetic mechanism underlying glucosinolate metabolism and transport and suggest novel approaches for improving the nutritional quality of Brassica crops through breeding cultivars with lower glucosinolate contents.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
12.1 Introduction
Glucosinolates (GSLs) are a group of sulfur-rich, nitrogen-containing plant secondary metabolites mainly found in the Brassicales order (Fahey et al. 2001), which includes many economically and nutritionally important crops and condiments, such as oilseed rape (Brassica napus), broccoli (Brassica oleracea var. italica), cabbage (B. oleracea var. capitata), turnip (Brassica rapa), mustard (Brassica juncea L. Czern), and wasabi (Wasabia japonica), as well as the model plant Arabidopsis thaliana. In addition, GSLs have also been identified in the genus Drypetes of the family Euphorbiaceae (Fahey et al. 2001). GSLs share an identical core structure containing a ß-thioglucose group linked to a sulfonated aldoxime moiety, plus a variable aglycone side chain (R) derived from one of eight amino acids (Halkier and Du 1997). Based on the amino acid precursors and the type of modification to the R group, GSLs can be divided into three major classes: aliphatic, indole, and aromatic GSLs (Halkier and Gershenzon 2006). Aliphatic GSLs are derived from alanine, leucine, isoleucine, valine, and methionine, while indole GSLs and aromatic GSLs are derived from tryptophan and phenylalanine or tyrosine, respectively. By 2000, at least 120 different GSLs were reported in 16 families of the order Capparales, and the Brassicaceae family alone was found to contain at least 96 of these. A more recent review elucidated and documented the discovery of additional natural GSL structures, citing around 132 unique GSLs from nature (Agerbirk and Olsen 2012). The structural diversity of these compounds is mainly caused by extensive modification of the variable side chain by elongation of the amino acid precursors and from a wide variety of side-chain modifications, including hydroxylation, oxidation, methylation, glucosylation, desaturation, and sulfation (Halkier and Gershenzon 2006).
Though most GSLs are not bioactive in their intact form, they are rapidly hydrolyzed by an endogenous family of plant enzymes called myrosinases (thioglucoside glucohydrolases (TGGs); EC 3.2.1.147), β-glucosidases that are compartmentalized in the vacuoles of myrosin cells, a location separate from that of GSLs. Once plant tissues are damaged by wounding, herbivore or pathogen attack, freezing, or grazing (Bones and Rossiter 2006; Fahey et al. 2001), the myrosinases are mixed with GSLs, resulting in hydrolysis of the thioglycoside bond to yield glucose and an unstable aglucone. The latter compound is either spontaneously rearranged into bioactive isothiocyanate or is converted into alternative hydrolysis products such as simple nitriles, epithionitriles, or organic thiocyanates (Wittstock and Burow 2010). The types of breakdown products of the GSL–myrosinase system depend mainly on the chemical nature of the side chain of the parent GSL, the reaction conditions, and the cofactors that are present (Fahey et al. 2001; Halkier and Gershenzon 2006).
GSLs and their degradation products have been recognized for their roles in plant defense and their distinctive effects on human health and on the flavor of cruciferous vegetables. Glucoraphanin (4-methylsulfinylbutyl, GRA), which is known to reduce the risk of aggressive prostate cancer (Halkier and Gershenzon 2006), is the most widely studied GSL. Despite the importance of certain GSLs and their metabolites to human health, most GSLs are also undesirable substances in Brassica crops for animal feed, due to the deleterious effects of their breakdown products on animal growth and reproductive performance. To reduce the levels of GSLs in Brassica crops, oilseed rape breeders have devoted much effort to developing genetically improved varieties with lower amounts of GSLs. Significant progress has been made toward this goal through classical breeding approaches, and several varieties with low levels of seed GSLs (less than 30 μmol/g in defatted meal) and erucic acid (less than 2% of the total fatty acids present in the oil) have been released in Canada and marketed under the name “canola.” While processed canola meal has been widely accepted in the feed industry as a high-quality feedstuff for livestock and poultry, a number of reports have documented reduced performance in farm animals fed diets containing significant amounts of canola meal (Khajali and Slominski 2012; Leeson et al. 1987).
The completion of genome sequencing of B. napus and its parental species B. rapa and B. oleracea provided the Brassica scientific community with a valuable tool for further improving seed quality through regulating and controlling secondary metabolism pathways (Chalhoub et al. 2014; Liu et al. 2014; Parkin et al. 2014; Wang et al. 2011b). In Arabidopsis, the close relative of Brassica crops, most genes responsible for GSL biosynthesis, breakdown, and transport have been characterized using biochemical and reverse genetics approaches (Halkier and Gershenzon 2006). Based on this research in Arabidopsis, orthologous genes involved in GSL metabolism and transport in Brassica crops have been identified, allowing for the manipulation of these genes and the development of Brassica vegetables with high levels of anticancer GSLs and B. napus varieties containing much lower levels of undesirable GSLs.
This chapter presents an overview of the genes responsible for GSL biosynthesis, transport, and breakdown in Brassica crops, with special emphasis on elucidating the evolutionary processes that resulted in the variation in GSL profiles of Brassica crops. Based on this information, we present perspectives for further research aimed at modifying and reducing different kinds of GSLs in B. napus.
12.2 Variation in GSL Composition and Content in Brassica Crops
The chemical composition of many Brassicaceae genera has been studied, with a focus on identifying variations in oil content and seed fatty acid and GSL composition (Warwick 2011). Comparative studies of GSL profiles indicate that the type of GSLs present and their concentrations vary considerably between species in the Brassicaceae family, as well as between cultivars of the same species, and within different organs or developmental stages of the same plant (Daxenbichler et al. 1991; Fahey et al. 2001). In addition, the total GSL content and the relative proportion of individual GSLs are also influenced by the genotype and by agronomic and environmental factors (such as growth stage, harvest time, soil moisture, and temperature) (Gu et al. 2012; Padilla et al. 2007; Wang et al. 2012; Yang and Quiros 2010).
Numerous studies have described the GSL contents and composition in representative Brassica species, and these data have been compiled in several reviews (Daxenbichler et al. 1991; Fahey et al. 2001; Ishida et al. 2014; Jeffery et al. 2003; Padilla et al. 2007). As many as 20 kinds of GSLs have been identified in commercial Brassica crops (Table 12.1), which possess substantially different GSL profiles, and usually only 3 or 4 predominant kinds of GSLs occur in the same plant (Rosa 1997). Comparison of the GSL profiles and concentrations in different tissues during different growth stages from four Brassica crops of the “triangle of U” (Brassica carinata, Brassica nigra, B. juncea, and B. rapa) revealed that sinigrin (2-propenyl, SIN) is the dominant GSL in three mustards (B. carinata, B. nigra, and B. juncea) (Table 12.2), where it represents more than 90% of the total GSL concentration in ripe seeds and over 50% of the total GSL concentration in green tissues (Bellostas et al. 2007). B. carinata contains other GSLs, including gluconapin (3-butenyl, NAP), 4-hydroxyglucobrassicin (4-hydroxyindol-3-ylmethyl, 4HGBS), gluconasturtiin (2-phenylethyl, GST), and progoitrin (2-hydroxy-3-butenyl, PRO), the last of which is ultimately decomposed into oxazolidine-2-thiones, which are considered to be goitrogenic compounds in monogastric animals (Bellostas et al. 2007; Fahey et al. 2001). The GSL profile of B. rapa is quite distinct from that of the aforementioned three mustards (Table 12.2). In B. rapa, 16 GSLs have been identified. Among these, the aliphatic GSLs, NAP, and glucobrassicanapin (4-pentenyl, GBN), and their hydroxylated forms, PRO and gluconapoleiferin (2-hydroxy-4-pentenyl, GNL), were found to be the most abundant, while the concentrations of indolic and aromatic GSLs were low and showed the fewest differences among the different varieties (Cartea and Velasco 2008). Most B. rapa varieties had a proportion of NAP of between 70 and 95% of the total GSL content and a proportion of GBN of below 20% of the total GSL content, while other minor GSLs, such as glucoiberin (3-methylsulfinilpropyl, GIV), PRO, glucoalyssin (5-methylsulphinylpentyl, GAL), and GST, accounted for less than 20% of the total GSL content (Padilla et al. 2007).
Diversity in the concentration and type of GSLs is much higher in B. oleracea than in B. rapa species (Ishida et al. 2014). All B. oleracea types and cultivars contain high concentrations of glucobrassicin (3-indolymethyl, GBS) and GIV and most contain substantial amounts of SIN. For example, SIN accounts for most of the GSLs in kale (B. oleracea var. acephala), while GBS and GIB account for most of those in cabbage (B. oleracea var. capitata) leaves (Cartea et al. 2008). The most common GSLs found in broccoli (B. oleracea var. italica) are GRA, SIN, PRO, NAP, and the indole GSLs GBS and neoglucobrassicin (1-methoxy-3-indolylmethyl, NGBS) (Kushad et al. 1999). The predominant GSL GRA (accounting for more than 50% of the total GSLs and the precursor of sulforaphane) is the most important health-promoting compound in broccoli, but only trace amounts of GRA are present in most B. rapa, B. napus, and B. juncea vegetables and oilseeds (Liu et al. 2012; Tian et al. 2005). GRA was not detected in several B. oleracea crops, including cabbage, Brussels sprouts (B. oleracea var. gemmifera), and cauliflower (B. oleracea var. botrytis). In cauliflower, SIN and GIB are the major aliphatic GSLs present (together occurring at a concentration of 0.42 μmol/g FW), and GBS (1.5 μmol/g FW) and 4-methoxyglucobrassicin (4-methoxy-3-indolylmethyl, 4MGBS, 0.4 μmol/g FW) are the major indole GSLs (Tian et al. 2005). Broccoli sprouts and Brussels sprouts contain higher amounts of total GSLs than do broccoli and cauliflower. The major GSLs detected in broccoli sprouts are 4MBGS, GRA, GER, and GIB (0.385, 1.33, 1.02, and 0.599 μmol/g FW, respectively; Tian et al. 2005; West et al. 2002). GBS (3.74 μmol/g FW) is the most abundant GSL in Brussels sprouts, while the concentration of SIN, PRO, and NAP (1.55, 1.33, and 1.08 μmol/g FW, respectively) is also relatively higher than that of other GSLs (Tian et al. 2005). In Chinese kale (B. oleracea var. alboglabra), the total and individual GSL contents varied extensively among the different edible parts, and NAP was the most abundant GSL in the edible plant parts (Sun et al. 2011).
Due to their toxic and antinutritive effect on animals, GSLs have long since been regarded as unfavorable components of B. napus seeds. Hence, developing a double-low B. napus variety with seeds lacking erucic acid and containing only low levels of GSL has been an important objective of rapeseed breeding programs, and much research examining variation in GSL composition and content in B. napus has been conducted (Font et al. 2005; Sang et al. 1984). Based on the GSL content, the seeds of 499 B. napus accessions were divided into three types, containing high, medium, and low levels of GSLs, and the GSL components of each of these types were systemically analyzed by high-performance liquid chromatography (Li et al. 2005). In B. napus varieties containing high and medium levels of GSLs, but not in those containing low levels, the dominant and stable components are PRO and NAP. Although GST and 4HGBS are minor components of B. napus varieties containing high levels of GSL, they are major components of varieties containing low GSL levels (Li et al. 2005). Accurately measuring the GSL profiles and identifying the corresponding GSL biosynthetic, breakdown, and transport genes in different Brassica crops are of great importance for further improving the GSL profiles in given tissues and organs. For instance, ideal B. napus varieties would have high levels of GSLs in the vegetative tissues, but lack GSLs in the seeds.
12.3 Genes Involved in the Metabolism, Transport, and Regulation of GSLs
Substantial advances have recently been made in our understanding of the metabolism and regulation of GSLs in plants, particularly in Arabidopsis, where structural and regulatory genes involved in GSL biosynthesis, transport, and degradation pathways have been identified through in vitro biochemical assays and mutant studies (Burow et al. 2010; Radojčić Redovniković et al. 2008; Sønderby et al. 2010).
12.3.1 GSL Biosynthetic Genes
GSL biosynthesis is comprised of three independent stages: (i) amino acid chain elongation, in which additional methylene groups are inserted into the side chain of certain aliphatic and aromatic amino acids, (ii) conversion of the amino acid moiety to form the core structure of GSLs, and (iii) subsequent secondary modifications of side chains to generate chemical diversity (Grubb and Abel 2006; Halkier and Gershenzon 2006). Methionine undergoes a series of chain elongation cycles in which one methylene group is added per time prior to entering the core structure pathway. These chain elongation reactions include deamination by a branched-chain amino acid aminotransferase (BCAT), condensation with acetyl-CoA by a methylthioalkylmalate synthase (MAM), isomerization by an isopropylmalate isomerase (IPMI), and oxidative decarboxylation by an isopropylmalate dehydrogenase (IPM-DH) (Sønderby et al. 2010). The newly formed 2-oxo acid can either be transformed into the corresponding methionine derivative and enter the core GSL structure pathway or undergo another round of chain elongation (Radojčić Redovniković et al. 2008). In A. thaliana, three tandemly duplicated and functionally diverse MAM members were identified as being responsible for the condensation step of the chain elongation. Functional analysis demonstrated that AtMAM2 (absent in ecotype Columbia) and AtMAM1 catalyze the condensation reaction of the first and the first two elongation cycles, respectively, for the synthesis of aliphatic GSLs with short carbon chains (3C and 4C, respectively) (Benderoth et al. 2006; Kroymann et al. 2003; Textor et al. 2004), while AtMAM3 catalyzes all six additions of methylene groups and the formation of all aliphatic GSLs, especially long-chain GSLs (6C, 7C, and 8C) (Textor et al. 2007). Hence, the number and expression patterns of MAM genes in a plant determine variations in aliphatic GSLs during the earliest stages of GSL biosynthesis and have a fundamental impact on GSL composition and diversity in plant tissues.
The GSL core structure is formed from precursor amino acids via a series of reactions catalyzed by various cytochrome P450 (CYP) monooxygenases (Halkier and Gershenzon 2006). Briefly, the five characterized CYP79 homologs in Arabidopsis catalyze the conversion of amino acids to their corresponding aldoximes. CYP79F1 and CYP79F2 encode the enzymes that catalyze aldoxime production in the biosynthesis of the major GSLs derived from chain-elongated methionine derivatives. CYP79B2 and CYP79B3 catalyze the biosynthesis of indole-3-acetaldoxime from tryptophan, whereas CYP79A2 converts phenylalanine to phenylacetaldoxime, the precursor of benzyl GSL (Radojčić Redovniković et al. 2008). Biochemical studies identified differences in the substrate specificity of CYP79F1 and CYP79F2, showing that CYP79F1 metabolizes homomethionine and di-, tri-, tetra-, penta-, and hexahomomethionines, resulting in both short- and long-chain methionine derivatives, whereas CYP79F2 only catalyzes the production of long-chain penta- and hexahomomethionines (Chen et al. 2003; Radojčić Redovniković et al. 2008). The aldoximes are further metabolized to form S-alkylthiohydroximates by CYP83A1 and CYP83B1, cytochrome P450 of the CYP83 family (Bak and Feyereisen 2001). Both biochemical and transgenic lines of evidence show that CYP83A1 mainly metabolizes the aliphatic aldoximes to form aliphatic GSLs, whereas CYP83B1 mostly metabolizes indole-3-acetaldoxime and aromatic oximes to synthesize the corresponding substrates for indolic and aromatic GSLs, respectively (Bak and Feyereisen 2001; Naur et al. 2003). In a subsequent step, the resulting S-alkylthiohydroximates are cleaved to yield thihydroximates by a C-S lyase SUR1 (Mikkelsen et al. 2004). The second to last step in the formation of GSLs is the S-glycosylation of thihydroximates, a reaction that is catalyzed by glucosyltransferases of the UGT74 family. This reaction appears to be unique and catalyzes the formation of an S-glycosidic bond between glucose and the acceptor thiohydroximate, leading to the production of the corresponding desulfo-GSL (Grubb et al. 2004). The results of biochemical and genetic analyses demonstrated that UGT74C1 plays a key role in the biosynthesis of aliphatic GSLs and that UGT74B1 catalyzes the formation of aromatic GSLs (Grubb et al. 2004, 2014). Three sulfotransferase (SOT) proteins perform the final step of GSL biosynthesis. Biochemical characterization showed that SOT16 metabolizes tryptophan- and phenylalanine-derived desulfo-GSLs, whereas SOT17 and SOT18 metabolize long-chained aliphatic desulfo-GSLs (Piotrowski et al. 2004).
After parent GSL formation, a wide range of further modifications can occur on the methionine side chain and occasionally on the glucose moiety (Mikkelsen et al. 2002; Neal et al. 2010), giving rise to an enormous variety of GSL structures. These secondary modifications, which take place in an organ- and developmental stage-specific manner (Radojčić Redovniković et al. 2008; Sønderby et al. 2010), are particularly important as the structure of the side chain largely determines the nature of the products formed following GSL hydrolysis by myrosinases (Sønderby et al. 2010; Wittstock and Halkier 2002). For aliphatic GSLs, these modifications include oxidations, hydroxylations, alkenylations, and benzoylations, while for indole GSLs, they include hydroxylations and methoxylations.
The S-oxygenation of aliphatic GSLs is a common modification catalyzed by five flavin-monooxygenases, designated FMOGS-OX1 to FMOGS-OX5 (Li et al. 2008). FMOGS-OX5 shows substrate specificity for the long-chain 8-methylthiooctyl GSLs (8MTOs), whereas FMOGS-OX1 to FMOGS-OX4 exhibit broad chain length specificity and catalyze the conversion from methylthioalkyl (MT) GSL to the corresponding methylsulfinylalkyl (MS) independently of chain length (Li et al. 2008), resulting in the production of the potent cancer-preventive substances sulforaphane (4-methylsulfinylbutyl isothiocyanate, 4MSB ITC), which is derived from GRA, and the 7-methylsulfinylheptyl (7MSOH) and 8-methylsulfinyloctyl (8MSOO) isothiocyanates, derived from 7-methylthioheptyl GSL (7MTH) and 8MTO, respectively (Li et al. 2008). Hence, the five FMOGS-OX genes could potentially be used in genetic engineering strategies to optimize the GSL profiles of Brassica crops. Substantial variation in Arabidopsis GSL profiles between different genotypes has expedited the identification of the GS-AOP locus, which encodes the two tandemly duplicated 2-oxoglutarate-dependent dioxygenases, AOP2 and AOP3 (Kliebenstein et al. 2001). AOP2 directly catalyzes the conversion of methylsulfinylalkyl GSLs to the alkenyl GSLs NAP or GBN (n = 2–3), and the GS-OH locus can further convert NAP to PRO (Hansen et al. 2008). AOP3 controls the production of hydroxyalkyl GSLs (n = 2) from methylsulfinylalkyl GSLs. When both AOPs are non-functional, the plant accumulates the precursor methylsulfinyl alkyl GSLs (Liu et al. 2014). Secondary modifications of indole GSLs mainly include hydroxylation by CYP81F2, which is essential for the 4-hydroxylation of unmodified indolyl-3-methyl (I3M), and catalyzes the formation of 4-hydroxy I3M (4OH-I3M) and 4-methoxy I3M (4M-I3M) from I3M (Bednarek et al. 2009; Pfalz et al. 2009; Sønderby et al. 2010).
12.3.2 Regulatory Genes of GSL Biosynthesis
Biosynthesis of GSLs is tightly regulated by six R2R3-MYB transcription factors (TFs) belonging to subgroup 12 of the R2R3 MYB family, which has a conserved “[L/F]LN[K/R]VA” motif (Dubos et al. 2010). In Arabidopsis, MYB28, MYB29, and MYB76 positively regulate the biosynthesis of aliphatic GSLs with partial functional redundancy (Hirai et al. 2007). During aliphatic GSL biosynthesis, AtMYB28 acts as the major positive regulator and AtMYB29 as an accessory factor in the response to methyl jasmonate signaling in the trans-activation of the aforementioned aliphatic GSL biosynthetic genes, i.e., AtMAM1, AtMAM3, AtCYP79F1, AtCYP79F2, AtCYP83A1, AtAOP2, AtSOT17, and AtSOT18 (Gigolashvili et al. 2008a; Hirai et al. 2007). Arabidopsis mutants defective in MYB28 function had decreased amounts of both long- and short-chain aliphatic GSLs, whereas the myb29 or myb76 mutant contained significantly reduced levels of short-chained aliphatic GSLs, indicating that MYB28 regulates the biosynthesis of all methylsulfinyl GSLs, whereas MYB29 and MYB76 regulate the biosynthesis of short-chained GSLs (Gigolashvili et al. 2008b). The total aliphatic GSLs but not indolic GSLs were significantly increased in the leaves of plants overexpressing AtMYB28, AtMYB29, or AtMYB76 (Gigolashvili et al. 2008b; Hirai et al. 2007). Overexpression of both AtMYB28 and AtMYB29 significantly repressed the expression of the indolic GSL pathway genes, indicating that a reciprocal antagonistic relationship exists between the aliphatic and indolic GSL biosynthetic pathways (Gigolashvili et al. 2008a).
Conversely, AtMYB34, AtMYB51, and AtMYB122, which were identified as important regulators of the indolic GSL biosynthetic pathway, significantly reduced the transcript levels of AtCYP79B2, AtCYP79B3, AtCYP83B1, AtUTG74B1, AtSOT16, and 3’-phosphoadenosine 5’-phosphosulphate transporter (PAPST1) genes, which are involved in the indolic GSL biosynthetic pathway (Frerigmann and Gigolashvili 2014; Guo et al. 2013; Sønderby et al. 2010). The three MYB transcription factors exhibit both additive and epistatic interactions in the regulation of indolic GSL biosynthesis (Frerigmann and Gigolashvili 2014). Lines lacking the two main regulators of indolic GSL biosynthesis, MYB34 and MYB51, exhibit a significant reduction in total indolic GSLs, demonstrating the importance of these two genes for indolic GSL biosynthesis. Previous research also showed that MYB34 and MYB51 have distinct roles in indolic GSL production, functioning in different tissues or under different environmental conditions. MYB51 is the central regulator of indolic GSL biosynthesis in shoots and is activated by salicylic acid (SA) and ethylene (ET) treatments. By contrast, MYB34 regulates indolic GSL biosynthesis mainly in the roots and functions in abscisic acid (ABA) and methyl jasmonate (MeJA) signaling. Interestingly, MYB51 appears to regulate indolic GSL biosynthesis in roots in the myb34 mutant. MYB122 only plays an accessory role in indolic GSL biosynthesis and in JA/ET-induced GSL biosynthesis (Frerigmann and Gigolashvili 2014).
In addition to the MYB transcription factors, some other regulators of GSL biosynthesis have also been characterized. Arabidopsis CaM-binding protein IQ-DOMAIN1 (IQD1) binds calmodulin in a Ca2+-dependent manner and is a positive regulator of total GSL accumulation during biotic stress responses, with a gain-of-function IQD1 mutation resulting in elevated levels of both indole and aliphatic GSLs and a reduction in insect herbivory and infestation (Laluk et al. 2012; Levy et al. 2005). Another CaM-binding transcription factor SIGNAL RESPONSIVE1 (AtSR1) also proved to be a key regulator of GSL levels through transcriptional regulation of several genes involved in GSL metabolism, including AtIQD1, AtMYB51, and AtSOT16, and is a negative regulator for herbivory tolerance in Arabidopsis (Laluk et al. 2012). AtSLIM1 was identified as a central transcription factor that negatively regulates both aliphatic and indolic biosynthesis under sulfur-limiting conditions and downregulates AtMBY34 transcription (Maruyama-Nakashita et al. 2006). Another characterized regulator of GLS biosynthesis is DNA-binding-with-one-finger (DOF) transcription factor AtDof1.1 (also known as AtOBP2), which is induced by wounding and herbivore attack and MeJA treatment, and specifically upregulates CYP83B1 expression and promotes indolic GSL accumulation (Skirycz et al. 2006). Although AtDof1.1 does not seem to regulate the expression of CYP79F1 and CYP79F2, the aliphatic GSL content was altered in AtDof1.1 overexpression lines (Skirycz et al. 2006). Loss-of-function mutations of Arabidopsis TERMINAL FLOWER2 (TFL2, also known as LHP1 or TU8) significantly increased the abundance of four long-chain aliphatic GSLs in the seeds, whereas indolyl-3-methyl GSL levels were significantly reduced relative to the wild type, leading to a reduction in symptoms resulting from infection by the obligate biotrophic fungus Plasmodiophora brassicae, which causes clubroot disease, a damaging disease in Brassicaceae (Kim et al. 2004; Le Roux et al. 2014). In addition, TFL2 regulates heterochromatin formation and represses the expression of genes involved in flowering time, floral organ identity, meiosis, and seed maturation (Nakahigashi et al. 2005).
12.3.3 GSL Transport Genes
The GSLs are believed to be synthesized mainly in rosette leaves and silique walls and then to be relocated to embryos through phloem by specific transporters (Lu et al. 2014). In Arabidopsis, GSLs have successfully been eliminated from the seeds by silencing two recently identified nitrate/peptide transporter family members, GTR1 and GTR2, which suggests that manipulation of these two transporters may increase the nutritional value of crops and be used in biotechnological approaches to control the allocation of GSLs to seeds in Brassica crops (Nour-Eldin et al. 2012). The gtr2 single mutant exhibited a significant reduction in total GSL levels in seeds and a threefold increase in aliphatic GSLs in source tissues (i.e., senescent leaves and silique walls), but no significant changes in GSL content in the seeds (Jorgensen et al. 2015; Nour-Eldin et al. 2012). In the gtr1 gtr2 double mutant, aliphatic and indolic GSLs were absent in the seeds, but exhibited a more than tenfold increase in source tissues, demonstrating that both plasma membrane-localized transporters are essential for long-distance GSL transport to the seeds and are responsible for loading GSLs from the apoplasm into the phloem, and finally for determining the tissue-specific distribution of GSLs in plants (Nour-Eldin et al. 2012). Identifying these two GSL transporters provides a strategy for breeding Brassica varieties that contain extremely low levels of total GSLs in the seeds but high levels in the green tissues by reducing functional GTR activity and blocking the translocation of GSLs.
12.3.4 GSL Breakdown Genes
Numerous studies to date have focused on the beneficial effects of GSLs and their breakdown products on human health and plant defense, and on their negative effects on animal nutrition. In the well-studied GSL–myrosinase-specifier protein system, myrosinases hydrolyse GSLs in the presence of water, producing a series of degradation products (Wittstock and Burow 2010). The types of products of myrosinase hydrolysis depend on the structure of the parent GSLs, reaction conditions, and availability of epithiospecifier proteins (ESPs) and nitrile-specifier proteins (NSPs) (Kissen and Bones 2009).
In Arabidopsis, six genes (TGG1-TGG6) encoding classical myrosinases have been identified on two chromosomes (Xu et al. 2004). Among these genes, TGG1 and TGG2 were tandem duplicates of TGG3, while TGG5 and TGG6 were tandem duplicates of TGG4. These duplicated genes share the same gene structure as their parent genes. Although TGG3 and TGG6 are predominantly expressed in specific tissues (Xu et al. 2004), both are probably pseudogenes that encode non-functional proteins due to multiple frameshift mutations (Wang et al. 2009). TGG1 is expressed in myrosin cells, stomatal guard cells, and phloem cells of all the aboveground organs except the seeds (Barth and Jander 2006; Xue et al. 1995). Similar to TGG1, TGG2 is also highly expressed in the aboveground tissues (Xu et al. 2004), but is much less abundant in the rosette leaves than is TGG1, and was not detected in guard cells (Zhao et al. 2008). TGG4 and TGG5 are primarily expressed in the roots. Despite the distinct expression patterns and the difference in vitro myrosinase activities of TGG1 and TGG2, GSL breakdown in the crushed leaves of TGG1 or TGG2 single mutants is basically unchanged, indicating that the two myrosinases may have redundant functions (Barth and Jander 2006). Leaf extracts of TGG1 TGG2 double mutants had no detectable in vitro myrosinase activity on exogenously applied aliphatic GSLs, and endogenous aliphatic GSLs were no longer broken down in disrupted leaf material of the double mutant (Barth and Jander 2006). However, myrosinase-independent breakdown of indolic GSLs still slowly proceeds, indicating the presence of a breakdown pathway for these GSLs that is independent of TGG1 and TGG2.
Several specifier proteins, such as ESPs and NSPs, myrosinase-associated proteins (MyAPs), such as EPITHIOSPECIFIER-MODIFIER1 (ESM1), MODIFIED VACUOLE PHENOTYPE1 (MVP1), and enzymes involved in further metabolism, such as nitrilases, have been shown to be involved in the generation of diversified GSL metabolic products in Arabidopsis (Wittstock and Burow 2010). Specifier proteins do not exhibit hydrolytic activity on GSLs, but affect the outcome of GSL hydrolysis products. In the absence of specifier proteins, ITCs are typically formed at neutral pH (Bones and Rossiter 2006). ESPs and the related thiocyanate-forming proteins (TFPs) catalyze the formation of epithionitrile, in the presence of GSLs with terminal double bonds in the side chain and ferrous ions, while the formation of thiocyanate purely depends on TFPs (Wittstock and Burow 2010). NSPs are involved in simple nitrile formation at acidic pH values, but do not catalyze epithionitrile or thiocyanate formation. The simple nitrile can be further converted by nitrilases (NITs) to a carboxylic acid in the presence of a specifier protein (Vorwerk et al. 2001; Wittstock and Burow 2010). ESP function is inhibited by ESM1, leading to decreased simple nitrile formation and increased ITC production for benzyl and alkyl GSLs, but not for alkenyl GSLs (Zhang et al. 2006). Cloning and sequence analysis of ESM1 revealed that it encodes a putative endoplasmic reticulum (ER) binding protein and that allelic variation in this gene contributes to the variation in GSL breakdown among different Arabidopsis accessions (Zhang et al. 2006). MVP1 is expressed ubiquitously and encodes another MyAP-like protein that is closely related to ESM1. The mvp1 mutant is impaired in endomembrane protein trafficking and shows a significant increase in simple nitrile production from allyl GSLs (Agee et al. 2010). Interestingly, MVP1 interacts with TGG2 and the PYK10 complex, but not with TGG1 in vitro, suggesting that MVP1 functions in the quality control of GSL hydrolysis by contributing to the proper tonoplast localization of TGG2 and in ER body-related defense systems by regulating the PYK10 complex (Agee et al. 2010; Nakano et al. 2012). An atypical myrosinase gene, PEN2, which may be limited to indole GSL hydrolysis and is required for pathogen resistance, was recently identified in Arabidopsis (Bednarek and Osbourn 2009).
12.4 Evolution of GSL-Related Genes in B. Napus and Its Parental Species
12.4.1 Identification of GSL-Related Genes from B. Napus and Its Parental Species
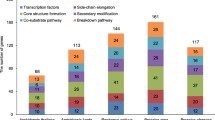
To identify GSL-related genes from B. napus and its parental species B. rapa and B. oleracea, we used the sequences of 58 GSL biosynthesis, 3 GSL transport, and 17 GSL breakdown genes characterized in A. thaliana as queries against the four publicly available genomes of Brassica crops based on a combination of syntenic and nonsyntenic homology analyses (Table 12.3). We identified 119, 119, 134, and 240 GSL biosynthetic genes in B. rapa, B. oleracea var. capitata, B. oleracea var. italica, and B. napus (both 120 genes in A and C subgenomes), respectively (Fig. 12.1). The fact that more GSL biosynthetic genes were identified in B. oleracea var. italica than in the other three Brassica crops is mainly a consequence of the expansion of genes responsible for core structure formation and side-chain modification. For three Arabidopsis GSL transporters, there are 8 orthologs in both B. rapa and two subgenomes of B. napus, while only 7 and 6 orthologs exist in B. oleracea var. capitata and B. oleracea var. italica, respectively. The number of GSL breakdown genes is almost identical among B. rapa, B. oleracea var. capitata, and two subgenomes of B. napus, while B. oleracea var. italica contains many more.

(Figure reprinted, with modifications, from Liu et al. (2014) under a CC BY license (Creative Commons Attribution 4.0 International License))
Comparison of aliphatic and indolic glucosinolate biosynthetic and breakdown genes in A. thaliana, B. rapa, B. oleracea var. capitata, and B. napus. The copy number of GSL biosynthetic genes in A. thaliana, B. rapa, B. oleracea var. capitata and B. napus is listed in square brackets. Potential anticancer substances/precursors are highlighted in blue bold. The most important transcription factors, amino acid chain elongation and side-chain modification loci MYB28 (HAG1), MAMs, and AOP2, are highlighted in red bold, with the number in parentheses (green) representing the number of non-functional genes. 1MOI3 M: 1-methoxyindol-3-ylmethyl GSL; 1OHI3 M: 1-hydroxyindol-3-ylmethyl GSL; 3 MSOP: 3-methylsulfinylpropyl GSL; 3 MTP: 3-methylthiopropyl GSL; 3PREY: 2-Propenyl GSL; 4BTEY: 3-butenyl GSL; 4BzOB: 4-benzoyloxybutyl GSL; 4MOI3 M: 4-methoxyindol-3-ylmethyl GSL; 4OHB, 4-hydroxybutyl GSL; 4OHI3M: 4-hydroxyindol-3-ylmethyl GSL; 4MSOB: 4-methylsulfinylbutyl GSL; 4MTB, 4-methylthiobutyl GSL; AITC: allyl isothiocyanate; DIM: 3,3’-diindolymethane; ESP: epithiospecifier protein; I3C: indole-3-carbinol; IAA: indole-3-acetaldehyde; IAN: indole-3-acetonitile; I3M: indolyl-3-methyl GSL; NSP: nitrile-specifier protein; TFP: thiocyanate-forming protein; and TGG: thioglucoside glucohydrolase
After the split with Arabidopsis, the Brassica progenitor species experienced a whole-genome triplication (WGT) and subsequently diverged into three diploid Brassica species, B. rapa, B. oleracea, and B. nigra. As a young allopolyploid species, B. napus was formed from multiple independent hybridization events between ancestors of the diploids B. rapa (A genome donor) and B. oleracea (C genome donor) (Nagaharu 1935). Hence, we found that most multi-copy genes might have originated from WGT events and that several gene families involved in GSL metabolism or transport also experienced homeologous gene loss events after the WGT, leading to the formation of 13 conserved single-copy GSL biosynthesis genes and single copies of GSL transport (PEN3) and breakdown (PEN2) genes in B. rapa, B. oleracea, and two subgenomes of B. napus. The 78 GSL-related genes present in Arabidopsis represent 0.28% of all Arabidopsis genes, while the GSL-related genes in B. rapa, B. oleracea var. capitata, B. oleracea var. italica, and B. napus represent 0.40, 0.36, 0.33, and 0.33% of all predicted genes in the corresponding species, indicating that the expansion levels and total numbers of GSL-related genes in Brassica crops are similar to the whole-genome gene expansion levels of the corresponding crops (P-value > 0.05).
To reveal the retention status of the GSL-related genes after the WGT, we determined the ratio of single- to multi-copy paralogous genes involved in various steps of GSL metabolism (Table 12.4). The proportion of total paralogous sets with different copy numbers over the whole genome was used as background, and we found that the expansion levels of transcription factors, side-chain modification, and breakdown genes in B. rapa were significantly higher than those of their backgrounds (P < 0.05). The same trends were observed for GSL breakdown genes in two B. oleracea genomes and for transcription factors in B. oleracea var. italica, indicating that a specific subset of GSL-related genes was retained in B. oleracea. Over-retention of GSL transcription factors occurred in the C subgenome of B. napus, while those associated with side-chain modification and breakdown were only over-retained in the A subgenome of B. napus. It seems that the GSL-related genes responsible for chain elongation, core structure formation, co-substrate pathways, and transport did not experience significant expansion, since they showed no significant difference from the background (Table 12.4). However, the GSL-related genes were significantly retained in all four studied Brassica crops, since the ratio of single- to multi-copy paralogous genes was significantly smaller than the background (P-value < 0.05), suggesting that GSL-related genes expanded in B. rapa and B. oleracea and were retained in the two subgenomes of B. napus. Tandem duplication (TD) also contributed greatly to the evolution of GSL-related genes in both Arabidopsis and Brassica species. We identified 11 TD events in Arabidopsis GSL-related genes, including 8 and 3 events associated with GSL biosynthesis and breakdown, respectively. We found that 21 pairs of paralogous genes had undergone more recent TD events after WGT in two B. oleracea crops and two subgenomes of B. napus. For example, SOT18 consists of 10 copies in B. rapa, B. oleracea var. capitata, and the C subgenome of B. napus, and 9 and 8 copies in B. oleracea var. italica and A subgenome of B. napus, respectively. At least six SOT18 genes originated from three TD events in all of these Brassica species, implying that these ancient TD events might have occurred after the Arabidopsis–Brassica split and before divergence of B. rapa and B. oleracea.
Similar to the findings of a previous study in B. rapa (Wang et al. 2011a), we found that a total of 11 GSL-related genes in Arabidopsis have no orthologs in the studied Brassica genomes, including a transcription factor (MYB76), two amino acid side-chain elongation genes (IPMDH3 and IPMI SSU3), one core structure formation gene (CYP79F2) for long-chain aliphatic GSL, four side-chain modification genes (FMOGS-OX1, FMOGS-OX3, FMOGS-OX4 and AOP3), and three GSL breakdown genes (NSP3, NIT1, and NIT3). It seems that the loss of these genes is not indispensable for GSL biosynthesis and breakdown, as paralogs with similar functions are present in the Brassica species.
12.4.2 Evolution of GSL Biosynthesis Genes Influencing Variation in GSL Profiles in B. napus and Its Parental Species
To date, more than 20 kinds of GSLs have been identified in commercial Brassica crops. The diversity of GSL types and variation in GSL profiles in these Brassica species are largely due to the evolution of GSL-related genes. In our study, we mainly focused on the evolution of MAM and AOP gene families in the four Brassica crops.
The MAM genes encode methylthioalkylmalate synthase, which is involved in amino acid chain elongation, and gave rise to GSLs with diverse chain lengths during the biosynthesis of methionine-derived GSLs (Zhang et al. 2015a, b, b). The phylogenetic and synteny relationships of MAM genes from 13 sequenced Brassicaceae species indicated that the MAM genes taken two independent lineage-specific evolution routes after the divergence from Aethionema arabicum. In the lineage I species such as A. thaliana, the MAM loci evolved three tandem genes encoding enzymes responsible for the biosynthesis of aliphatic GSLs with different carbon chain lengths, while in lineage II species such as Brassica crops, the MAM loci encode enzymes responsible for the biosynthesis of short-chain aliphatic GSLs (Zhang et al. 2015). In Arabidopsis, the MAM family contains three tandemly duplicated and functionally diverse members, MAM1, MAM2, and MAM3 (MAM-L). Functional analysis demonstrated that MAM2 and MAM1 catalyze the condensation of the first and the first two elongation cycles for the synthesis of short-chain Met-derived aliphatic GSLs (3C and 4C), respectively, while MAM3 catalyzes the formation of all aliphatic GSLs, especially long-chain GSLs (6C, 7C, and 8C) (Textor et al. 2007).
In B. rapa, B. oleracea var. capitata, and B. oleracea var. italica, MAM1/MAM2 genes experienced independent TD after WGT to produce 6, 7, and 6 orthologs, respectively (Fig. 12.1). Due to gene loss that occurred after the formation of B. napus from the fusion of two parental species, only 5 and 3 orthologs were retained in the A and C subgenomes of B. napus. The greatest diversity of GSL side-chain structures in Brassica is observed within B. oleracea. The main GSLs in this species (i.e., PRO, NAP, GRA, and SIN) are restricted to either 3C or 4C side-chain lengths (Liu et al. 2014). In contrast to the diversity observed in B. oleracea, B. nigra and the amphidiploid B. carinata only have the 3C GSL and SIN, and B. juncea mainly has 3C and 4C GSLs (SIN and NAP). B. rapa and B. napus lack 3C GSLs and predominately possess a mixture of 4C GSLs (NAP and PRO and their hydroxylated homologs), with small amounts 5C GSL GBS. Thus, all of these Brassica species can be considered to have functional alleles at the MAM1/MAM2 loci, while some variation occurred at the MAM3 locus, which led to the existence of 5C GSL in B. rapa and B. napus. Based on our analyses of expression patterns and phylogenetic and syntenic relationships, we identified a pair of genes, Bol017070 and Bra013007, which are the only orthologs with high expression in B. oleracea var. capitata, but are silenced in B. rapa (Liu et al. 2014). Their two descendant orthologs in B. napus, BnaA03g39720D and BnaCnng21190D, both showed weak expression in roots and silenced in siliques simultaneously, implying that Bol017070 might greatly promote the accumulation of the 3C GSL anticancer precursor SIN in B. oleracea. At the MAM3 locus, one orthologous group of genes, Bra008532, Bol040636, BnaA02g36350D, and BnaC02g27590D, showed no expression due to pseudogenization. In another MAM3 orthologous group, expression of Bra018524 is much higher than that of Bol016496, BnaA02g20830D, and BnaC02g26810D. Expression differences of MAM3 genes among Brassica crops most likely resulted in the increased biosynthesis of the 5C GSLs GBN and GNL in B. rapa.
In addition to MAM genes, AOPs are other crucial regulators of variation in aliphatic GSL profiles in Brassicaceae species (Hasan et al. 2008). Previous phylogenetic analyses showed that the core Brassicaceae species have retained AOP1, while AOP2 is retained by most of the lineage II species (excluding Sisymbrium irio and Raphanus sativus), and AOP3 by lineage I species. The variation in AOP2/AOP3 has led to different aliphatic GSL profiles in each lineage (Al-Shehbaz and Al-Shammary 1987). While the function of GSL-AOP1 is currently unknown, AOP2 catalyzes the conversion of methylsulfinylalkyl GSLs (GRA and GIB) to alkenyl GSLs (NAP and SIN), and the GS-OH locus can further convert NAP to PRO. AOP3 is associated with the production of hydroxyalkyl GSL, a compound not found in Brassica crops. When both AOPs are non-functional, the plant accumulates the methylsulfinylalkyl GSL precursor (Liu et al. 2014). Genetic variation at AOP2 is also linked to increased GSL accumulation, since its expression promotes the transcription of most GSL biosynthetic genes and two R2R3 domain MYB transcription factors (MYB28 and MYB29) of the pathway, suggesting that AOP2 plays a role in the positive feedback loop controlling aliphatic GSL biosynthesis (Burow et al. 2015).
Phylogenetic and BLASTN analysis indicated that the genomes of B. rapa, B. oleracea var. capitata, and B. napus possess 3, 3, and 5 orthologs of AOP2 and contain 3, 2, and 7 orthologs of AOP1, respectively (Fig. 12.2). Not all Brassica species have an ortholog of AtAOP3, and such species are unable to produce hydroxyalkyl GSLs. Similar to our results, a natural frameshift mutation resulting from a 2-bp deletion was identified in broccoli, which accumulates GRA by ceasing downstream biosynthesis of other 4C aliphatic GSLs (Li and Quiros 2003). In our previous study, we found that 2 non-functional AOP2 genes contributing to the accumulation of GRA due to the presence of premature stop codons (Liu et al. 2014). Hence, it would be a useful strategy to enhance the GRA concentrations in Brassica crops by blocking the side-chain modification pathway downstream of GRA through silencing of all orthologs of AOP2. Recently, this strategy has been successfully applied in the metabolic engineering for increasing the anticancer compound GRA by suppressing AOP2 gene family in both B. juncea and B. napus (Liu et al. 2012; Augustine and Bisht. 2015). In B. rapa, all three BrAOP2 paralogs have been proved to be active but functionally diverged (Zhang et al. 2015). Expression patterns of five AOP2 genes in B. napus are quite different, BnaA09g01260D and BnaC09g00410D showed the highest expression in siliques, while the rest AOP2 paralogs showed higher expression in flower and stem (Fig. 12.2), implying that these Bna.AOP2 genes might be functional. These results provide insight into the relationship between observed GSL profiles and the evolution of GSL biosynthesis genes and explain why anticancer compound GRA is abundant in B. oleracea, but not in B. rapa and B. napus. The AOP2 genes in B. rapa and B. napus are functional, reflecting the fact that the dominant GSLs are NAP and PRO in both B. rapa and B. napus.
Phylogenetic analysis of three AtAOP genes and orthologs in B. rapa, B. oleracea var. capitata, and B. napus. Full-length sequences of AOP proteins from Arabidopsis, B. rapa, B. oleracea var. capitata, and B. napus were aligned using ClustalW2. The phylogenetic tree (left panel) was constructed using MEGA 6.0 and the neighbor-joining method (1000 bootstrap replicates). Expression levels of Brassica AOP genes were derived from Tong et al. (2013) and Liu et al. (2014) and are presented as the log2-transformed (FPKM + 1) values
12.4.3 Evolution of Major Genes Controlling the Seed GSL Content in B. napus
Quantitative trait locus (QTL) mapping and association mapping (AM) are powerful methods for analyzing the genetic structure of quantitative traits and have been widely used to characterize the total seed GSL contents and profiles in different populations of B. napus (Fu et al. 2015; Hasan et al. 2008; Li et al. 2014; Uzunova et al. 1995). Recently, the orthologs of HAG1 (MYB28), which controls aliphatic GSL biosynthesis in Arabidopsis, were suggested as candidates for major QTLs on A09, C02, C07, and C09 of B. napus. These QTLs were detected independently in different studies using different methods, including conventional QTL mapping, AM, and associative transcriptomic analysis (Li et al. 2014; Lu et al. 2014; Zhao and Meng 2003). Howell et al. (2003) detected four QTLs that together accounted for at least 76% of the phenotypic variation in the accumulation of GSLs in B. napus seeds and revealed that the QTLs on A09, C02, and C09 were homoeologous loci (Howell et al. 2003). Harper et al. (2012) revealed that the HAG1 transcription factor gene family was a candidate in the quantitative control of GSL content of B. napus and that the orthologous genes on C02 and A09 had been lost from the low-GSL accessions (Harper et al. 2012). In our study, we identified three copies of HAG1 genes (BnaA03g40190D, BnaCnng43220D, and BnaC09g05300D) from the genome sequence of the French homozygous B. napus winter line “Darmor-bzh,” which is a double-low B. napus cultivar lacking detectable levels of erucic acid in the seed oil and with a low seed GSL content (Chalhoub et al. 2014). We found that the AtHAG1 orthologs on A09 and C02 were deleted from the double-low B. napus cultivar “Darmor-bzh,” leading to a reduction in seed GSL accumulation. The expression patterns of the three Bna.HAG1 genes were investigated in an elite semi-winter double-low B. napus cultivar “Zhongsuang No. 11,” which is widely cultivated in the Yangtze River region of China (Fig. 12.3). Among the three retained Bna.HAG1 genes, neither BnaA03g40190D nor BnaCnng43220D was expressed in siliques, indicating that the proteins encoded by these two genes probably lost DNA-binding activity for seed GSL accumulation. BnaC09g05300D exhibited the highest transcription levels in the root, followed by the stem and flower, and was expressed at very low levels in the leaf and siliques. Sequence alignment revealed that the BnaC09g05300D coding sequence is only 420 bp long, much shorter than that of AtHAG1 and other members of the HAG1 gene family in Brassica crops, but the intact MYB DNA-binding domain (PF00249) was still predicted to exist in the BnaC09g05300D protein sequence. These data suggest that the Bna.HAG1 gene family experienced not only gene loss due to segment deletion, but also loss of most function in the seeds during the breeding of low-GSL B. napus. In current low-GSL B. napus accessions, BnaC09g05300D, which controls the biosynthesis of aliphatic GSLs, might be the only functional Bna.HAG1 gene. Therefore, it is possible to further reduce the seed GSL content in low-GSL B. napus lines by silencing BnaC09g05300D expression.
Phylogenetic analysis of AtHAG1 and orthologs in B. rapa, B. oleracea var. capitata, and B. napus. Full-length sequences of AtHAG1 (MYB28), AtMYB29, AtMYB76, and three Bra.HAG1, four Bol.HAG1, and two Bna.HAG1 proteins were aligned using ClustalW2. The phylogenetic tree (left panel) was constructed using MEGA 6.0 and the neighbor-joining method (1000 bootstrap replicates). The BnaC09g05300D protein sequence was too short to be excluded in the phylogenetic analysis. Expression levels of Brassica HAG1 genes were derived from Tong et al. (2013) and Liu et al. (2014) and are presented as the log2-transformed (FPKM + 1) values
12.4.4 Evolution of GSL Transport Genes in B. napus
The GSLs are believed to be synthesized mainly in the roots and vegetative tissues and accumulate abundantly in the embryos, where no de novo synthesis occurs (Nour-Eldin and Halkier 2013). Therefore, there must be specific transporters that are responsible for the relocation of GSLs from source tissues to embryos through the phloem. Recently, two members of the nitrate/peptide transporter family in Arabidopsis, GTR1 and GTR2, were identified as high-affinity plasma membrane-localized, GSL-specific proton symporters in a screen of an in vitro library of Arabidopsis transporters (Nour-Eldin et al. 2012). Previous studies suggested that GTR2 is essential for loading GSLs into the phloem, while GTR1 additionally may be involved in distributing GSLs within the leaf. Importantly, GTR1 and GTR2 are essential for the long-distance transport of both aliphatic and indole GSLs to seeds, because the gtr1 gtr2 double mutant had only trace levels of GSLs in seeds and a concomitant increase in rosettes and silique walls (Nour-Eldin et al. 2012). However, it is notable that indole GSLs are transported between rosettes and roots in the absence of GTRs, suggesting the existence of an indole glucosinolate-specific transporter besides GTR1 and GTR2 (Jorgensen et al. 2015).
We identified 32 orthologs of AtGTR in the four Brassica crops we investigated, including 15 GTR1 and 17 GTR2 genes. Phylogenetic analysis and tissue-specific expression detection showed that the transcription levels of most Bna.GTR genes are lower than those of orthologs in the parental species B. rapa or B. oleracea var. capitata (Fig. 12.4). For example, Bra029248 and Bol020699 showed higher expression than BnaA02g33530D and BnaC02g42260D. The expression of GSL-related genes was determined in the Chinese double-low B. napus cultivar “Zhongsuang No. 11.” This analysis indicated that the expression of Bna.GTR genes is reduced in this cultivar, suggesting that the reduced transport of GSLs from source tissues to seeds accounts for the hypo-accumulation of GSLs in the seeds of this low-GSL content variety. For each AtGTR gene, we identified at least one Bna.GTR ortholog with high expression (Fig. 12.4). For instance, BnaA09g06190D, BnaC09g05810D, BnaC03g51560D, and BnaC03g75950D which might be the major GTR members responsible for the long-distance transport of GSL in the B. napus cultivar “Zhongsuang No. 11,” were expressed at higher levels than other members. Lu et al. (2014) reported that the transcript abundance in the leaves of the candidate gene involved in GSL transport, BnaA.GTR2a, located on chromosome A02, was correlated with seed GSL content, accounting for 18.8% of the phenotypic variation in seed GSL content between B. napus cultivars (Lu et al. 2014). Recently, we also found that Bna.GRT2 on chromosome A09 is a candidate GSL transporter and is associated with seed GSL content based on AM analysis of seed GSL content using the 60K Brassica Infinium SNP array in 520 B. napus accessions. These results strongly suggest that transport engineering can be used to eliminate antinutritional GSLs in seeds by silencing GTR transporters in B. napus.
Phylogenetic analysis of two AtGTR genes and orthologs in B. rapa, B. oleracea var. capitata, and B. napus. Full-length sequences of GTR proteins from Arabidopsis, B. rapa, B. oleracea var. capitata, and B. napus were aligned using ClustalW2. The phylogenetic tree (left panel) was constructed using MEGA 6.0 and the neighbor-joining method (1000 bootstrap replicates). Expression levels of Brassica GTR genes were derived from Tong et al. (2013) and Liu et al. (2014) and are presented as the log2-transformed (FPKM + 1) values
Indole GSL 4HGBS is the major GSL present in the low-GSL B. napus varieties. Whether the total GSL content can be further reduced by silencing all of the Bna.GTR genes merits further investigation. In addition, the major GSL transporter, GTR1, is multifunctional and may be involved in the transport of structurally distinct compounds, including GSLs, jasmonoyl-isoleucine, and gibberellin, and may positively regulate stamen development by mediating gibberellin transport in Arabidopsis (Saito et al. 2015). The gtr1 mutants are severely impaired in filament elongation and anther dehiscence, resulting in reduced fertility, and hence, it is uncertain whether silencing of all of the Bna.GTR genes would produce normal B. napus plants that lack GSLs in the seeds. Although there are potential limitations in genetic engineering applications, the Bna.GTR genes represent the most promising regulation loci among the GSL-related genes and have potential applications in molecular breeding efforts to further reduce GSL levels in the seeds and increase them in the vegetative tissues and roots, where they play important roles in enhancing biotic and/or abiotic resistance in B. napus.
References
Agee AE, Surpin M, Sohn EJ, Girke T, Rosado A, Kram BW et al (2010) MODIFIED VACUOLE PHENOTYPE1 is an Arabidopsis myrosinase-associated protein involved in endomembrane protein trafficking. Plant Physiol 152:120–132
Agerbirk N, Olsen CE (2012) Glucosinolate structures in evolution. Phytochemistry 77:16–45
Al-Shehbaz IA, Al-Shammary KI (1987) Distribution and chemotaxonomic significance of glucosinolates in certain Middle-Eastern cruciferae. Biochem Syst Ecol 15:559–569
Augustine R, Bisht NC (2015) Biofortification of oilseed Brassica juncea with the anti-cancer compound glucoraphanin by suppressing GSL-ALK gene family. Sci Rep 5:18005
Bak S, Feyereisen R (2001) The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol 127:108–118
Barth C, Jander G (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46:549–562
Bednarek P, Osbourn A (2009) Plant-microbe interactions: chemical diversity in plant defense. Science 324:746–748
Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M et al (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323:101–106
Bellostas N, Sørensen JC, Sørensen H (2007) Profiling glucosinolates in vegetative and reproductive tissues of four Brassica species of the U-triangle for their biofumigation potential. J Sci Food Agr 87:1586–1594
Benderoth M, Textor S, Windsor AJ, Mitchell-Olds T, Gershenzon J, Kroymann J (2006) Positive selection driving diversification in plant secondary metabolism. Proc Nat Acad Sci USA 103:9118–9123
Bones AM, Rossiter JT (2006) The enzymic and chemically induced decomposition of glucosinolates. Phytochemistry 67:1053–1067
Burow M, Atwell S, Francisco M, Kerwin RE, Halkier BA, Kliebenstein DJ (2015) The Glucosinolate biosynthetic gene AOP2 mediates feed-back regulation of jasmonic acid signaling in Arabidopsis. Mol Plant 8:1201–1212
Burow M, Halkier BA, Kliebenstein DJ (2010) Regulatory networks of glucosinolates shape Arabidopsis thaliana fitness. Curr Opin Plant Biol 13:348–353
Cartea ME, Velasco P (2008) Glucosinolates in Brassica foods: bioavailability in food and significance for human health. Phytochem Rev 7:213–229
Cartea ME, Velasco P, Obregón S, Padilla G, de Haro A (2008) Seasonal variation in glucosinolate content in Brassica oleracea crops grown in northwestern Spain. Phytochemistry 69:403–410
Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X et al (2014) Plant genetics. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science 345:950–953
Chen S, Glawischnig E, Jørgensen K, Naur P, Jørgensen B, Olsen CE, Hansen CH, Rasmussen H, Pickett JA, Halkier BA (2003) CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J 33:923–937
Daxenbichler ME, Spencer GF, Carlson DG, Rose GB, Brinker AM, Powell RG (1991) Glucosinolate composition of seeds from 297 species of wild plants. Phytochemistry 30:2623–2638
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
Fahey JW, Zalcmann AT, Talalay P (2001) The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56:5–51
Font R, del Rio-Celestino M, Cartea E, de Haro-Bailon A (2005) Quantification of glucosinolates in leaves of leaf rape (Brassica napus ssp. pabularia) by near-infrared spectroscopy. Phytochemistry 66:175–185
Frerigmann H, Gigolashvili T (2014) MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol Plant 7:814–828
Fu Y, Lu K, Qian L, Mei J, Wei D, Peng X et al (2015) Development of genic cleavage markers in association with seed glucosinolate content in canola. Theor Appl Genet 128:1029–1037
Gigolashvili T, Berger B, Flügge U-I (2008a) Specific and coordinated control of indolic and aliphatic glucosinolate biosynthesis by R2R3-MYB transcription factors in Arabidopsis thaliana. Phytochem Rev 8:3–13
Gigolashvili T, Engqvist M, Yatusevich R, Muller C, Flugge UI (2008b) HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol 177:627–642
Grubb CD, Abel S (2006) Glucosinolate metabolism and its control. Trends Plant Sci 11:89–100
Grubb CD, Zipp BJ, Ludwig-Müller J, Masuno MN, Molinski TF, Abel S (2004) Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J 40:893–908
Grubb CD, Zipp BJ, Kopycki J, Schubert M, Quint M, Lim EK, Bowles DJ, Pedras MS, Abel S (2014) Comparative analysis of Arabidopsis UGT74 glucosyltransferases reveals a special role of UGT74C1 in glucosinolate biosynthesis. Plant J 79:92–105
Gu Z-X, Guo Q-H, Gu Y-J (2012) Factors influencing glucoraphanin and sulforaphane formation in Brassica plants: a review. J Integr Agr 11:1804–1816
Guo R, Qian H, Shen W, Liu L, Zhang M, Cai C, Zhao Y, Qiao J, Wang Q (2013) BZR1 and BES1 participate in regulation of glucosinolate biosynthesis by brassinosteroids in Arabidopsis. J Exp Bot 64:2401–2412
Halkier BA, Du L (1997) The biosynthesis of glucosinolates. Trends Plant Sci 2:425–431
Halkier BA, Gershenzon J (2006) Biology and biochemistry of glucosinolates. Annu Rev Plant Biol 57:303–333
Hansen BG, Kerwin RE, Ober JA, Lambrix VM, Mitchell-Olds T, Gershenzon J et al (2008) A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol 148:2096–2108
Harper AL, Trick M, Higgins J, Fraser F, Clissold L, Wells R et al (2012) Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat Biotechnol 30:798–802
Hasan M, Friedt W, Pons-Kuhnemann J, Freitag NM, Link K, Snowdon RJ (2008) Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus). Theor Appl Genet 116:1035–1049
Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A et al (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104:6478–6483
Howell PM, Sharpe AG, Lydiate DJ (2003) Homoeologous loci control the accumulation of seed glucosinolates in oilseed rape (Brassica napus). Genome 46:454–460
Ishida M, Hara M, Fukino N, Kakizaki T, Morimitsu Y (2014) Glucosinolate metabolism, functionality and breeding for the improvement of Brassicaceae vegetables. Breed Sci 64:48–59
Jeffery EH, Brown AF, Kurilich AC, Keck AS, Matusheski N, Klein BP et al (2003) Variation in content of bioactive components in broccoli. J Food Compos Anal 16:323–330
Jorgensen ME, Nour-Eldin HH, Halkier BA (2015) Transport of defense compounds from source to sink: lessons learned from glucosinolates. Trends Plant Sci 20:508–514
Khajali F, Slominski B (2012) Factors that affect the nutritive value of canola meal for poultry. Poultry Sci 91:2564–2575
Kim JH, Durrett TP, Last RL, Jander G (2004) Characterization of the Arabidopsis TU8 glucosinolate mutation, an allele of TERMINAL FLOWER2. Plant Mol Biol 54:671–682
Kissen R, Bones AM (2009) Nitrile-specifier proteins involved in glucosinolate hydrolysis in Arabidopsis thaliana. J Biol Chem 284:12057–12070
Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T (2001) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate–dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13:681–693
Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T (2003) Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Nat Acad Sci USA 100:14587–14592
Kushad MM, Brown AF, Kurilich AC, Juvik JA, Klein BP, Wallig MA et al (1999) Variation of glucosinolates in vegetable crops of Brassica oleracea. J Agri Food Chem 47:1541–1548
Laluk K, Prasad KV, Savchenko T, Celesnik H, Dehesh K, Levy M et al (2012) The calmodulin-binding transcription factor SIGNAL RESPONSIVE1 is a novel regulator of glucosinolate metabolism and herbivory tolerance in Arabidopsis. Plant Cell Physiol 53:2008–2015
Le Roux C, Del Prete S, Boutet-Mercey S, Perreau F, Balagué C, Roby D et al (2014) The hnRNP-Q protein LIF2 participates in the plant immune response. PLoS ONE 9:e99343
Leeson S, Atteh J, Summers J (1987) The replacement value of canola meal for soybean meal in poultry diets. Can J Anim Sci 67:151–158
Levy M, Wang Q, Kaspi R, Parrella MP, Abel S (2005) Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J 43:79–96
Li G, Quiros CF (2003) In planta side-chain glucosinolate modification in Arabidopsis by introduction of dioxygenase Brassica homolog BoGSL-ALK. Theor Appl Genet 106:1116–1121
Li P, Zhao Y, Zhang W, Ding X, Yang M, Wang X et al (2005) Analysis of glucosinolate components and profiles in Brassica napus. Sci Agri Sin 38:1346–1352
Li J, Hansen BG, Ober JA, Kliebenstein DJ, Halkier BA (2008) Subclade of flavin-monooxygenases involved in aliphatic glucosinolate biosynthesis. Plant Physiol 148:1721–1733
Li F, Chen B, Xu K, Wu J, Song W, Bancroft I et al (2014) Genome-wide association study dissects the genetic architecture of seed weight and seed quality in rapeseed (Brassica napus L.). DNA Res 21:355–367
Liu Z, Hirani AH, McVetty PB, Daayf F, Quiros CF, Li G (2012) Reducing progoitrin and enriching glucoraphanin in Braasica napus seeds through silencing of the GSL-ALK gene family. Plant Mol Biol 79:179–189
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA et al (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5:3930
Lu G, Harper AL, Trick M, Morgan C, Fraser F, O’Neill C et al (2014) Associative transcriptomics study dissects the genetic architecture of seed glucosinolate content in Brassica napus. DNA Res 21:613–625
Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18:3235–3251
Mikkelsen MD, Petersen B, Olsen C, Halkier B (2002) Biosynthesis and metabolic engineering of glucosinolates. Amino Acids 22:279–295
Mikkelsen MD, Naur P, Halkier BA (2004) Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J 37:770–777
Nagaharu U (1935) Genome-analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japan J Bot 7:389–452
Nakahigashi K, Jasencakova Z, Schubert I, Goto K (2005) The Arabidopsis heterochromatin protein1 homolog (TERMINAL FLOWER2) silences genes within the euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol 46:1747–1756
Nakano RT, Matsushima R, Nagano AJ, Fukao Y, Fujiwara M, Kondo M et al (2012) ERMO3/MVP1/GOLD36 is involved in a cell type-specific mechanism for maintaining er morphology in Arabidopsis thaliana. PLoS ONE 7:e49103
Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE et al (2003) CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol 133:63–72
Neal CS, Fredericks DP, Griffiths CA, Neale AD (2010) The characterisation of AOP2: a gene associated with the biosynthesis of aliphatic alkenyl glucosinolates in Arabidopsis thaliana. BMC Plant Biol 10:170
Nour-Eldin HH, Halkier BA (2013) The emerging field of transport engineering of plant specialized metabolites. Curr Opin Biotechnol 24:263–270
Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jorgensen ME, Olsen CE et al (2012) NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488:531–534
Padilla G, Cartea ME, Velasco P, de Haro A, Ordas A (2007) Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry 68:536–545
Parkin IAP, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE et al (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15:R77
Pfalz M, Vogel H, Kroymann J (2009) The gene controlling the indole glucosinolate modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21:985–999
Piotrowski M, Schemenewitz A, Lopukhina A, Müller A, Janowitz T, Weiler EW et al (2004) Desulfoglucosinolate sulfotransferases from Arabidopsis thaliana catalyze the final step in the biosynthesis of the glucosinolate core structure. J Biol Chem 279:50717–50725
Radojčić Redovniković I, Glivetić T, Delonga K, Vorkapić-Furač J (2008) Glucosinolates and their potential role in plant. Period Biol 110:297–309
Rosa EA (1997) Glucosinolates from flower buds of Portuguese Brassica crops. Phytochemistry 44:1415–1419
Saito H, Oikawa T, Hamamoto S, Ishimaru Y, Kanamori-Sato M, Sasaki-Sekimoto Y et al (2015) The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat Commun 6:6095
Sang J, Minchinton I, Johnstone P, Truscott R (1984) Glucosinolate profiles in the seed, root and leaf tissue of cabbage, mustard, rapeseed, radish and swede. Can J Plant Sci 64:77–93
Skirycz A, Reichelt M, Burow M, Birkemeyer C, Rolcik J, Kopka J et al (2006) DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J 47:10–24
Sønderby IE, Geu-Flores F, Halkier BA (2010) Biosynthesis of glucosinolates–gene discovery and beyond. Trends Plant Sci 15:283–290
Sun B, Liu N, Zhao Y, Yan H, Wang Q (2011) Variation of glucosinolates in three edible parts of Chinese kale (Brassica alboglabra Bailey) varieties. Food Chem 124:941–947
Textor S, Bartram S, Kroymann J, Falk KL, Hick A, Pickett JA et al (2004) Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta 218:1026–1035
Textor S, De Kraker J-W, Hause B, Gershenzon J, Tokuhisa JG (2007) MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol 144:60–71
Tian Q, Rosselot RA, Schwartz SJ (2005) Quantitative determination of intact glucosinolates in broccoli, broccoli sprouts, brussels sprouts, and cauliflower by high-performance liquid chromatography–electrospray ionization–tandem mass spectrometry. Anal Biochem 343:93–99
Tong C, Wang X, Yu J, Wu J, Li W, Huang J et al (2013) Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genomics 14:689
Uzunova M, Ecke W, Weissleder K, Röbbelen G (1995) Mapping the genome of rapeseed (Brassica napus L.). I. construction of an RFLP linkage map and localization of QTLs for seed glucosinolate content. Theor Appl Genet 90:194–204
Vorwerk S, Biernacki S, Hillebrand H, Janzik I, Müller A, Weiler EW et al (2001) Enzymatic characterization of the recombinant Arabidopsis thaliana nitrilase subfamily encoded by the NIT2/NIT1/NIT3-gene cluster. Planta 212:508–516
Wang H, Wu J, Sun S, Liu B, Cheng F, Sun R et al (2011a) Glucosinolate biosynthetic genes in Brassica rapa. Gene 487:135–142
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S et al (2011b) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039
Wang M, Sun X, Tan D, Gong S, Meijer J, Zhang J (2009) The two non-functional myrosinase genes TGG3 and TGG6 in Arabidopsis are expressed predominantly in pollen. Plant Sci 177:371–375
Wang J, Gu H, Yu H, Zhao Z, Sheng X, Zhang X (2012) Genotypic variation of glucosinolates in broccoli (Brassica oleracea var. italica) florets from China. Food Chem 133:735–741
Warwick SI (2011) Brassicaceae in agriculture. In: Schmidt R, Bancroft I (eds) Genetics and genomics of the Brassicaceae. Springer, New York, pp 33–65
West L, Tsui I, Haas G (2002) Single column approach for the liquid chromatographic separation of polar and non-polar glucosinolates from broccoli sprouts and seeds. J Chromatogr A 966:227–232
Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7:263–270
Wittstock U, Burow M (2010) Glucosinolate breakdown in Arabidopsis: mechanism, regulation and biological significance. Arabidopsis Book 8:e0134
Xu Z, Escamilla-Treviño L, Zeng L, Lalgondar M, Bevan D, Winkel B, Mohamed A, Cheng C-L, Shih M-C, Poulton J, Esen A (2004) Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol Biol 55:343–367
Xue J, Jørgensen M, Pihlgren U, Rask L (1995) The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol Biol 27:911–922
Yang B, Quiros CF (2010) Survey of glucosinolate variation in leaves of Brassica rapa crops. Genet Resour Crop Ev 57:1079–1089
Zhang Z, Ober JA, Kliebenstein DJ (2006) The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18:1524–1536
Zhang J, Liu Z, Liang J, Wu J, Cheng F, Wang X (2015) Three genes encoding AOP2, a protein involved in aliphatic glucosinolate biosynthesis, are differentially expressed in Brassica rapa. J Exp Bot 66:6205–6218
Zhang J, Liu Z, Liang J, Wu J, Cheng F, Wang X (2015a) Three genes encoding AOP2, a protein involved in aliphatic glucosinolate biosynthesis, are differentially expressed in Brassica rapa. J Exp Bot 66:6205–6218
Zhang J, Wang X, Cheng F, Wu J, Liang J, Yang W et al (2015b) Lineage-specific evolution of Methylthioalkylmalate synthases (MAMs) involved in glucosinolates biosynthesis. Front Plant Sci 6:18
Zhao J, Meng J (2003) Detection of loci controlling seed glucosinolate content and their association with Sclerotinia resistance in Brassica napus. Plant Breed 122:19–23
Zhao Z, Zhang W, Stanley BA, Assmann SM (2008) Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20:3210–3226
Acknowledgements
This work was supported by grants from the National Science Foundation of China (31571701, 31171619, U1302266, and 31401412), the 973 Project (2015CB250100), the 863 Project (2013AA102602), the 111 Project (B12006) and the Fundamental Research Funds for the Central Universities (XDJK2012A009 and XDJK2013C031).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Lu, K., Snowdon, R., Li, J. (2018). Case Study for Trait-Related Gene Evolution: Glucosinolates. In: Liu, S., Snowdon, R., Chalhoub, B. (eds) The Brassica napus Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-319-43694-4_12
Download citation
DOI: https://doi.org/10.1007/978-3-319-43694-4_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-43692-0
Online ISBN: 978-3-319-43694-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)