Abstract
Breeding of oilseed rape (Brassica napus ssp. napus) has evoked a strong bottleneck selection towards double-low (00) seed quality with zero erucic acid and low seed glucosinolate content. The resulting reduction of genetic variability in elite 00-quality oilseed rape is particularly relevant with regard to the development of genetically diverse heterotic pools for hybrid breeding. In contrast, B. napus genotypes containing high levels of erucic acid and seed glucosinolates (++ quality) represent a comparatively genetically divergent source of germplasm. Seed glucosinolate content is a complex quantitative trait, however, meaning that the introgression of novel germplasm from this gene pool requires recurrent backcrossing to avoid linkage drag for high glucosinolate content. Molecular markers for key low-glucosinolate alleles could potentially improve the selection process. The aim of this study was to identify potentially gene-linked markers for important seed glucosinolate loci via structure-based allele-trait association studies in genetically diverse B. napus genotypes. The analyses included a set of new simple-sequence repeat (SSR) markers whose orthologs in Arabidopsis thaliana are physically closely linked to promising candidate genes for glucosinolate biosynthesis. We found evidence that four genes involved in the biosynthesis of indole, aliphatic and aromatic glucosinolates might be associated with known quantitative trait loci for total seed glucosinolate content in B. napus. Markers linked to homoeologous loci of these genes in the paleopolyploid B. napus genome were found to be associated with a significant effect on the seed glucosinolate content. This example shows the potential of Arabidopsis-Brassica comparative genome analysis for synteny-based identification of gene-linked SSR markers that can potentially be used in marker-assisted selection for an important trait in oilseed rape.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oilseed rape (Brassica napus ssp. napus; genome AACC, 2n = 38) is the most important source of vegetable oil in Europe and the second most important oilseed crop in the world after soybean. Brassica napus is a relatively young species that originated in a limited geographic region through spontaneous hybridisations between turnip rape (B. rapa; AA, 2n = 20) and cabbage (B. oleracea; CC, 2n = 18) genotypes (Kimber and McGregor 1995). The gene pool of elite oilseed rape breeding material has been depleted by breeding for specific oil and seed quality traits, with particularly strong bottleneck selection for zero seed erucic acid (C22:1) and low seed glucosinolate content (so-called double-low, 00 or canola quality). The first erucic acid-free variety, derived from a spontaneous mutant of the German spring rapeseed cultivar “Liho”, was released in Canada in the early 1970s. In 1969 the Polish spring rape variety “Bronowski” was identified as a low-glucosinolate form, and this cultivar provided the basis for an international backcrossing program to introduce this polygenic trait into high-yielding erucic acid-free breeding lines. The result was the release in 1974 of the first 00-quality spring rapeseed variety, “Tower”. Today the overwhelming majority of modern spring and winter oilseed rape varieties have 00-quality. However, residual segments of the “Bronowski” genotype in modern cultivars are believed to cause reductions in yield, winter hardiness, and oil content (Sharpe and Lydiate 2003). Furthermore, the restricted genetic variability in modern 00-quality oilseed rape (Hasan et al. 2006) is particularly relevant with regard to the development of genetically diverse heterotic pools of adapted genotypes for hybrid breeding. For this purpose B. napus genotypes containing high levels of erucic acid and seed glucosinolates (so-called ++ seed quality) represent a comparatively genetically divergent source of germplasm (Röbbelen 1975; Thompson 1983; Schuster 1987).
Glucosinolates are secondary plant metabolites synthesized by species in the family Brassicaceae, which includes a large number of economically important Brassica crops and the model plant Arabidopsis thaliana. The various glucosinolate compounds are designated aliphatic, aromatic and indole glucosinolates depending on whether they originate from aliphatic amino acids (methionine, alanine, valine, leucine, isoleucine), aromatic amino acids (tyrosine, phenylalanine) or tryptophan, respectively. Together with the myrosinase enzymes (also known as thioglucosidases) glucosinolates form the glucosinolate-myrosinase system (Wittstock and Halkier 2002), which is generally believed to be part of the plant’s defence against insects and possibly also against pathogens (Rask et al. 2000). When plant tissue is damaged the glucosinolates are hydrolysed by the myrosinases to release a range of defence compounds from substrate cells (Mithen et al. 2000).
After oil extraction from the seeds of oilseed rape the residual meal, which contains 38–44% of high quality protein, is used in livestock feed mixtures. However, high intakes of glucosinolates and their degradation products in rapeseed-based meals can cause problems of palatability and are associated with goitrogenic, liver and kidney abnormalities (Walker and Booth 2001). This particularly limits the use of the rich-protein meal as a feed supplement for monogastric livestock. Seed-specific optimisation of the glucosinolate content and composition would help to improve the nutritional value of rapeseed meal without compensating the disease and pest resistance properties in the crop (Wittstock and Halkier 2002). Genetic control of glucosinolate accumulation is polygenic, and the biosynthesis pathways for different glucosinolate compounds are well characterised in A. thaliana. Furthermore, Howell et al. (2003) demonstrated through comparative mapping that high-glucosinolate rapeseed genotypes often carry low-glucosinolate alleles at one or more of the major quantitative trait loci (QTL) controlling seed glucosinolate accumulation. With effective molecular markers for marker-assisted selection these genotypes could be used to introduce new genetic variation for low seed glucosinolate content into breeding programs. A number of studies have described detection of QTL for total seed glucosinolate content in different oilseed rape crosses (Uzunova et al. 1995; Howell et al. 2003; Sharpe and Lydiate 2003; Zhao and Meng 2003; Basunanda et al. 2007). Four QTL on B. napus chromosomes N9, N12, N17, and N19 were detected independently in different studies, indicating that these QTL represent major loci that influence seed glucosinolate content in different materials. The QTL on N9, N12 and N19 were found by Howell et al. (2003) to be homoeologous loci.
Markers for QTL detected by classical genetic mapping in individual crosses are not necessarily transferable to other material, and the utility of QTL-linked markers for marker-assisted selection is limited by the relative effects of individual loci on the trait of interest (Snowdon and Friedt 2004). On the other hand, detection of marker-trait associations based on linkage disequilibrium in genetically diverse materials can identify alleles with direct linkage to genes showing significant effects on the trait. In plant breeding populations the technique has seldom been used for marker development (Breseghello and Sorrells 2006), although association approaches can be particularly suitable for identification of useful allelic variation in genetically diverse genotype collections (Flint-Garcia et al. 2003). To date association studies in plants have mainly been performed in species for which extensive sequence data is available. For example, genome-wide analysis was used by Aranzana et al. (2006) to confirm trait associations of flowering time and disease resistance genes in A. thaliana, and sequence diversity in trait-relevant candidate genes has also been used to uncover allele-trait associations in Arabidopsis (Hagenblad and Nordborg 2002; Balasubramanian et al. 2006; Ehrenreich et al. 2007), rice (Bao et al. 2006; Iwata et al. 2007) and maize (Thornsberry et al. 2001; Wilson et al. 2004; Yu et al. 2006). On the other hand, genome-wide and candidate gene association studies have also been successful in crops with less well-characterised genomes, for example potato (Gebhardt et al. 2004). Oesterberg et al. (2002) identified associations with flowering time in sequence variants of the COL1 gene in Brassica nigra, but to date this remains the only report of an association study in a brassica crop.
In recent years considerable progress in the accumulation and distribution of Brassica genome data has been made by participants in the Multinational Brassica Genome Project (see http://www.brassica.info/). With the increasing amount of Brassica-Arabidopsis comparative genomics data it is becoming possible to navigate between and among the chromosomes of A. thaliana and B. napus. In some cases this can enable the map positions of B. napus QTL for traits of agronomic importance to be compared with the positions of potential candidate genes in the model genome. Brassica sequences with homology to the corresponding A. thaliana regions can then potentially be used for database-oriented identification of new markers for fine mapping, association studies or marker-assisted selection towards trait improvement. Moreover, it is also potentially possible to identify relevant candidate genes for important traits in oilseed rape, based on their positions in syntenic maps compared to important QTL.
According to Peleman and van der Voort (2003), distinguishing as many alleles as possible at loci of interest and determining phenotypic values for these alleles should greatly improve the predictive power of selection markers and enable marker-assisted combination of positive alleles for different loci. Because B. napus is a facultative outcrosser, a high degree of heterozygosity would be expected in natural populations. However, cultivars and gene bank collections of this amphipolyploid species are maintained as pure-breeding lines by self-pollination, so that genetically diverse genotype collections are effectively homozygous inbred lines and therefore ideal for allele-trait association studies. In this study we performed structure-based association studies for seed glucosinolate content in two divergent sets of B. napus genotypes. For the association studies a set of new simple-sequence repeat (SSR) markers was developed whose closest orthologs in A. thaliana are physically closely linked to promising candidate genes for seed glucosinolate biosynthesis. In order to incorporate information on the population structure into the association analysis, the potentially gene-linked markers were supplemented with a large set of SSR markers distributed throughout the genome. Furthermore, we also tested trait associations of previously mapped SSR markers for which homologous loci were localised near major QTL for seed glucosinolate content. This research tests the utility of association studies based on gene-linked and QTL-linked markers to detect seed glucosinolate content in B. napus. At the same time we describe a technique for synteny-based identification of gene-linked SSR markers for marker development in oilseed rape.
Materials and methods
Plant materials
Two different sets of genetically diverse B. napus genotypes were used for the allele-trait association studies (Table 1). The primary genotype set comprised 94 genetically diverse B. napus gene bank accessions from a B. napus “core collection” which spans the genetic diversity present in European gene bank collections of winter and spring oilseed, fodder and vegetable rape varieties. The core collection was selected based on phenotypic descriptors that were assessed during a European project on genetic diversity in Brassica crop species (Lühs et al. 2003; Poulsen et al. 2004), in combination with available pedigree information. The genetic diversity within the core collection has been described previously (Hasan et al. 2006). A second set of genotypes was used to further investigate markers that showed significant associations with glucosinolate content in the gene bank accessions. The second set of material comprised 46 winter-type, predominantly oilseed rape genotypes that were chosen based on pedigree knowledge to cover as broadly as possible the genetic and phenotypic variation present in current western European cultivars. Thirty-two of the 46 genotypes were cultivars or breeding lines with low seed glucosinolate content.
The gene bank accessions were grown in field trials in Rauischholzhausen, Germany, in 2003 and 2004, while the second set of genotypes were grown in Einbeck, Germany, from 2003 to 2005. Seeds were harvested from five to six self-pollinated plants per genotype and mean total seed glucosinolate content was estimated by near infrared reflectance spectroscopy (NIRS). Approximately 2 g seeds per sample were measured by monochromator analysis in a spinning cell at all wavelengths between 1,100 and 1,800 nm. For the molecular marker analyses genomic DNA samples were extracted from young leaves of five pooled plants per genotype using a standard CTAB extraction protocol (Doyle and Doyle 1990).
Potentially gene-linked SSR markers identified by comparative genome analysis
Twelve new Brassica SSR primer combinations were identified in sequences with homology to A. thaliana chromosome regions containing relevant candidate genes for glucosinolate content. First, interesting Arabidopsis chromosome regions with putative associations to glucosinolate QTL in B. napus were identified by in silico localisation of the closest A. thaliana orthologs for RFLP marker sequences from three major homoeologous B. napus glucosinolate QTL. Sequences for the RFLP probes CA72, pO119, pW141, pW200, and pW157, which were reported by Howell et al. (2003) to label loci belonging to homoeologous QTL on B. napus chromosomes N9, N12 and N19, were obtained from the EMBL database of the European Bioinformatics Institute (http://www.ebi.ac.uk/embl/). Four A. thaliana chromosome regions containing orthologous sequences to one or more of the abovementioned markers were identified based on the BLASTn annotations reported by Parkin et al. (2005). By searching the biological process “glucosinolate biosynthesis” in the gene ontology database of the Arabidopsis Information Resource (TAIR: http://www.arabidopsis.org/) the genes cytochrome P450 monooxygenase 83B1 (CYP83B1: At4g31500), cytochrome P450 79A2 (CYP79A2: At5g05260), methylthioalkylmalate synthase (MAM1/MAML: tandem duplication At5g23010/At5g23020) and altered tryptophan regulation (ATR1: At5g60890) were identified as the physically closest potential candidates to the QTL-marker orthologs in the four relevant chromosome regions on A. thaliana chromosomes 4 and 5, respectively.
The “SSR Search” tool of the Brassica ASTRA database from the Plant Genetics and Genomics platform of Primary Industries Research Victoria, Australia (http://hornbill.cspp.latrobe.edu.au/cgi-binpub/brassica/index.pl) was used to search A. thaliana genome regions up to 500 kbp upstream and downstream of the four selected candidate genes for potentially gene-linked SSR sequences. A total of thirty-two putative Brassica SSR primer combinations were identified in the different candidate gene regions and all primers were tested for suitability in B. napus. Twelve of the primer pairs gave clear, reproducible and polymorphic amplification products at one or more loci in B. napus and were used to screen for allelic polymorphisms in the 94 gene bank accessions. Sequences for these new SSR primers are available in Supplementary Table 1. All of the four putative candidate genes were represented by these potentially gene-linked SSR markers.
Four publicly-available Brassica SSRs (BRAS014, CB10425, Ol10-D03 and Ol11-C02) were also included in the association analysis in the primary genotype set. These four primers amplify SSR markers that are known to be linked to the seed glucosinolate QTL on B. napus N17 (Basunanda et al. 2007; F. Lipsa and R. Snowdon, unpublished results), for which no tightly-linked RFLP markers with clear synteny to Arabidopsis regions containing putative candidate genes were available.
Genome-wide SSR markers
Population structure among the 94 gene bank accessions was analysed using allelic data from 46 publicly available Brassica SSR primer combinations that amplify loci dispersed throughout the entire B. napus genome. Thirty of these primer combinations were also used previously to screen the genetic diversity in these genotypes (Hasan et al. 2006). For population structure analysis in the 46 winter oilseed rape genotypes, allelic data from a total of 104 SSR primer combinations that amplified 559 marker alleles were kindly provided by the breeding companies KWS Saat AG, SW Seed GmbH and Saaten-Union Resistenzlabor GmbH. This data was generated as part of the project GABI-BRIDGE: Brassica napus allelic diversity in candidate genes.
SSR analyses
PCR reactions were performed in a GeneAmp PCR System 9700 thermal cycler in a volume of 15 μL containing 20 ng of DNA template, 0.75 pmol of each primer, 0.2 mM dNTP mix, 1×PCR reaction buffer containing 15 mM MgCl2, a further 1 mM MgCl2 and 0.25 units of Taq DNA polymerase (Qiagen, Hilden, Germany). To reduce primer-labelling costs, PCR products were labelled with the M13-tailing technique described by Berg and Olaisen (1994). In this method the fluorescently labelled universal M13 primer 5′-AGGGTTTTCCCAGTCACGACGTT-3′ is added to the PCR reaction, and the forward primer of each SSR is appended with the sequence 5′-TTTCCCAGTCACGACGTT-3′. After the first round of amplification the PCR fragments are subsequently amplified by the labelled universal primer. A touch-down PCR cycle was modified from the procedure described by Xu et al. (2005) as follows: An initial denaturation was performed at 95°C for 2 min, followed by five cycles of denaturation for 45 s at 95°C, annealing for 5 min beginning at 68°C and decreasing by 2°C in each subsequent cycle, and extension for 1 min at 72°C. Then five cycles were performed with 45 s denaturation at 95°C, 1 min annealing beginning at 58°C and decreasing 2°C in each subsequent cycle, and 1 min of extension at 72°C. The PCR was then completed with an additional 27 cycles of 45 s denaturation at 94°C, 2 min of annealing at 47°C, and 30 s of extension at 72°C, with a final extension at 72°C for 10 min. The SSR polymorphisms were separated and visualised using a LI-COR GeneReadir 4200 (MWG Biotech, Ebersberg, Germany). Allele sizes including the 23 bp labelled M13 tail primer were scored with the software RFLP-SCAN (Version 2.01, Scanalytics Inc., Fairfax, VA, USA) based on a labelled length standard.
Analysis of population structure
A potential problem for every population-based association study is the presence of undetected population structure that can mimic the signal of association and lead to false positives or to missed real effects (Marchini et al. 2004). We analysed the population structure with the model-based Bayesian clustering approach in the software STRUCTURE 2.1 (Pritchard et al. 2000) using allelic data from genome-wide SSR markers. Many Brassica SSR primer combinations amplify different marker alleles at multiple loci in the paleopolyploid B. napus genome, and homoplasic alleles may be amplified at different loci. This means it can be difficult or impossible to assign the different marker alleles to individual loci in genotypes with high allelic diversity. Hence all SSR alleles were scored dominantly as present or absent in each genotype, and no information on marker linkage could be included in the population structure model. Therefore the model of no admixture was applied for the analysis of population structure, as stipulated by the user instructions for STRUCTURE 2.1. The basis of the Bayesian clustering method is the allocation of individual genotypes to groups in such a way that Hardy–Weinberg equilibrium and linkage equilibrium are valid within clusters, whereas these forms of equilibrium are absent between clusters. For each of the two genotype sets the optimum number of clusters (K) was selected after ten independent runs of a burn-in of 100,000 iterations, followed by 100,000 iterations using a model allowing for no admixture and correlated allele frequencies. We tested for K = 1–10 in the gene bank accessions and K = 1–5 in the set of winter rapeseed genotypes. A summary of the average logarithm of the probability of data likelihoods (LnP(D)) for both sets of genotypes is given in Table 2.
Structure-based association analysis
Due to the high allelic diversity, the clear population structure and an expectation of low familial relatedness due to the way the genotype collections were selected, we performed structured association tests rather than using a mixed-model approach (Yu et al. 2006) to control for false positives (type I errors) caused by the population structure. Associations between the marker data and the total seed glucosinolate content were tested using the logistic regression approach of Pritchard et al. (2000), as modified by Thornsberry et al. (2001) in order to deal with quantitative traits. This procedure is implemented in the software package TASSEL 2 (http://www.maizegenetics.net/). The response variable was the presence or absence of the SSR polymorphism, while the quantitative trait (total seed glucosinolate content) and the population structure (Q-matrix) were used as independent variables. In the null hypothesis, candidate polymorphisms are independent of the seed glucosinolate content (only the Q-matrix is included in the model), whereas in the alternative hypothesis the candidate polymorphisms are associated with the seed glucosinolate content (the quantitative trait and the Q-matrix are both included in the model). The test statistic Λ derives from the ratio between these two likelihoods and indicates the degree of association between individual polymorphisms and the quantitative trait. The null distribution of random markers was simulated by 1,000 permutations of the quantitative trait data over all genotypes. The P value for individual polymorphisms was calculated as the proportion of observed Λ greater then the maximal permuted Λ. This approach enables evaluation of associations involving quantitative traits while controlling for population structure. Only markers with an allele frequency of 5% or greater were included in the association analysis. In order to account for type I error bias the P values were adjusted for multiple tests using a procedure proposed by Whitt and Buckler (2003) based on the permuted P values of random markers. The rescaled P value accounts for the proportion of random markers with a permuted P value less than or equal to 0.05. According to Thornsberry et al. (2001) the true test statistic probably lies somewhere between the rescaled P value and P(Λ), since some of the random markers are probably truly associated with the trait. Therefore P(Λ) provides an overview of markers with potential association to trait, while the rescaled P value is a conservative test to reduce the likelihood of false-positive associations.
Map positions of markers with significant associations to seed glucosinolate content
Where possible the map positions of markers with significant associations to seed glucosinolate content were identified in existing B. napus genetic maps. For SSR primers where the allele sizes were not given in published maps, the positions of all known loci were recorded. Annotations of public Brassica SSR markers to the A. thaliana genome were obtained from the public microsatellite database at http://brassica.bbsrc.ac.uk/cgi-bin/ace/searches/browser/BrassicaDB. Glucosinolate-associated SSR markers from the set of new, synteny-based markers were screened for polymorphisms among the parents of three different doubled-haploid (DH) mapping populations and integrated into the maps of these populations where possible. The genetic mapping procedure followed Basunanda et al. (2007). Markers that deviated significantly (P > 0.01) from the expected 1:1 segregation in the DH populations were presumed to represent two or more homoeologous loci with identical allele sizes and hence could not be mapped.
Linkage disequilibrium
In order to gain information about the putative map positions of the gene-linked SSR markers in cases where these markers could not be directly mapped in available mapping populations, we used TASSEL to analyse linkage disequilibrium (LD) based on the parameter r 2 (the squared allele frequency correlation). The significance of the LD between marker pairs was determined by Fisher’s exact test. Due to the pre-selection for the association analysis only markers with a minimum allele frequency of 0.05 were included in the LD analysis, as recommended by Thornsberry et al. (2001). In a first step the LD was calculated among all markers that were significantly associated with seed glucosinolate content, in order to identify previously mapped markers with high LD to new, unmapped markers. Subsequently, the LD was recalculated within groups of markers with significant LD. Levels of LD were expected to be somewhat underestimated by the available SSR allele data, because in a paleopolyploid like B. napus it is known that identical alleles can be amplified by multiple loci. Therefore no presumption was made that two markers amplified by the same primer combination must necessarily belong to the same locus, even when these showed high LD.
Results
As expected from previous investigations (Hasan et al. 2006) a significant population structure was observed among the 94 gene bank accessions. As seen in Table 2, the highest average likelihoods for the population structure in this set of material were observed with K values between 3 and 7, whereby the most stable prediction (standard deviation = 18.35) was obtained with three groups (K = 3). These groups comprised (1) twenty genotypes of predominantly spring-type oilseed rape, (2) twenty genotypes of mainly fodder or vegetable rape, and (3) fifty-four predominantly winter oilseed rape genotypes, respectively. The most stable and high average likelihoods for population structure amongst the 46 winter rapeseed genotypes were obtained with K = 2 or 3. Seventeen oilseed genotypes were strongly assigned to the same group in both cases, while the remaining 29 genotypes were divided with K = 3 into another group of 20 oilseed types and a group of nine exotic genotypes, including fodder rape varieties and resynthesised (RS) rapeseed lines. Such material is known to represent a divergent B. napus gene pool in comparison to oilseed B. napus genotypes (Seyis et al. 2003), and since most of these exotic genotypes also exhibited high glucosinolate content this grouping was expected to be particularly relevant for the association analysis with glucosinolate content. We therefore used the respective Q-matrix outputs of the three-subpopulation runs (K = 3) for the structure-based association analyses in both sets of genotypes. A broad range in total seed glucosinolate content was observed among the 94 gene bank accessions, whereas the winter rapeseed set included 32 genotypes with low seed glucosinolate content (<25 μmol/mg dry weight). Details of the groupings of the accessions along with the mean total seed glucosinolate data used for the association analyses are given in Table 1.
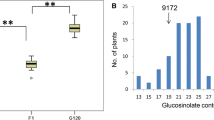
Using the complete set of 62 polymorphic SSR primer combinations, a total of 348 polymorphic SSR marker alleles were amplified in the 94 gene bank accessions. Of these, a total of 51 marker alleles from 29 SSR primer combinations were found to exhibit a significant association (P ≤ 0.05) to total seed glucosinolate content in the 94 B. napus gene bank accessions. Ten of the markers also exhibited significant association using the rescaled P values, indicating that these associations are not likely to be caused by type I errors. All markers with significant associations to seed glucosinolate content are described in detail in Table 3, including information (where available) on map positions and annotations to the A. thaliana genome. Positions of glucosinolate-associated markers with known physical linkage to relevant candidate genes in A. thaliana are shown in Fig. 1. The phenotypic distributions of the genotypes with the 51 marker alleles showing significant associations to seed glucosinolate content in the gene bank accessions are illustrated by box-plots in Fig. 2. Allelic data for all SSR markers with significant associations to total seed glucosinolate content are available in Supplementary Table 2.
Chromosomal positions in Arabidopsis thaliana (numbers in Mbp) of orthologs for potentially gene-linked Brassica SSR markers (italics) in comparison to potential candidate genes for glucosinolate biosynthesis (bold italics) and RFLP markers (non-italic, non-bold) located at major seed glucosinolate QTL (Uzunova et al. 1995; Howell et al. 2003). Markers with the prefix Gi are new SSR sequences identified by synteny studies in candidate gene regions. Primer sequences of the new SSR markers are available in Supplementary Table 1
Boxplots showing distributions of total seed glucosinolate content within 94 genetically diverse B. napus gene bank accessions for 51 SSR marker alleles with significant association (P ≤ 0.05) to glucosinolate content. Boxes cover the inter-quartile range around the mean (horizontal lines), while the vertical whiskers cover the remaining variation with the exception of outliers (open circles) and extreme values (open squares)
In order to get an idea of the abundance of these glucosinolate-associated markers in European winter rapeseed, and particularly in material with low seed glucosinolate content, we re-screened all of the significantly associated SSRs in the set of 46 winter rapeseed genotypes. Interestingly, many of the significantly associated marker alleles were only found at very low frequencies (<5%) in the winter rapeseed set, and only three markers with frequencies of greater than 5% also showed significant association to seed glucosinolate content among these 46 genotypes. All three of these markers were associated with low glucosinolate content. In two cases (Na12-G04 and Ol10-D02), different marker alleles amplified by the same SSR primer combinations showed significant associations in the two different sets of materials. The marker Gi31_387 was the only marker allele that was found to be significantly associated to seed glucosinolate content in both sets of materials. As seen in Table 3, the sequence of Gi31 is located in A. thaliana only 736 bp downstream of the gene CYP83B1. In the gene bank accessions, the two marker alleles amplified by this primer combination, Gi31_385 and Gi31_387, are associated with significantly increased and decreased total seed glucosinolate content, respectively. The allele-trait association of both alleles together with the very short physical distance to the candidate gene strongly support the potential involvement of homoeologous CYP83B1 copies in biosynthesis of seed glucosinolates in B. napus. Two of four marker alleles from the SSR sequence Gi30, which is located somewhat further away from CYP83B1, were also significantly associated with total seed glucosinolate content in the gene bank accessions.
For the three other candidate genes we were also able to identify putatively linked SSR markers with significant associations to seed total glucosinolate content (Table 3). The SSR Gi24, located 166 kbp from CYP79A2 in A. thaliana, amplified a single band whose presence in the gene bank accessions was associated with an increased glucosinolate content. The SSR Gi12, although located 382 kbp away from ATR1 in A. thaliana, amplified a single band that was associated with a mean decrease in total glucosinolate content. Two of three bands amplified by the SSR Gi28, which is derived from a sequence near the duplicated MAM1/MAML gene locus in A. thaliana, were associated with increased and reduced total seed glucosinolate content, respectively, in the gene bank accessions. Both of the latter markers, along with Gi31_387, also showed significant associations with rescaled P-values, meaning that a type I error is unlikely.
Where sequence and annotation information were available, the glucosinolate-associated SSRs from the genome-wide marker set were also compared with the Arabidopsis genome to establish further potential physical linkages to candidate genes. For example, Ol11-G11 amplifies a marker allele with significant association to glucosinolate content, although this SSR maps to two loci on B. napus N03 and N13 (Basunanda et al. 2007; Rygulla et al. 2008) where no major QTL for total seed glucosinolate content are known. As shown in Fig. 1, the sequence of Ol11-G11 is annotated in A. thaliana to the sequence At3g10040 (data from BrassicaDB), which is located on A. thaliana chromosome 3 only 117 kbp downstream from the gene IQ-DOMAIN 1 (IQD1: At3g09710). A further glucosinolate-associated marker, Ra3-E05, also annotates nearby on A. thaliana chromosome 3. This chromosome regions shows no obvious homology to Brassica regions involved in seed glucosinolate QTL, however IQD1 is nevertheless a further interesting candidate gene for this trait because it is known to modulate expression of numerous glucosinolate pathway genes. Gain-of-function and loss-of-function IQD1 alleles in A. thaliana are correlated with increased and decreased total glucosinolate accumulation, respectively (Levy et al. 2005).
Knowledge about homoeologous B. napus map positions of the SSR markers that showed significant associations to seed total glucosinolate content in the gene bank accessions allowed these to be sorted into groups of markers that might putatively be linked to the same responsible gene or genes, either in the same chromosome region or in common homoeologous chromosome regions. For gene-linked SSR markers with unknown map positions, significant LD to a marker with a known map position was used to infer the putative map positions of the new marker. Results of LD analysis for three groups of markers showing significant LD within their respective group are shown in Fig. 3. The relatively low LD within the groups was reflected by generally low correlation coefficients among the putative neighbouring alleles; however, weak LD patterns could nevertheless be observed. For example one group of markers (Fig. 3a) showed linkage disequilibrium flanking two presumably allelic SSR markers (Gi28_442 and Gi28_444, respectively) derived from sequences near the duplicated MAM1/MAML locus in A. thaliana. Since all of these markers are known to have loci on B. napus chromosome N17 (see Table 3), and in the case of BRAS014_162 and CB10425_327 are known to map near the seed glucosinolate QTL on this chromosome (Basunanda et al. 2007), this indicates that orthologs of MAM1 and/or MAML might be among the responsible genes influencing variation in total seed glucosinolate at this locus. On the other hand the groups of markers exhibiting significant LD did not always derive from the same linkage group. This is not unexpected, since homoeologous SSR loci in B. napus often exhibit homoplasic alleles. The consequence is that it is apparently inconclusive to use this method to infer map positions, since it is very difficult to relate specific SSR alleles from a diversity study to unique chromosome locations.
Linkage disequilibrium (LD) around the gene-linked SSR markers a Gi28, b Gi30, and c Gi24 in the 94 B. napus gene bank accessions. Cells above the diagonal show the squared allele frequency correlation r 2, while the cells below the diagonal represent the significance level of the LD determined by Fisher’s exact test
A description of LD might nevertheless be useful to detect potential copies of candidate genes contributing to different QTL. For example, four markers showing LD on either side of the putative gene-linked markers Gi30_385 and Gi30_390 (Fig. 3b) map to different chromosomes: Na10_C01-282 on N14, Na10-D03_176 on N13, Ol9-A06_109 on N12 and BRAS020_260 on N9. The Gi30 sequence is separated in A. thaliana by a physical distance of only 30 kb from the glucosinolate biosynthesis gene CYP83B1, and the neighbouring RFLP marker pW157 has been mapped in B. napus to loci on N1, N11, N9 and N19. Neighbouring markers in the latter two chromosomes have homology to N12. A similar example is shown in Fig. 3b for the trait-associated markers that show LD around the marker Gi24_247, whose sequence in A. thaliana is located near the gene CYP79A2. These markers have known homoeologous loci on a number of different B. napus chromosomes including N1, N4, N6, N9, N11, N13, N14, N17 and N19. Again this suggests that a considerable number of homoeologous chromosome regions might contain copies of CYP79A2, along with different copies of the linked marker loci. The supposition of homologous gene loci associated to glucosinolate content was supported by the fact that many of the new, potentially gene-linked markers deviated significantly from the expected 1:1 segregation in different DH populations (data not shown), which in turn also prevented us from genetically mapping these markers.
Discussion
The aim of this work was to investigate the potential use of Brassica-Arabidopsis comparative genomics data for marker and gene identification in oilseed rape based on sequence orthology to A. thaliana. Using marker sequences from important B. napus QTL for seed total glucosinolate content, along with comparative mapping data, we were able to navigate to potential orthologous genome regions in A. thaliana. This enabled us to identify four promising candidate genes with putative physical linkage to homoeologous Brassica genome regions involved in seed glucosinolate biosynthesis. Through in silico screening of neighbouring Brassica genomic and EST sequences a number of new Brassica SSR sequences with putative close physical linkage to these four genes were identified, and hom(oe)ologous markers for many of these SSR primers showed significant associations with seed glucosinolate content when screened in B. napus gene bank accessions. A number of the trait-associated markers showed skewed segregation in DH mapping populations, indicating the presence of two or more homologous copies of the markers and their putatively linked genes.
In addition, whole-genome association analyses were performed with SSR markers dispersed throughout the B. napus genome. This approach also led to the identification of numerous markers with significant associations to glucosinolate content. In some cases the markers were mapped in available B. napus genetic maps, and numerous markers showed no apparent relationship to known QTL regions for seed glucosinolate content. This indicates that we may have identified novel allelic variation for this important trait, which should be of considerable interest for breeding purposes. The orthologous sequences in A. thaliana for two of the genome-wide SSR markers are closely physically linked to a further promising candidate gene for glucosinolate biosynthesis, IQD1. The successful identification of new markers associated to an important seed quality trait underlines the great promise of in silico mapping data for gene discovery in oilseed rape based on intergenomic comparisons to A. thaliana. Marker sequences, QTL and association data from the crop plant can potentially be used to discover or confirm potential candidate genes in the model species. Furthermore, with the growing resource of Brassica genomic sequence data and its alignment to the A. thaliana genome, it is now also possible to identify new molecular markers in linkage disequilibrium to genes of interest in Arabidopsis. Of particular interest for practical plant breeding is the possibility to identify gene-linked SSR markers: SSRs are robust, highly polymorphic markers that are relatively cheap and easy to use, and hence predestined for use in marker-assisted selection. On the other hand, recent developments in high-throughput sequencing technologies may soon enable large-scale re-sequencing of candidate gene orthologs. At present it is still difficult to develop locus-specific assays for single-nucleotide polymorphisms (SNPs) in polyploid species like B. napus, which can contain many orthologous and paralogous gene copies. However in the near future it is likely that high-throughput SNP discovery will become an important tool for gene discovery and association genetics in oilseed rape. High-density SNP maps will considerably improve our knowledge of LD in B. napus and enable much more accurate use of Arabidopsis-Brassica comparative genomics data. At present little is known about the extent of LD in B. napus.
Seed glucosinolate content in B. napus is governed by complex biochemical interactions that make it difficult to predict the actions of individual genes. Because specific pathway branches control the synthesis of different aliphatic, aromatic and indole glucosinolates, dissection of QTL for total glucosinolate content into the individual components is desirable to gain more information about which QTL may involve global pathway genes and which QTL might be more specific for individual compounds. The five candidate genes we identified in this study are well characterised in A. thaliana, and the use of selection markers with putative linkage to these genes might enable selection for specific glucosinolate pathway chains. For example, the cytochrome P450 monooxygenase enzyme CYP83B1 (Hoecker et al. 2004) catalyses the N-hydroxylation of tryptophan-derived indole-3-acetaldoxime, an intermediate in the biosynthesis of indole glucosinolates (Bak et al. 2001; Hansen et al. 2001). The enzyme encoded by CYP79A2 catalyses the conversion of L-phenylalanine to phenylacetaldoxime, a precursor of the aromatic benzyl-glucosinolates in A. thaliana (Wittstock and Halkier 2000), whereas ATR1 encodes a transcription factor which activates the expression of tryptophan synthesis genes as well as the tryptophan-metabolizing genes CYP79B2, CYP79B3, and CYP83B1; ATR1 therefore plays a central regulatory role in the production of indole-3-acetic acid and indole glucosinolates (Celenza et al. 2005). On the other hand, the two tandemly duplicated loci MAM1 and MAML encode genes that catalyze the condensation reactions of the first two cycles in methionine side-chain elongation in A. thaliana, therefore they play a vital role in methionine chain elongation and the biosynthesis of aliphatic glucosinolates (Kroymann et al. 2001; Textor et al. 2004). At present it is not known if different homoeologous methylthioalkamate synthase loci in B. napus also carry the gene duplication seen in A. thaliana.
All but a few of the the glucosinolate-associated markers we identified in the 94 B. napus gene bank accessions were found at only very low frequencies in the set of winter rapeseed genotypes. This appears to indicate that the glucosinolate-associated alleles we identified represent novel allelic diversity for this trait that is not present in current European 00-quality oilseed rape cultivars. On the other hand, these results underline the finding of Howell et al. (2003) that most low-glucosinolate cultivars still contain alleles at some loci that in fact are associated with increased total glucosinolate content. The markers we identified will potentially help to further reduce glucosinolate content in existing elite 00-quality oilseed rape, and to introduce new genetic diversity into the comparatively narrow gene pool of 00-quality rapeseed.
Overall the results of this study give strong indications that genetically linked homologous copies of a small number of key biosynthetic and regulatory genes play a major role in the accumulation of aliphatic, aromatic and indole glucosinolates in B. napus seeds. By identifying gene-linked SSR markers with significant associations to total seed glucosinolate content in genetically diverse oilseed rape germplasm, we hope to provide a simple molecular tool for marker-assisted combination of positive alleles in new, low-glucosinolate genotypes. This has considerable interest for breeding because the markers should enhance the identification of high-glucosinolate accessions carrying desirable alleles that until now have been largely ignored in breeding of 00-quality oilseed rape. Inter-crossing of different high-glucosinolate genotypes that contain complementary marker alleles associated with reduced total glucosinolate content at different gene loci should result in transgressive segregation with the possibility for marker-assisted pyramiding of positive alleles at all major loci. Ultimately this could open the way for the development of new, genetically diverse heterotic pools for hybrid breeding.
References
Aranzana MJ, Kim S, Zhao K, Bakker E, Horton M, Jakob K, Lister C, Molitor J, Shindo C, Tang C, Toomajian C, Traw B, Zheng H, Bergelson J, Dean C, Marjoram P, Nordborg M (2006) Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLOS Genet 1:531–539
Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R (2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13:101–111
Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D (2006) The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat Genet 38:711–715
Bao JS, Corke H, Sun M (2006) Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor Appl Genet 113:1171–1183
Basunanda P, Spiller TH, Hasan M, Gehringer A, Schondelmaier J, Lühs W, Friedt W, Snowdon RJ (2007) Marker-assisted increase of genetic diversity in a double-low seed quality winter oilseed rape genetic background. Plant Breed 126:581–587
Berg ES, Olaisen B (1994) Hybrid PCR sequencing—sequencing of PCR products using a universal primer. Biotechniques 17:896–901
Breseghello F, Sorrells ME (2006) Association analysis as a strategy for improvement of quantitative traits in plants. Crop Sci 46:1323–1330
Celenza JL, Quiel JA, Smolen GA, Merrikh H, Silvestro AR, Normanly J, Bender J (2005) The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol 137:253–262
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Ehrenreich IM, Stafford PA, Purugganan MD (2007) The genetic architecture of shoot branching in Arabidopsis thaliana: A comparative assessment of candidate gene associations vs. quantitative trait locus mapping. Genetics 176:1223–1236
Flint-Garcia SA, Thornsberry JM, Buckler ES IV (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Gebhardt C, Ballvora A, Walkemeier B, Oberhagemann P, Schüler K (2004) Assessing genetic potential in germplasm collections of crop plants by marker-trait association: a case study for potatoes with quantitative variation of resistance to late blight and maturity type. Mol Breed 13:93–102
Hagenblad J, Nordborg M (2002) Sequence variation and haplotype structure surrounding the flowering time locus Fr1 in Arabidopsis thaliana. Genetics 161:289–298
Hansen CH, Du L, Naur P, Olsen CE, Axelsen KB, Hick AJ, Pickett JA, Halkier BA (2001) CYP83B1 is the oxime-metabolizing enzyme in the glucosinolate pathway in Arabidopsis. J Biol Chem 276:24790–24796
Hasan M, Seyis F, Badani AG, Pons-Kuhnemann J, Lühs W, Friedt W, Snowdon RJ (2006) Analysis of genetic diversity in the Brassica napus L. gene pool using SSR markers. Genet Resour Crop Evol 53:793–802
Hoecker U, Toledo-Ortiz G, Bender J, Quail PH (2004) The photomorphogenesis-related mutant red1 is defective in CYP83B1, a red light-induced gene encoding a cytochrome P450 required for normal auxin homeostasis. Planta 219:195–200
Howell PM, Sharpe AG, Lydiate DJ (2003) Homoeologous loci control the accumulation of seed glucosinolates in oilseed rape (Brassica napus). Genome 46:454–460
Iwata H, Uga Y, Yoshioka Y, Ebana K, Hayashi T (2007) Bayesian association mapping of multiple quantitative trait loci and its application to the analysis of genetic variation among Oryza sativa L. germplasms. Theor Appl Genet 114:1437–1449
Kimber DS, McGregor DI (1995) The species and their origin, cultivation and world production. In: Kimber D, McGregor DI (eds) Brassica oilseeds: production and utilization. CABI Publishing, Wallingford, pp 1–9
Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T (2001) A gene controlling variation in Arabidopsis thaliana glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol 127:1077–1088
Levy M, Wang Q, Kaspi R, Parrella MP, Abel S (2005) Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defence. Plant J 43:79–96
Lühs W, Seyis F, Frauen M, Busch H, Frese L, Willner E, Friedt W, Gustafsson M, Poulsen G (2003) Development and evaluation of a Brassica napus core collection. In: Knüpffer H, Ochsmann J (eds) Rudolf Mansfeld and Plant Genetic Resources. Proceedings of a symposium dedicated to the 100th birthday of Rudolf Mansfeld, Gatersleben, Germany, 8–9 October 2001. Schriften zu Genetischen Ressourcen 19. ZADI/IBV, Bonn, pp 284–289 (http://www.genres.de/infos/rei-bd22.htm)
Marchini J, Cardon LR, Phillips MS, Donnelly P (2004) The effects of human population structure on large genetic association studies. Nat Genet 36:512–517
Mithen RF, Dekker M, Verkerk R, Rabot S, Johnson IT (2000) The nutritional significance, biosynthesis and bioavailability of glucosinolates in human foods. J Sci Food Agric 80:967–984
Oesterberg MK, Shavorskaya O, Lascoux M, Lagercrantz U (2002) Naturally occurring indel variation in the Brassica nigra COL1 gene is associated with variation in flowering time. Genetics 161:299–306
Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765–781
Peleman JD, van der Voort JR (2003) Breeding by design. Trends Plant Sci 8:330–334
Poulsen G, Busch H, Frauen M, Frese L, Friedt W, Gustafsson M, Ottosson F, Seyis F, Stemann G, Ulber B, Willner E, Lühs W (2004) The European Brassica napus core collection—characterisation, evaluation and establishment. Cruciferae Newsl 25:115–116
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rask L, Andréasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J (2000) Myrosinase: gene family evolution and herbivory defense in Brassicaceae. Plant Mol Biol 42:93–113
Röbbelen G (1975) Totale Sortenumstellung bei Körnerraps. Bericht über die Arbeitstagung der Arbeitsgemeinschaft der Saatzuchtleiter, Gumpenstein. Arbeitsgemeinschaft der Saatzuchtleiter, Austria, pp 119–146
Rygulla W, Snowdon RJ, Friedt W, Happstadius I, Cheung WY, Chen D (2008) Identification of quantitative trait loci for resistance against Verticillium longisporum in oilseed rape (Brassica napus). Phytopathology 98:215–221
Schuster W (1987) Die Entwicklung des Anbaues und der Züchtung von Ölpflanzen in Mitteleuropa. Fat Sci Technol 89:15–27
Seyis F, Snowdon RJ, Lühs W, Friedt W (2003) Molecular characterisation of novel resynthesised rapeseed (Brassica napus L.) lines and analysis of their genetic diversity in comparison to spring rapeseed cultivars. Plant Breed 122:473–478
Sharpe AG, Lydiate DJ (2003) Mapping the mosaic of ancestral genotypes in a cultivar of oilseed rape (Brassica napus) selected via pedigree breeding. Genome 46:461–468
Snowdon RJ, Friedt W (2004) Molecular markers in Brassica oilseed breeding: current status and future possibilities. Plant Breed 123:1–8
Textor S, Bartram S, Kroymann J, Falk KL, Hick A, Pickett J.A, Gershenzon J (2004) Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain elongation cycle. Planta 218:1026–1035
Thompson KF (1983) Breeding winter oilseed rapeseed Brassica napus. Adv Appl Biol 7:1–104
Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES IV (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28:286–289
Uzunova M, Ecke W, Weissleder K, Röbbelen G (1995) Mapping the genome of rapeseed (Brassica napus L.). I. Construction of an RFLP linkage map and localization of QTLs for seed glucosinolate content. Theor Appl Genet 90:194–204
Walker KC, Booth EJ (2001) Agricultural aspects of rape and other Brassica products. Eur J Lipid Sci Technol 103:441–446
Whitt SR, Buckler ES IV (2003) Using natural allelic diversity to evaluate gene function. In: Grotewald E (ed) Plant functional genomics: methods and protocols. Humana Press, New York, pp 123–140
Wilson LM, Whitt SR, Ibáñez AM, Rocheford TR, Goodman MM, Buckler ES (2004) Dissection of maize kernel composition and starch production by candidate gene association. Plant Cell 16:2719–2733
Wittstock U, Halkier BA (2000) Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of l-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J Biol Chem 275:14659–14666
Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7:263–270
Xu XY, Bai GH, Carver BF, Shaner GE (2005) Mapping of QTLs prolonging the latent period of Puccinia triticina infection in wheat. Theor Appl Genet 110:244–251
Yu J, Pressior G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB, Kresovich S, Buckler ES (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38:203–208
Zhao J, Meng J (2003) Detection of loci controlling seed glucosinolate content and their association with Sclerotinia resistance in Brassica napus. Plant Breed 122:19–23
Acknowledgments
The authors thank Fatih Seyis, Wilfried Lühs, and KWS Saat AG for providing seed material for this study. We also thank Liane Renno and Petra Degen for valuable technical assistance. Some of the SSR primers were kindly provided by Wolfgang Ecke, University of Göttingen, and Jörg Schondelmaier, SaatenUnion Resistenzlabor GmbH, Leopoldshöhe. Genome-wide SSR data for the 46 winter rapeseed genotypes genotypes was made available by KWS Saat, SaatenUnion Resistenzlabor and SW Seed GmbH as part of the BMBF-funded project GABI-BRIDGE. We thank two anonymous reviewers and the subject editor for constructive comments that led to considerable improvements in the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Sillanpää.
Electronic supplementary material
Below is the link to the electronic supplementary material.
122_2008_733_MOESM1_ESM.xls
Supplementary Table 1. PCR primers for twelve new simple sequence repeat (SSR) sequences whose othologs in Arabidopsis thaliana are closely physically linked to glucosinolate biosynthesis genes. Five of the primer combinations amplify SSR loci in Brassica napus that were found to be significantly associated (P ≤ 0.05) with total seed glucosinolate content. (XLS 15 kb)
122_2008_733_MOESM2_ESM.xls
Supplementary Table 2. Allelic data for SSR markers showing significant allele-trait association to total seed glucosinolate content in two sets of genetically diverse B. napus genotypes: (a) 94 genetically diverse gene bank accessions; (b) 46 genetically diverse winter rapeseed cultivars and breeding lines. Cells with a green background indicate the presence of the marker allele in the particular genotype. Markers are sorted from left (low) to right (high) based on the mean total seed glucosinolate content of all genotypes containing the given marker allele, while each of the two sets of genotypes is also sorted in ascending order according to the mean total seed glucosinolate content. Hence the green cells in the above left corner of each table represent marker alleles associated with very low glucosinolate content, while the green cells at the bottom right of each table represent marker alleles associated with very high glucosinolate content. (XLS 95 kb)
Rights and permissions
About this article
Cite this article
Hasan, M., Friedt, W., Pons-Kühnemann, J. et al. Association of gene-linked SSR markers to seed glucosinolate content in oilseed rape (Brassica napus ssp. napus). Theor Appl Genet 116, 1035–1049 (2008). https://doi.org/10.1007/s00122-008-0733-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-008-0733-3