Abstract
Sarcomas comprise a heterogeneous group of rare malignancies of mesenchymal origin including more than 70 subtypes. They may arise in muscle, bone, cartilage and other connective tissues. Their high histological and genetic heterogeneity makes diagnosis and treatment very challenging. Deregulation of heparanase has been found in several sarcoma subtypes and high expression levels have been correlated with poor prognosis in Ewing’s sarcoma and osteosarcoma. Altered expression of specific heparan sulfate proteoglycans and heparan sulfate biosynthetic enzymes has also been observed. Advances in molecular pathogenesis of sarcomas have evidenced the critical role of several heparan sulfate binding growth factors and receptor tyrosine kinases, highly interconnected with the microenvironment, in sustaining tumor growth and progression. Interference with heparanase/heparan sulfate functions represents a potential therapeutic approach in sarcoma. In this chapter, we summarize the current knowledge about the biological significance of heparanase expression and its potential as a therapeutic target in subtypes of both soft tissue and bone sarcomas. Particular emphasis is given to the involvement of heparan sulfate proteoglycans and their synthesizing and modifying enzymes in bone physiology and disorders leading up to the pathobiology of bone sarcomas. The chapter also describes the cooperation between exostin loss-of-function and heparanase upregulation in hereditary Multiple Osteochondroma syndrome as a paradigmatic example of constitutive alteration of the heparanase/heparan sulfate proteoglycan system which may contribute to progression to malignant secondary chondrosarcoma. Preclinical evidence of the role of heparanase as a promising therapeutic target in various sarcoma subtypes is finally resumed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Heparanase
- Heparan sulfate
- Rhabdomyosarcoma
- Chondrosarcoma
- Osteosarcoma
- Synovial sarcoma
- Ewing’s sarcoma
- Multiple Osteochondroma
- Heparanase inhibitor
- Heparan sulfate mimetics
- Cancer therapy
1 Sarcomas
Sarcomas are rare mesenchymal tumors accounting for about 1% of all cancers in adults and 15–20% of pediatric tumors. They constitute a heterogeneous family of bone and soft tissue malignancies that comprises more than 70 subtypes [1]. Molecular classification distinguishes two categories:( 1) genetically simple sarcomas characterized by a tumor-specific chromosomal translocation or point mutation and a near-diploid karyotype; and (2) genetically complex sarcomas that lack consistent specific genetic changes but present unbalanced translocations, changes in chromosome number, genetic deletions and amplifications characteristic of unstable genome. The current view is that sarcomas should be considered as a collection of histologically and genetically distinct malignancies, a feature that, together with rarity, makes treatment and diagnosis in several cases, particularly challenging.
A huge variety of genetic alterations has been described in sarcomas. Nevertheless, a few general events driving sarcomagenesis can be recognized [2]. Most genetically simple sarcomas harbor pathognomonic chromosomal translocations. The resulting fusion genes, encoding chimeric transcription factors (e.g., EWS-FLI1 in Ewing’s sarcoma, PAX3/7-FOXO1A in alveolar rhabdomyosarcomas) or chromatin remodeling proteins (e.g., SS18-SSX1/2 in synovial sarcoma), induce transcriptional dysregulation of target genes. Epigenetic control of the transcriptome can also be subverted by genetic alterations that change the composition of chromatin remodeling complexes (e.g., loss of SMARCB1 in rhabdoid tumors). Other genetic changes directly alter cell signaling components [e.g., COL1A1-PDGFBB fusion gene in dermatofibrosarcoma protuberans, KIT or PDGFR mutations in gastrointestinal stromal tumors (GIST)]. Among sarcomas with complex genetic profiles , a category characterized by intermediate complexity harbors few recurrent amplifications leading to oncogene co-amplifications (e.g., CDK4 and MDM2 co-amplified with 12q chromosome in well-differentiated/dedifferentiated liposarcomas). Highly complex sarcomas, including, among others, osteosarcomas and embryonal rhabdomyosarcomas, harbor multiple numerical and structural chromosomal aberrations with no specific pattern. In these tumors, recurrent genomic alterations identified with some frequency include inactivating mutations of tumor suppressor genes (e.g., TP53, NF1, RB1, PTEN) [3].
Traditionally, the different types of sarcomas have been treated in the same manner despite differences in histology and biology. Surgery, with or without radio- and chemo-therapy, is the critical management for local control. Treatment of metastatic disease, which develops in 40–50% of patients, remains a challenge. Systemic doxorubicin-based cytotoxic regimens have been the gold standard since early seminal observations by Bonadonna et al. of the anthracycline clinical activity in sarcomas in the late 1960s [4,5,6]. First-line treatment of GIST and dermatofibrosarcoma protuberans represents an exception as they have shown a peculiar sensitivity to the tyrosine kinase inhibitor imatinib which is able to block the oncogenic activation of KIT and PDGFRβ, characteristic of these tumors [7]. Although treatment response varies among the different histologies, a substantial proportion of patients derives no benefit from first-line chemotherapy or experiences recurrence. Over the last years, the treatment options in second-line and beyond have expanded for soft tissue sarcomas, being increasingly subtype-directed [8, 9]. In fact, recent clinical trials evidenced a selected activity of various drugs in specific histotypes with progression-free survival benefit. From these studies, a few drugs received approval for use in sarcoma subtypes. These included the tyrosine kinase inhibitors sunitinib, regorafenib (GIST, dermatofibrosarcoma protuberans) and pazopanib (soft tissue sarcomas), the DNA binding and multi-tasking trabectedin (liposarcoma and leiomyosarcoma) and the microtubule-targeting eribulin (liposarcoma). Several other histology-driven therapies are currently under investigation [5, 10, 11].
Next-generation sequencing technologies are increasingly applied in sarcoma translational research. These potent tools offer now the opportunity to discover molecular abnormalities in the different sarcomas subtypes improving our knowledge of the biology of these challenging diseases and potentially identifying new actionable alterations and genome-based drug targets [3, 12]. A few studies reported targetable pathways in genetically complex sarcomas. For instance, gain of function of IGF1R and PI3K/mTOR signaling pathways have been described in a subset of patients with osteosarcomas [13, 14] and mutations along the receptor tyrosine kinase/RAS/PI3K pathway have been identified as frequent in embryonal rhabdomyosarcomas [15]. Although clinical validation will be needed to assess safety and efficacy of new treatments derived from these studies, there is great hope that implementation of next-generation sequencing to guide therapeutic treatments will improve the outcome of patients with bone and soft tissue tumors in the next future [16].
Advances in understanding the pathogenesis of sarcomas have evidenced a crucial role for the tumor microenvironment. As in other solid tumors, the complex interactions between tumor cells and components of the microenvironment are essential for sarcoma growth and dissemination and influence the response to therapies [17]. Vascular invasion by tumor cells, as well as VEGF expression and circulating VEGF levels, have been identified as prognostic factors in several studies [18]. Elevated expression of other pro-angiogenic factors, such as PDGFB and FGF2, has been associated with a worse prognosis [19]. Mechanisms underlying the enhanced expression of angiogenic factors are tumor-type specific. In addition to the increased expression of hypoxia-inducible factor (HIF1α) that activates the transcription of VEGF during the angiogenic switch [20], specific genetic alterations have been associated with elevated expression of growth factors. In osteosarcomas, VEGF pathway genes have been found amplified [21] and high levels of the growth factor correlated with progression and poor survival [22, 23]. In Ewing’s sarcomas, VEGF-A and PDGF-C have been shown to be upregulated by the specific fusion oncoproteins EWS-ETS and EWS-FLI, respectively [24, 25]. Growth factors such as VEGF, PDGF, and FGF2 activate receptor tyrosine kinase pathways in sarcoma and stromal cells driving proliferation, survival, motility, and angiogenesis through paracrine/autocrine loops. In fact, most receptor tyrosine kinase inhibitors approved and under investigation in sarcomas are thought to exert their effects by acting on the stroma and directly on tumor cells [18].
Effects of sarcoma therapies on the innate immune system have also been described. For instance, imatinib was shown to induce NK cell response in GIST patients, and trabectedin was found to induce depletion of monocytes, including tumor-associated macrophages, in soft tissue sarcoma patients [26, 27]. The sarcoma immune microenvironment is still poorly characterized and, not surprisingly, appears to be highly variable. Inflammation, T cell infiltration, and checkpoint proteins expression are dependent on the tumor histotype [28, 29]. Immunotherapy , an area of intense investigation that has already revolutionized the standard of care in other tumors, is still in an early phase of clinical development for sarcomas. The main approaches that are being investigated involve checkpoint inhibitors and adoptive T cell therapy. Encouraging responses have been observed only in selected sarcoma sub-groups so far. However, studies exploring new immune system enhancing approaches are ongoing. In addition, a variety of combination strategies, aimed at improving efficacy and assessing the safety of immune-modulating therapies in different sarcoma subtypes are under clinical evaluation [11, 30].
2 Heparanase in Sarcomas
Heparanase enzymatic activity was first described in murine sarcoma cell lines, before its gene cloning, in the late 1980s. Early papers reporting heparan sulfate (HS) endoglycosidase activity in sarcoma cells also described a relationship with the cell metastatic potential (Table 15.1). In highly spontaneously metastasizing mouse cell lines of Rous sarcoma virus-induced fibrosarcoma, the enzyme activity was found 20 fold higher compared to non-metastasizing or normal counterparts [31]. Similarly, extracellular matrix (ECM) degradation by tumor cell lines derived by nickel-induced rat rhabdomyosarcomas was characterized by partial hydrolysis of HS. The ECM degrading activity of subclones representative of various metastatic degrees correlated with the ability to spontaneously metastasize to the lung from the primary s.c. tumor site, but not after i.v. injection. These findings suggested that additional tumor cell capabilities, such as adhesion to biologic supports, are relevant in determining lung homing and colonization [32].
Accumulating evidence indicates that secreted heparanase can exert local effects in the tumor microenvironment as well as systemic effects. Thanks to the latter feature, Shafat and colleagues demonstrated the possibility to quantify heparanase protein in human biological fluids by an ELISA method [33]. Elevated levels of heparanase were found in the plasma of 64 pediatric patients with hematological and solid tumors, including 15 sarcomas (7 osteosarcomas, 4 rhabdomyosarcomas, 4 Ewing’s sarcomas), compared with healthy controls. Evaluation of plasma levels after chemotherapy showed a correlation with response to treatment, although with a trend not statistically significant in the heterogeneous sarcoma subgroup, suggesting that heparanase could represent a potential tumor marker. The positive immunostaining in 5 out of 8 Ewing’s sarcoma biopsy specimens showed for the first time heparanase expression in human sarcoma [34]. The same group subsequently extended the immunohistochemical analysis of heparanase to a cohort of 69 Ewing’s sarcoma patients. Positive staining was found in all specimens. Notably, the intensity of staining, which was scored as strong in 51% of cases, correlated with patient age and tumor size, two parameters associated with worse prognosis in Ewing’s sarcoma. Correlation with metastasis, the main disease prognostic factor, could not be analyzed in this cohort due to low case number [35].
Heparanase expression was confirmed for the first time in rhabdomyosarcomas by Masola and colleagues. Human cell lines of both the alveolar and embryonal subtypes were found to express heparanase mRNA and protein, while enzyme activity was assessed in conditioned media. On the other hand, real-time PCR revealed a higher heparanase expression in 12 rhabdomyosarcoma biopsies compared to fetal skeletal muscle, and enzyme activity in plasma from 15 patients was significantly higher compared to healthy controls. The involvement of heparanase in rhabdomyosarcoma cell invasiveness was shown by gene silencing [36]. Subsequently, heparanase expression was confirmed in several cell lines from both soft tissue- and bone-sarcomas [37, 38].
Kazarin et al. [39] examined the expression of heparanase in biopsies from a heterogeneous cohort of 101 adult soft tissue sarcoma patients. Samples from primary tumors and metastases included malignant fibrous histiocytomas and sarcomas with no defined subtype histology, which together represented 50% of cases. Other histologies included liposarcomas, leiomyosarcoma, angiosarcomas, chondrosarcomas, and synovial sarcomas. Heparanase immunohistochemical staining indicated a large extent (> 50% of cells) in more than 95% of samples and overexpression in nearly 50% of cases including all subgroups. No correlation was found, however, with the risk of disease recurrence evaluated in 55 patients, or between the primary tumor and metastasis from the same patient evaluated in 10 cases. Unfortunately, the sample size was too small for any statistical analysis related to specific sarcoma sub-types. These findings highlighted the need to address the clinical significance of heparanase, and likely any tumor biomarker, in homogeneous sarcoma subtypes due to the high histological and molecular heterogeneity of these tumors [39].
As widely described in other sections of this Book, heparanase has multiple functions. Through its HS degradation activity, heparanase modulates structural and biochemical functions of HS proteoglycans (HSPGs) working in concert with them so that together they have been referred to as the heparanase/HSPG axis [40, 41]. As an endo-β-glucuronidase , heparanase participates in the complex biosynthetic/catabolic machinery, also including glycosyltransferases, sulfotransferases, and endosulfatases, which allow cells to finely control HS composition and sequence. Alterations of these HS modifying enzymes may profoundly affect the ability of HSPGs to interact with hundreds of growth factors, cytokines, chemokines, and several other structural and regulatory proteins, thereby influencing their multiple functions [42, 43]. Indeed, HS deregulation and alterations in HSPGs expression have been reported in several pathological conditions [44]. In cancer, they have been shown to influence both initiation and progression, regulating growth and survival, differentiation, angiogenesis, immune response, metastasis and response/resistance to a given drug treatment [reviewed in 45,46,47,48]. Several preclinical studies, focusing on cell-surface associated HSPGs, i.e., glypicans and syndecans, or HS metabolizing enzymes, evidenced subtype-specific roles in sarcoma pathobiology in keeping with the high histological and molecular heterogeneity of these tumors [41]. In most cases, however, the connection with heparanase expression has not yet been elucidated. For instance, glypican-5, overexpressed in rhabdomyosarcomas, was found to promote cell proliferation by enhancing signaling of heparin-binding growth factors such as FGF2, HGF, Wnt and Hedgehog (Hh) ligands [49, 50]. Hh signaling is thought to play an oncogenic role in rhabdomyosarcomas [51, 52]. Li and collaborators [50] demonstrated that glypican-5 participates in activation of Hh signaling by promoting the interaction of the Sonic Hh ligand with its receptor Patched (Ptc1). By using a non glycanated glypican-5 mutant, the authors demonstrated that the HS chains are essential for binding of both the ligand and receptor [50]. Interestingly, another member of the glypican family, glypican-3, also expressed in rhabdomyosarcomas [53], exerts an opposite role in the regulation of Hh signaling by competing with Ptc1 for Sonic Hh binding. Differently, from glypican-5, glypican-3 binds only the Hh ligand, mostly at the core protein [54]. The cooperation between glypican-5 and the Hh signaling in supporting sarcomagenesis also emerged in a comprehensive transcriptome analysis of a human mesenchymal stem cell line performed at various stages during the gradual transformation to sarcoma upon prolonged culture. At late stages, both glypican-5 and Ptc1 were found significantly overexpressed and co-localized. Moreover, silencing of the HSPG by RNA interference reduced cell proliferation [55]. In another study, heparanase and Hh pathway components, Ptc1, Smoothened, and glioma-associated oncogene homolog-1, were analyzed in a series of 23 alveolar orbital rhabdomyosarcomas by immunohistochemistry and nested RT-PCR. Consistent results with both techniques showed elevated expression of either heparanase or the Hh signaling components compared with normal muscle. In contrast, expression levels in samples from patients that underwent preoperative chemoradiotherapy were not significantly different from the normal tissue [56]. Although a role for heparanase in regulating Hh signaling has already been proposed in another tumor, i.e., medulloblastoma [57], mechanistic links with deregulated expression and functions of Hh components and glypican-5 have yet to be elucidated in rhabdomyosarcomas.
In the next sections, we review the literature reporting on the biological significance of heparanase expression and function in specific sarcoma sub-types .
3 Bone Sarcomas
Bone-forming tumors are benign or malignant neoplasms defined by neoplastic cells that differentiate along the lines of osteoblasts, and able to secrete the organic components of bone, which in turn may or may not mineralize [58]. They are heterogeneous tumors characterized by a broad spectrum of biological behaviors ranging from indolent to very aggressive with a rapidly fatal outcome. The three most common forms of primary bone tumors are osteosarcoma, Ewing’s sarcoma, and chondrosarcoma. Whereas osteosarcoma and Ewing’s sarcoma, mainly affecting adolescents and young adults, exhibit a high propensity to metastasize to the lungs, chondrosarcoma, more frequently observed after the age of 40, is characterized by a high frequency of local recurrence. The combination of chemotherapy, surgical resection and radiotherapy have contributed to improving patients’ outcome. Nonetheless, refractory and metastatic bone sarcomas remain lethal.
The occurrence of bone sarcomas in the context of rare hereditary disorders has provided unequivocal evidence of the relevance of mutations of genes coding for HSPGs (e.g., Glypican 3 in Simpson-Golabi-Behmel syndrome) or HS biosynthetic enzymes (e.g., exostins (EXTs) in Multiple Osteochondroma (MO) syndrome) in promoting and sustaining neoplastic growth. Emerging evidence indicates that the machinery involved in bone development and homeostatic processes, including angiogenesis which is intimately coupled with osteogenesis through reciprocal crosstalk [59], can be recruited and hijacked by neoplastic cells. Here, we summarize studies addressing the involvement of HSPGs and their synthesizing and modifying enzymes, with particular reference to heparanase, in bone physiology and disorders, focusing on the pathobiology of chondrosarcoma and osteosarcoma.
3.1 HSPGs and Heparanase in Bone Development and Biology
Bone is a specialized connective tissue composed of bone forming cells , the osteoblasts, deriving from mesenchymal stem cells, and bone resorbing osteolytic cells, the osteoclasts, considered as highly specialized macrophages derived from the monocyte lineage [60,61,62]. The formation of bone proceeds broadly via two types of processes. The intramembranous ossification, characteristic of flat bones, occurs through the differentiation of mesenchymal progenitor cells that proliferate and then differentiate into osteoblasts producing an osteoid matrix which undergoes calcification. The endochondral ossification , characteristic of appendicular skeleton and vertebral column, develops through an intermediate cartilaginous process. The progenitor cells in the growth plate, a highly organized structure driving long bone elongation, differentiate into chondrocytes that secrete a cartilaginous matrix. Then, the chondrocytes undergo hypertrophy and secrete proangiogenic factors to promote blood vessel formation and influx of mesenchymal progenitors which differentiate into chondroclasts, osteoclasts and osteoblasts. The cartilage template is then degraded by chondroclasts and replaced by a mineralized matrix synthesized by osteoblasts. The development of bone requires coordination between cell-cell, cell-matrix, and growth factor-mediated signaling to achieve ossification and mineralization [63]. In particular, osteoblastic differentiation requires ordered presentation and balance of several growth promoting elements including circulating molecules (e.g. growth factors, cytokines) and tissue architecture-related signals (cell-cell contact and cell adhesion) which share HS as a major co-factor [63,64,65]. In turn, osteoblasts produce many crucial mitogenic and adhesion factors that bind extracellular HS chains. The bone presents a highly specialized microenvironment and, although collagen is the prevalent organic component, HSPGs represent the most bioactive elements of the developing matrix. Actually, during osteogenesis, expression and temporal changes in HSPG structure (e.g. HS sequence and length variation, critical positioning of sulfate groups) are instrumental in the concerted signaling flow of molecules coordinating mesenchymal stem cells growth/commitment and, ultimately, the osteoblast phenotype [63, 66]. In fact, HSPGs interact with a wide number of bioactive molecules with a central role in osteogenesis including Hhs, FGFs and their receptors, bone morphogenic proteins (BMPs), as well as collagens, laminins, and fibronectins. As HS interacting abilities “follow HS structure”, the activity HSPG biosynthetic and modifying enzymes, including heparanase, may critically influence the signaling triggered by HS-binding molecules [63, 65,66,67].
Several lines of evidence support a relevant, although not yet fully elucidated, role of heparanase in bone formation and remodeling . Depending on the cellular context, the cell differentiation status and the surrounding microenvironment, heparanase has been associated with the osteogenic or osteolytic process. In the bone microenvironment , it has emerged as a relevant endogenous factor playing crucial functions in cell-cell communication and cell differentiation through modification of HSPGs and modulation of gene expression.
Saijo et al. [68] described sequential changes of heparanase and VEGF expression during endochondrial ossification in a model of fracture repair in mice. Heparanase, highly expressed in osteo(chondro)clasts at the chondro-osseous junction in the growth plate (physiological condition) and in the fracture callus (pathological condition), was suggested to promote fracture repair by recruiting VEGF into the local microenvironment and then osteoclast precursors and osteoprogenitors. Kram et al. [69] described the expression of heparanase in osteoblastic cells and its ability to stimulate bone formation and mass. Progressive increasing expression of heparanase mRNA was observed in murine bone marrow stromal pre-osteoblast MC3T3-E1 cells undergoing osteoblastic differentiation in osteogenic medium, whereas heparanase was undetectable in MC3T3-E1 cells incubated in non-osteogenic medium. In contrast, heparanase transcript, abundantly present at the monocytic stage of osteoclastogenic cultures, was found markedly decreased in cultures at an advanced stage of differentiation, suggesting downregulation of the enzyme during osteoclastogenesis. Notably, ex vivo bone marrow stromal cells derived from transgenic mice overexpressing human heparanase (hpa-tg mice), or MC3T3-E1 cells exposed to soluble human heparanase, spontaneously underwent osteogenic differentiation even in absence of osteogenic medium. These findings demonstrated the ability of heparanase to directly induce osteogenic differentiation and stimulate osteoblast activity. Moreover, observation of the skeletal phenotype of wt vs hpa-tg mice supported a positive regulation of bone formation by the heparanase-HSPG system as the transgene caused a marked increase of trabecular bone mass and cortical thickness. In this model, stimulation of bone formation was independent of the proangiogenic function of heparanase but likely related to its ability to regulate availability and activity of HS-binding proteins (e.g. VEGFs, FGFs) directly implicated in the control of osteoblast number and functions. Conversely, hpa-tg mice-derived bone marrow cells that underwent osteoclastic differentiation following stimulation with M-CSF and RANK, displayed an increased osteolytic activity with respect to the cells derived from wt animals [70]. These findings highlighted the relevance of the microenvironment in influencing heparanase functions and bone marrow cell behavior. Studies examining the expression of HSPGs and related enzymes in MC3T3-E1 cells undergoing osteoblastic differentiation provided insights into the temporal, structural and functional changes in HSPGs during osteogenesis [66, 71]. Proliferating cells (day 5) displayed a high level of HSPGs, mainly glypican-3 known to promote FGF- and BMP-mediated mitogenic signaling [62]. In this experimental model, the active production of HSPGs was associated with increased expression of HS synthetic enzymes (i.e. glycosyltransferases EXTs, HS N-deacetylase/N-sulfotransferases NDSTs, 2- and 6-O sulfotransferases), highlighting the need for longer, more sulfated and complex HS chain bound to a variety of HSPG core proteins to sustain the growth process. During the shift of MC3T3-E1 cells from a proliferative to a differentiated status (day 14), a progressive reduction of HS chain complexity was observed. Indeed, in cells fully committed to osteogenic differentiation, the production of short and highly sulfated HS chains correlated with increased expression of NDST-1 and glypican-3 protein core. Thus, osteogenically committed cells likely need the production of fewer, short and homogeneous, but more highly sulfated HS side chains to mediate specific growth factor signals to switch from proliferation to differentiation [71]. Mineralizing MC3T3-E1 cells (day 20) were characterized by the presence of short and less sulfated HS and high expression levels of heparanase. Moreover, these cells exhibited increased expression of syndecan-2, a HSPG involved in bone ECM deposition and tissue consolidation [66, 72]. Overall, these findings indicated a different HSPG profile and a systematic HS variability with more complex sugars made during the MC3T3-E1 cell growth process compared to the subsequent phases of osteogenic differentiation characterized by intense HSPG turnover likely bolstered by higher levels of heparanase.
Glypican-3 was demonstrated to mediate MC3T3-E1 cell commitment toward osteogenesis by inducing the osteogenic transcription factor Runx2 [71]. Complex crosstalk has been described between Runx2 and the FGF2/HSPG axis which forms an ECM-regulated feedback loop controlling osteoblast proliferation and differentiation [73]. Signaling mediated by FGFs is fundamental for bone development [74]. Indeed, disruption of the FGF2 gene in knock-out mice resulted in decreased bone mass whereas mutations in FGFRs are responsible for several clinically distinct craniosynostosis syndromes in humans [75, 76]. Reintroduction of Runx2 in mouse calvaria Runx2-null osteoprogenitor cells was reported to markedly increase expression of genes related to FGF2/HSPGs axis (e.g. FGFR2 and FGFR3, syndecan −1, −2, −3, glypican-1) [73]. In addition, the transcription factor increased expression of EXT1 and heparanase and altered the relative expression of NDSTs and O-sulfotransferases. As HS structural diversities determined by saccharide sequence, sulfation degree and pattern are known to affect FGF/FGFR signaling outcome [67, 77], Runx2 indirectly changed osteoprogenitor responsiveness to FGF2 during the transition from active proliferation to growth arrest. In turn, FGF2 and HS from differentiating MC3T3-E1 cells stimulated Runx2 expression [78].
By a combination of ex vivo and in vitro approaches, along with pharmacological inhibition of heparanase by the phospho-sulfo-mannan PI88, Brown et al. [79] investigated the contribution of the enzyme in long bone formation in developing mice. High expression levels of heparanase mRNA and protein were detected in perichondrium, periosteum and at the chondroosseous junction, sites of crucial signaling events regulating bone length and width. Moreover, experiments performed in the murine chondrogenic cell line ATDC5, suggested that heparanase activity was strictly titrated at the transition from chondrogenesis to osteogenesis. A biphasic pattern of heparanase expression was also observed during the osteogenic differentiation timeframe (0–21 days) of rat marrow stromal cells [80]. Protein and mRNA expression levels reached a peak on days 10 and 14, respectively, followed by a gradual decline. Notably, consistent with a declined osteogenic differentiation ability over the age, heparanase expression in osteogenic differentiated marrow stromal cells from aged rats was weaker compared with that from young rats.
3.2 HSPGs and Heparanase in Bone Disorders
Smith et al. [81] described for the first time the expression and function of heparanase in human primary osteoblasts and found lower levels of expression and activity in human osteoporotic osteoblasts from bone fragments compared to the cells from healthy subjects. The significant correlation found between the decrease in heparanase mRNA expression and the activity of the bone turnover marker alkaline phosphatase in osteoporotic osteoblasts was consistent with the downregulation of several osteogenic genes (e.g. VEGFA, FGFR2, COL15A1, BMP3). Osteoblasts exposed to exogenous heparanase displayed increased levels of histone H3 phosphorylation at Ser 28, a modification coupled with the induction of transcription of immediate-early genes [82]. These findings suggested a direct involvement of HPSE in human osteoblastogenesis through histone H3 modulation and epigenetic regulation of osteogenic gene expression.
Heparanase has also been implicated in cartilage disruption and subchondral bone remodeling occurring in human osteoarthritis, a characteristic adult disease state of cartilage. Gibor et al. [83] described heparanase expression and enzymatic activity in adult human osteoarthritic cartilage and suggested a contribution of the enzyme in the pathologic interactions between the chondrocytes and their pericellular matrix. In fact, the addition of exogenous heparanase to cultured human primary chondrocytes induced the expression of the metalloproteinases MMP13 and ADMTS4, acting as ECM catabolic enzymes, and downregulated anabolic genes (i.e., aggrecan core ACAN and COL2A1). The effect on catabolic gene products, partially mediated by FGF2 signaling, was reverted by treatment with the heparanase inhibitor PG545. This observation is consistent with findings in multiple myeloma models evidencing that secretion of heparanase, along with other molecules promoting matrix degradation, enhances bone destruction within the tumor microenvironment [84] (Sanderson et al., Chap. 12 in this volume).
Recently, in apparent contrast with previous findings, Chanalaris et al. [85] did not find differential expression of heparanase mRNA in human knee cartilage from osteoarthritic donors with respect to specimens from normal subjects. Nonetheless, osteoarthritic cartilage samples showed a marked dysregulation of the expression of HS biosynthetic and modifying enzymes with increased expression of the EXT glycosyltransferases, the glucuronyl epimerase GLCE, and the sulfotransferase HS6ST1. Also, HS6ST1 was demonstrated to boost FGF2-ERK signaling in human chondrocytes. Overall, these findings support the involvement of highly dynamic modulation of HS structure and function in the regulation of bone formation under both physiological and pathological conditions .
3.3 HSPGs and Heparanase in Osteochondromas and Chondrosarcomas
Chondrosarcomas constitute a heterogeneous group of malignant bone tumors, characterized by the production of cartilage matrix and displaying different histopathology and clinical behaviors. Following osteosarcoma, chondrosarcoma is the second most frequent primary malignancy of the bone [86]. Conventional chondrosarcomas are typically low or intermediate grade and are characterized by indolent clinical behavior and low metastatic potential, whereas high-grade chondrosarcomas (5–10%) are associated with high metastatic potential and poor prognosis. Localized chondrosarcomas are generally well managed by surgery. Chondrosarcomas are inherently resistant to chemo- and radio-therapy due to low mitotic fraction, activation of multidrug resistance pumps, and limited drug penetration into the tumor microenvironment characterized by poor vascularity and abundant hyaline-dense ECM. Conventional chondrosarcomas occur either de novo in the bone medulla or arise, as secondary tumors, from preexisting benign cartilage lesions, named enchondromas and osteochondromas, during periods of bone growth in a site adjacent to the growth plate [87, 88]. Enchondromas can develop central chondrosarcoma whereas osteochondromas can be precursors of peripheral chondrosarcomas. Enchondromas arise within the metaphyseal portion of the bone. Osteochondromas that appear as cartilage-capped bony neoplasms on the outer surface of bones, can occur as sporadic/solitary or as multiple lesions in the context of hereditary Multiple Osteochondroma (MO) syndrome. MO is an autosomal dominant disorder characterized by short stature, skeletal deformities and the formation of osteochondromas (exostoses). This syndrome represents an interesting model of oncogenesis driven by complex deregulation of HSPG synthesis and metabolism [65, 87, 89, 90]. Loss-of-function mutations of the tumor suppressors EXT genes have been identified in both sporadic and MO osteochondromas , although associated with different gene alterations and mechanisms [87, 91, 92]. EXT1/2 glycosyltransferases function in hetero-oligomeric complexes to polymerize HS chain. Loss of either enzyme causes a total deficit of HS chains resulting in embryonic lethality. Specific EXT mutations are considered early-stage molecular alterations able to increase the proliferative capacity of normal chondrocytes. In fact, by causing reduction/lack of HS, or HSPG mislocation, EXT mutations produce a deep perturbation of signaling pathways tightly implicated in the regulation of chondrocyte proliferation/differentiation, such as Indian Hh, BMP, and FGF pathways. McCormick et al. [93] demonstrated that EXT1 mutation caused aberrant processing and cytoplasmic accumulation of HSPG resulting in abnormal diffusion of Hh ligands in the extracellular environment at the growth plate. Absence of HS and intracellular accumulation of syndecan-2 and CD44v3 HSPGs were also observed in the osteochondroma and peripheral chondrosarcoma cartilage [94]. In mice carrying EXT1 mutation, a reduced amount of HS potentiated Indian Hh signaling resulting in delayed hypertrophic differentiation and increased chondrocyte proliferation [95]. Moreover, an increased diffusion area of Indian Hh was supposed to produce a loss of polar organization allowing chondrocytes to growth in the wrong direction. Additional molecular insights revealed that a somatic “second hit”, likely complementing germline EXT mutations to further decrease HS production, is required for osteochondroma development. Actually, loss-of-heterozygosity, aneuploidy, and other large genomic changes can render local resident cells EXT1- or EXT2-null [96]. Further clinical observations and experimental data from mouse models add levels of complexity in the scenario of osteochondroma formation and its potential evolution towards peripheral chondrosarcoma. The observed heterogeneous distribution of HS-positive and -negative cells in murine and human osteochondromas paved the basis for a “niche-based” model of oncogenesis implicating both cells with homozygous inactivation of EXT genes and wild type cells in shaping osteochondroma [92, 97, 98]. The EXT-negative cells present in the osteochondromas would create an extracellular mutation-promoting environment favoring the acquisition of late-stage mutations (e.g., p53, Rb) in EXT-positive cells retaining one or both copies of EXT genes. Such alterations, occurring in EXT-positive cells likely endowed with stem-like genotype, would provide a proliferative advantage over the osteochondroma EXT-null cells [92, 99]. Thus, osteochondromas would serve as a niche which facilitates the committed stem cells/EXT wild type chondrocytes to acquire genetic changes to develop malignant secondary peripheral chondrosarcomas .
Heparanase has been recently defined as an important culprit coupled with EXT loss in Multiple Osteochondroma (MO) [92]. Early studies by Trebicz-Geffen et al. [100] provided the first evidence of higher levels of heparanase in specimens and cell cultures from MO patients compared with solitary exostoses and healthy subjects, suggesting that increased HS degradation, in addition to reduced synthesis by EXT loss of function , could contribute to HS low levels in MO. Increased expression of heparanase was also described in tumor cartilage from MO by Yang and colleagues [101]. Huegel et al. [102] evidenced, by immunohistochemical staining, the presence of heparanase in all chondrocytes within the exostoses and hypothesized that, in the MO syndrome context, the endoglycosidase upregulation results from a feedback mechanism triggered by EXT inactivation-induced modulations in HS levels [102]. Consistently, treatment of primary mesenchymal cells with the HS antagonist Surfen significantly increased heparanase level. Although somewhat paradoxical and counterintuitive, heparanase plays a concurrent role in stimulating chondrogenesis by further decreasing the levels of HS. Incubation of ATDC5 chondrogenic cells with human recombinant heparanase was found to promote cell migration, proliferation, and differentiation. Coherently, in vitro chondrogenesis was significantly counteracted by the heparanase inhibitor, glycol-split heparin, roneparstat (= SST0001) [102] (Noseda and Barbieri, Chap. 21 in this volume). These findings are in accordance with the inverse relationship between EXT and heparanase expression reported in several types of cancer cells [92]. Overexpression of heparanase was detected in 5/7 specimens from human chondrosarcoma patients although the tumor subtype was not reported [39].
An additional study from Presto et al. [103] showed that NDST1 directly interact with EXT2 during HS chain formation and that EXT mutations can modulate expression/function of NDST1 thus affecting both HS polymerization and sulfation. By combining different analytic methods, Veraldi and colleagues [104] investigated the structural complexity of HS from human EXT-mutant MO and peripheral chondrosarcoma specimens compared with HS from prepubescent growth plate and fetal cartilagineous samples. Most pathologic samples of both osteochondromas and peripheral chondrosarcomas displayed HS characterized by higher sulfation degree compared with other samples. In line with this observation, a positive correlation was found between enhanced expression of the 6-O sulfotransferases HS6ST1 and HS6ST2 and histological grade of chondrosarcoma, pointing to a relevant role for HS 6-O sulfation in disease progression [105, 106].
Interestingly, central chondrosarcomas, devoid of EXT alterations, are distinct genetic entities with respect to peripheral chondrosarcomas; they were shown to exhibit aberrant cytoplasmic accumulation of HSPG (e.g., CD44v3 and syndecan-2) and deregulated Indian Hh signaling [107]. Aberrant localization of HSPGs was also observed in low-grade variant of clear cell chondrosarcoma as well as in aggressive mesenchymal and dedifferentiated subtypes [108]. These observations highlight deregulation of HSPGs as a common feature in bone cartilage tumors although the underlying molecular mechanisms have not yet been elucidated .
3.4 HSPGs and Heparanase in Osteosarcomas
Osteosarcoma , the predominant form of bone cancer primarily occurring in children and adolescents, preferentially arises in the long bones near the metaphyseal growth plates [60, 109]. Osteosarcoma is mostly sporadic but a greater incidence is observed in subjects with Page’s disease of bone, after therapeutic radiation, and in certain cancer predisposition syndromes (e.g., Li-Fraumeni). Current therapies integrate surgery and combinatorial chemotherapy resulting in cures in about 70% of non-metastatic patients. Unfortunately, an overall 5-year survival rate of about 20% is reported for patients with metastatic or relapsed disease [60]. Etiological factors and pathogenic mechanisms underlying osteosarcoma development are very complex and not yet fully elucidated. The challenging genomic complexity and instability, along with intratumoral and intertumoral heterogeneity makes very hard the identification of drivers as well as vulnerabilities for novel effective therapeutic approaches. The “multiple drivers” hypothesis pointed out the contribution of both first drivers (e.g., p53, Notch1, Ptc1) and synergistic drivers (e.g., Rb1, PTEN) in osteosarcomagenesis. According to this hypothesis, the nature and the number of alterations deeply impact the onset, the latency, and progression of this malignancy [109]. Recently, novel types of genetic abnormalities were described including chromothripsis (i.e., a phenomenon in which a single catastrophic event results in massive genomic rearrangements and remodeling of chromosomes) and kataegis (i.e., a pattern of localized hypermutation colocalized with regions of somatic genome rearrangements) [109].
Osteosarcomas are composed of malignant osteoblasts producing immature bone and osteoid tissue, an organic mineralized matrix primarily composed of collagen I [60]. Various hypotheses have implicated among osteosarcoma originating cells, mutation-harboring mesenchymal stem cells, osteoblast-committed cells undergoing defective differentiation, and/or osteocytes [110]. In mesenchymal stem cells, the inactivation of p53, frequently disrupted in these tumors, was shown to promote early osteogenesis by accelerating osteoblastic differentiation while impairing osteocyte terminal maturation [111]. On the other hand, osteoblasts from pluripotent stem cells derived from Li-Fraumeni patients, harboring mutant p53, were able to recapitulate in vivo osteosarcoma features [112]. Also, murine osteocytes immortalized by SV-40, inactivating p53, were shown to originate osteosarcomas [113]. Taking into consideration the osteosarcoma high heterogeneity, it is likely that all three cell types can contribute to osteosarcomagenesis.
Defective osteogenic differentiation resulting from deregulation of Hh, Notch, Wnt, and BMP signaling pathways and overactivation of several growth factors/receptor tyrosine kinase axes (e.g. VEGF/VEGFR, IGF1/IGF1R, PDGF/PDGFR), have been involved in osteosarcoma development [60]. Deregulation of HSPGs and related enzymes that could greatly affect these signaling pathways contributing to osteosarcomagenesis, were described in several reports. For instance, a strong expression of syndecan-2 was found in mature osteoblasts, whereas low levels were observed in osteosarcoma cell lines [72, 114]. Syndecan-2 exogenous expression in U2OS osteosarcoma cells decreased migration/invasion and chemoresistance suggesting an oncosuppressive role for this HSPG. Consistently, syndecan-2 levels were found higher in bone tumors of patients responding to chemotherapy with respect to non-responders [114]. Conversely, increased expression of syndecan-4 in high-grade osteosarcomas was associated with large tumor size and distant metastases [115]. The HS 6-O-sulfatase SULF2 was shown to be a direct transcriptional target of p53 in several cancer cell lines including U2OS cells [116]. Importantly, p53 has been shown to directly bind heparanase promoter inhibiting its activity, whereas mutant p53 variants failed to exert an inhibitory effect [117]. Several preclinical and clinical studies have correlated heparanase expression with aggressive tumor phenotype [see Chap. 1]. High heparanase expression was detected by immunohistochemistry in 37/51 osteosarcoma specimens with protein expression levels correlating with poor response to chemotherapy, metastasis occurrence, and poor survival rate. Moreover, multivariate analyses revealed the protein overexpression as a significant independent risk factor for distant metastasis [118]. High levels of heparanase were also detected in plasma samples from pediatric cancer patients including 7 patients suffering from osteosarcomas [34]. Zeng et al. [119] confirmed the expression of heparanase in 51% of human osteosarcoma biopsies and found a significant correlation with tumor size. Moreover, these authors noted that 40% of the samples were positive for both heparanase and HIF1α. The expression of both proteins correlated with the presence of lung metastasis and poorer patients’ survival, suggesting functional cooperation in promoting angiogenesis and tumor progression. Actually, in U2OS cells, heparanase silencing by shRNA decreased expression of HIF1α and reduced cell proliferation and migration/invasion. Likewise, proliferation, adhesiveness, and invasiveness of the human osteosarcoma cell line MG63 were significantly inhibited by heparanase silencing [120]. In cell lines derived from FBJ virus-induced mouse osteosarcoma, heparanase expression was found associated with a high metastatic potential [121]. Cell surface expression of HS was found significantly higher in poorly metastatic FBJ-S1 cells with respect to the FBJ-LL highly metastatic cells consistently with lower levels of both heparanase and EXT1 expression. Moreover, the authors demonstrated by molecular approaches that FBJ-S1 cell motility was regulated by heparanase, under EXT1 control. In U2OS and SAOS osteosarcoma preclinical models, treatment of mice harboring tumor xenografts with the heparanase inhibitor Roneparstat induced a significant antitumor activity providing preclinical proof of principle that targeting heparanase could represent a valuable therapeutic approach in this malignancy [37].
4 Targeting Heparanase in Sarcomas
Several lines of evidence , discussed in details in other sections of this Book, highlight the implication of heparanase in critical processes of tumor biology (e.g., growth, angiogenesis, metastasis, drug resistance) and its upregulation in the vast majority of malignancies examined, including carcinomas and hematological tumors as well as sarcomas. Such evidence, and the favorable feature of being the only HS degrading endoglycosidase, not substitutable with other enzymes, has supported the idea that heparanase could be a suitable target and promoted the development of heparanase inhibitors as anticancer therapeutics [46, 122] (Chhabra and Ferro; Hammond and Dredge; Noseda and Barbieri, Naggi and Torri; Giannini et al., Chaps. 19, 20, 21, 22 and 23 in this volume). A few studies tested the potent heparanase inhibitor roneparstat (100NA,RO-H, ST0001) (Noseda and Barbieri, Naggi and Torri, Chaps. 20 and 21; in this volume), a chemically modified non-anticoagulant heparin, in preclinical models of human sarcomas [41]. The first sarcoma model applied in these studies was Ewing’s sarcoma [35], a natural choice as roneparstat had previously been shown effective in multiple myeloma models [123, 124]. Ewing’s sarcoma is an aggressive tumor that mainly develops in bones, sharing with multiple myeloma a functional microenvironment characterized by complex interactions between cellular components (tumor cells, osteoclasts and other stromal cells), humoral factors (growth factors and cytokines) and the ECM which provides a favorable “niche” for tumor growth and progression [125, 126]. The biological phenotype of both tumors has been shown to be influenced by signaling pathways mediated by growth factors (e.g. IGF-1, PDGF, bFGF, VEGF) some of which are transcriptional targets of the oncogenic fusion protein EWS-FLI1 which drives tumorigenesis in Ewing’s sarcoma [127, 128]. In studies on multiple myeloma, the cooperation between heparanase and the HSPG syndecan-1 was demonstrated to regulate the functions of several growth factors in the bone niche, promoting myeloma cell growth, angiogenesis, and metastasis, effects that were counteracted by roneparstat [123, 124, 129]. Similarly, the glycol-split heparin effectively inhibited TC71 Ewing’s sarcoma cell invasion stimulated by VEGF and bFGF through Matrigel, a reconstituted basement membrane highly rich in HSPGs. Moreover, roneparstat induced a strong antitumor effect in mice harboring TC71 tumor xenografts with 25% of cures noted in treated animals [35].
In a subsequent report, investigation of the effects of roneparstat was extended to a panel of six human pediatric sarcoma models including bone (osteosarcoma , Ewing’s sarcoma) and soft tissue (rhabdomyosarcomas, rhabdoid tumor) histotypes with simple or complex genotype [37]. The study confirmed the ability of the heparin derivative to abrogate cell invasion induced by heparin/HS-binding growth factors (PDGF, bFGF, VEGF, HGF). Moreover, a marked drug inhibitory effect on the release/secretion of several angiogenesis-related molecules was reported (e.g., VEGF, MMP-9). All sarcoma cell lines growing in mice as tumor xenografts were responsive to roneparstat antitumor effect with maximum tumor growth inhibition (around 90%) obtained in the genetically simple sarcoma models (i.e., TC71 Ewing’s sarcoma, RH30 alveolar rhabdomyosarcoma, and the A204 rhabdoid tumor previously misclassified as a rhabdomyosarcoma). Combination treatments with roneparstat and antiangiogenic agents, the anti-VEGF antibody bevacizumab and the tyrosine kinase inhibitor sunitinib, were shown to significantly increase the antitumor efficacy compared to single-agent therapies in the Ewing’s sarcoma model.
An additional study by Cassinelli and colleagues addressed the impact of roneparstat treatment on sarcoma cell signaling [130]. As HS mimetic, roneparstat can act as a multi-target agent inhibiting heparanase and competing with HS in their broad regulatory functions. Overall, these effects are expected to influence growth factor signaling in both tumor and stromal cells. Focusing on RTKs-mediated signaling, the authors applied a multiplexed phosphoproteomic approach to investigate the effects of drug treatment on receptor activation in sarcoma cells comprising various histotypes. Roneparstat was found to inhibit in a context-dependent manner growth factor/receptor tyrosine kinase axes implicated in sarcoma pathobiology, and inhibition was further validated by cellular functional assays. In vivo, reduced activation of EGFR, ERBB4, InsR, and IGF1R in tumor xenografts from treated mice confirmed the drug pharmacodynamic effect. The good tolerability of roneparstat evidenced in preclinical tumor models suggested that it could be used in combination with conventional cytotoxic drugs. The combination with the camptothecin irinotecan, a drug of clinical interest in pediatric sarcoma patients, was well tolerated and highly effective in the A204 rhabdoid tumor xenograft significantly enhancing tumor growth inhibition, complete responses, and cures as compared to single drugs administration. A204 cells are characterized by constitutive high activation of PDGFRα which support rhabdoid tumor growth but is not directly implicated as a driver of malignant transformation. Early characterization of roneparstat activity in tumor models evidenced its antimetastatic potential against experimental metastases induced by intravenously injected B16 murine melanoma cells [131]. Using the orthotopic A204 rhabdoid tumor that metastasizes from the primary xenograft site to the lung, Lanzi and colleagues confirmed the antimetastatic activity of the heparin derivative in a human sarcoma model of spontaneous dissemination (unpublished, Fig. 15.1). Notably, the heparin derivative was also able to counteract malignant transformation driven by the COL1A1/PDGFB fusion oncogene generated by chromosomal translocation in dermatofibrosarcoma protuberans [130]. In this sarcoma, the constitutive activation of PDGFRβ is induced through an autocrine loop supported by the functional PDGFBB produced by processing of the chimeric oncoprotein [132].
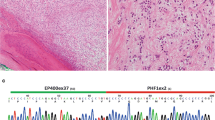
Inhibition of spontaneous lung micrometastases from orthotopic A204 rhabdoid tumor by the heparanase inhibitor roneparstat. A204 cells were injected i.m. in SCID mice. Roneparstat was administered s.c at 60 mg/kg (twice/day, 5 days/week) for 6 weeks. After treatment interruption, mice were sacrificed when primary tumors had similar volumes. Lungs were formalin fixed and paraffin embedded. Sections were subjected to immunohistochemistry with anti-human vimentin antibodies and positive spots were quantified. ∗, P < 0.05 by Student’t-test
Another heparin derivative, the supersulfated low molecular weight heparin ssLMWH with high anti-heparanase activity, was tested in human synovial sarcoma models [38]. ssLMWH inhibited anchorage-independent growth in soft agar and invasion in Matrigel of synovial sarcoma cells. Moreover, it downregulated the activation of several receptor tyrosine kinases. In cells with elevated constitutive activation of IGF1R, a strong synergistic effect was shown with the dual IGF1R/InsR tyrosine kinase inhibitor BMS754807. Previous studies have associated IGF1R expression with a high incidence of metastases in synovial sarcoma [133] while its activation has been shown to be promoted by the IGF2 ligand whose transcription is epigenetically induced by the SS18-SSX fusion oncoproteins peculiar of these sarcomas [134]. Despite a complete inhibition of IGF1R and InsR, BMS754807 did not achieve effective inhibition of downstream signaling pathways in synovial sarcoma cells, likely due to bypass resistance pathways. In contrast, the combined treatment with BMS754807 and ssLMWH enhanced inhibition of both AKT and ERK signaling which resulted in apoptosis induction and suppression of cell motility in vitro. An impressive effect was also obtained in vivo by the drug combination that abrogated the orthotopic growth of synovial sarcoma xenografts and their spontaneous dissemination to the lungs.
Similarly to heparin derivatives, DMBO, designed as mimetic of the pyranosidic ring structure of HS, was found to bind growth factors and cytokines (i.e. VEGF, HB-EGF, TNF-α) and to inhibit heparanase catalytic activity. In in vitro assays, the oxazine inhibited osteosarcoma cell proliferation, migration, and invasion. In vivo, it was able to inhibit liver experimental metastases induced by intravenously injected cells [135].
The dual nature of heparin derivatives and the oxazine DMBO, as heparanase inhibitors and HS mimetics, hampers a precise mechanistic interpretation of their biological effects [136]. Two recent reports described new small molecule inhibitors of heparanase enzymatic activity [137, 138] (Giannini et al., Chap. 23 in this volume). The best compounds in these series showed inhibitory effects similar to those observed with roneparstat and ssLMWH on invasion of rhabdoid tumor and synovial sarcoma cells and the expression of proangiogenic factors in osteosarcoma cells. These findings suggest that inhibition of heparanase endoglycosidase activity is shared by agents belonging to different chemical classes.
5 Concluding Remarks
Sarcomas are characterized by an aggressive phenotype, angiogenesis, and propensity to metastasize primarily to the lung. Identification of specific vulnerabilities has been successful only in a few histologies. For most patients with advanced disease, survival rates with available systemic therapies (i.e., conventional cytotoxic, new targeted and histology-driven) remain low, while immunotherapy is still in early clinical phases. New therapeutic approaches able to counteract sarcoma progression and improve patients’ outcome are highly desirable.
Since early reports describing the detection of an endoglycosidase able to produce biologically active HS fragments in sarcoma cells, heparanase was associated with the cell metastatic potential. The emerging role of heparanase in bone formation and remodeling during development suggests that its multiple functions in cooperation with HSPGs can be hijacked by bone sarcoma cells and exploited to promote cell signaling, angiogenesis, and dissemination. Likewise, aberrant cooperation of heparanase with other HS modifying enzymes appears to participate in the pathogenesis of cartilaginous tumors through a complex and still incompletely understood interconnection between HS, heparanase, the heparanome, and the transcription machinery. Several aspects of heparanase deregulation and pathological functions in sarcomas remain to be elucidated including the relationship with oncogenic players and molecular pathogenesis in the various histological subtypes.
Whereas most studies examining the clinical significance of heparanase in human malignancies have been carried out in hematological or epithelial tumors, investigation applying homogeneous cohorts of sarcoma patients is challenging because of the rarity and high heterogeneity of these malignancies. Only a few studies have correlated heparanase expression with poor prognosis in patients with Ewing’s sarcoma and osteosarcoma to date.
Nevertheless, studies addressing the effects of heparanase inhibitors in sarcoma models have provided preclinical proof-of-concept that heparanase represents a druggable vulnerability in either bone or soft tissue sarcomas. The potential of HS mimetics to improve current therapies was confirmed by the enhanced inhibition of sarcoma xenograft growth and spontaneous metastatic dissemination as well as the high rate of cures in combination regimens with cytotoxic and targeted agents. These findings provide a rational basis for including sarcomas in the evaluation of HS mimetics undergoing clinical development. It will be also interesting in future studies to target the heparanase/HSPG system with new heparanase targeting approaches by specific inhibitors (e.g., small molecules, antibodies) currently undergoing preclinical development.
Abbreviations
- BMP:
-
Bone Morphogenic Protein
- ECM:
-
Extracellular Matrix
- EXT:
-
Exostosin
- FGF:
-
Fibroblast Growth Factor
- FGFR:
-
Fibroblast Growth Factor Receptor
- GAG:
-
Glycosaminoglycan
- GIST:
-
Gastrointestinal Stromal Tumor
- Hh:
-
Hedgehog Ligand
- HIF1α:
-
Hypoxia-inducible Factor 1α
- HS:
-
Heparan Sulfate
- HSPG:
-
HS Proteoglycan
- IGF1R:
-
Insulin-like Growth Factor 1 Receptor
- InsR:
-
Insulin Receptor
- MO:
-
Multiple Osteochondroma
- NDST:
-
N-deacetylase/N-sulfotransferase
- PDGF:
-
Platelet Derived Growth Factor
- PDGFR:
-
Platelet Derived Growth Factor Receptor
- Ptc1:
-
Patched 1
- VEGF:
-
Vascular Endothelial Growth Factor
References
Fletcher, C. D. M., Bridge, J. A., Hogendoorn, P. C. W., et al. (Eds.). (2013). WHO classification of tumours of soft tissue and bone, 4th edn IARC WHO classification of tumours, no 5. Geneva: WHO Press.
Taylor, B. S., Barretina, J., Maki, R. G., et al. (2011). Advances in sarcoma genomics and new therapeutic targets. Nature Reviews. Cancer, 11(8), 541–557.
Barretina, J., Taylor, B. S., Banerji, S., et al. (2010). Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nature Genetics, 42(8), 715–721.
Bonadonna, G., Monfardini, S., De Lena, M., et al. (1970). Phase I and preliminary phase II evaluation of adriamycin (NSC 123127). Cancer Research, 30(10), 2572–2582.
Frezza, A. M., Stacchiotti, S., & Gronchi, A. (2017). Systemic treatment in advanced soft tissue sarcoma: What is standard, what is new. BMC Medicine, 15(1), 109.
Blay, J. Y. (2018). Getting up-to-date in the management of soft tissue sarcoma. Future Oncology, 14(10s), 3–13.
Koseła-Paterczyk, H., & Rutkowski, P. (2017). Dermatofibrosarcoma protuberans and gastrointestinal stromal tumor as models for targeted therapy in soft tissue sarcomas. Expert Review of Anticancer Therapy, 17(12), 1107–1116.
Radaelli, S., Stacchiotti, S., Casali, P. G., et al. (2014). Emerging therapies for adult soft tissue sarcoma. Expert Review of Anticancer Therapy, 14(6), 689–704.
Linch, M., Miah, A. B., Thway, K., et al. (2014). Systemic treatment of soft-tissue sarcoma-gold standard and novel therapies. Nature Reviews. Clinical Oncology, 11(4), 187–202.
Vos, M., & Sleijfer, S. (2018). EJC’s biennial report on metastatic soft tissue sarcoma: State of the art and future perspectives. European Journal of Cancer, 88, 87–91.
Pollack, S. M., Ingham, M., & Spraker, M. B. (2018). Emerging targeted and immune-based therapies in sarcoma. The Journal of Clinical Oncology, 36(2), 125–135.
Movva, S., Wen, W., & al, C. W. (2015). Multi-platform profiling of over 2000 sarcomas: Identification of biomarkers and novel therapeutic targets. Oncotarget, 6(14), 12234–12247.
Perry JA, Kiezun A, Tonzi P et al. (2014) Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proceedings of the National Academy of Sciences of the United States of America 111(51):E5564–E5573.
Behjati, S., Tarpey, P. S., Haase, K., et al. (2017). Recurrent mutation of IGF signalling genes and distinct patterns of genomic rearrangement in osteosarcoma. Nature Communications, 8, 15936.
Hingorani, P., Janeway, K., Crompton, B. D., et al. (2016). Current state of pediatric sarcoma biology and opportunities for future discovery: A report from the sarcoma translational research workshop. Cancer Genetics, 209(5), 182–194.
Lucchesi, C., Khalifa, E., Laizet, Y., et al. (2018). Targetable alterations in adult patients with soft-tissue sarcomas: Insights for personalized therapy. JAMA Oncology, 4(10), 1398–1404.
Ehnman, M., & Larsson, O. (2015). Microenvironmental targets in sarcoma. Frontiers in Oncology, 5, 248.
Versleijen-Jonkers, Y. M., Vlenterie, M., van de Luijtgaarden, A. C., et al. (2014). Anti-angiogenic therapy, a new player in the field of sarcoma treatment. Critical Reviews in Oncology/Hematology, 91(2), 172–185.
Rocchi, L., Caraffi, S., Perris, R., et al. (2014). The angiogenic asset of soft tissue sarcomas: A new tool to discover new therapeutic targets. Bioscience Reports, 34(6), e00147.
Shintani, K., Matsumine, A., Kusuzaki, K., et al. (2006). Expression of hypoxia-inducible factor (HIF)-1alpha as a biomarker of outcome in soft-tissue sarcomas. Virchows Archiv, 449(6), 673–681.
Yang, J., Yang, D., Sun, Y., et al. (2011). Genetic amplification of the vascular endothelial growth factor (VEGF) pathway genes, including VEGFA, in human osteosarcoma. Cancer, 117(21), 4925–4938.
Lammli, J., Fan, M., Rosenthal, H. G., et al. (2012). Expression of vascular endothelial growth factor correlates with the advance of clinical osteosarcoma. International Orthopaedics, 36(11), 2307–2313.
Yu, X. W., Wu, T. Y., Yi, X., et al. (2014). Prognostic significance of VEGF expression in osteosarcoma: A meta-analysis. Tumour Biology, 35(1), 155–160.
Fuchs, B., Inwards, C. Y., & Janknecht, R. (2004). Vascular endothelial growth factor expression is up-regulated by EWS-ETS oncoproteins and Sp1 and may represent an independent predictor of survival in Ewing’s sarcoma. Clinical Cancer Research, 10(4), 1344–1353.
Zwerner, J. P., & May, W. A. (2001). PDGF-C is an EWS/FLI induced transforming growth factor in Ewing family tumors. Oncogene, 20(5), 626–633.
Ménard, C., Blay, J. Y., Borg, C., et al. (2009). Natural killer cell IFN-gamma levels predict long-term survival with imatinib mesylate therapy in gastrointestinal stromal tumor-bearing patients. Cancer Research, 69(8), 3563–3569.
Germano, G., Frapolli, R., Belgiovine, C., et al. (2013). Role of macrophage targeting in the antitumor activity of trabectedin. Cancer Cell, 23(2), 249–262.
Pollack, S. M., He, Q., Yearley, J. H., et al. (2017). T-cell infiltration and clonality correlate with programmed cell death protein 1 and programmed death-ligand 1 expression in patients with soft tissue sarcomas. Cancer, 123(17), 3291–3304.
Cancer Genome Atlas Research Network. (2017). Comprehensive and integrated genomic characterization of adult soft tissue sarcomas. Cell, 171(4), 950–965.
Wisdom, A. J., Mowery, Y. M., Riedel, R. F., et al. (2018). Rationale and emerging strategies for immune checkpoint blockade in soft tissue sarcoma. Cancer, 124(19), 3819–3829.
Ricoveri, W., & Cappelletti, R. (1986). Heparan sulfate endoglycosidase and metastatic potential in murine fibrosarcoma and melanoma. Cancer Research, 46(8), 3855–3861.
Becker, M., Moczar, M., & al, P. M. F. (1986). Solubilization and degradation of extracellular matrix by various metastatic cell lines derived from a rat rhabdomyosarcoma. Journal of the National Cancer Institute, 77(2), 417–424.
Shafat, I., Zcharia, E., Nisman, B., et al. (2006). An ELISA method for the detection and quantification of human heparanase. Biochemical and Biophysical Research Communications, 341(4), 958–963.
Shafat, I., Barak, A. B., Postovsky, S., et al. (2007). Heparanase levels are elevated in the plasma of pediatric cancer patients and correlate with response to anticancer treatment. Neoplasia, 9(11), 909–916.
Shafat, I., Ben-Arush, M. W., Issakov, J., et al. (2011). Pre-clinical and clinical significance of heparanase in Ewing’s sarcoma. Journal of Cellular and Molecular Medicine, 15(9), 1857–1864.
Masola, V., Maran, C., Tassone, E., et al. (2009). Heparanase activity in alveolar and embryonal rhabdomyosarcoma: Implications for tumor invasion. BMC Cancer, 9, 304.
Cassinelli, G., Lanzi, C., Tortoreto, M., et al. (2013). Antitumor efficacy of the heparanase inhibitor SST0001 alone and in combination with antiangiogenic agents in the treatment of human pediatric sarcoma models. Biochemical Pharmacology, 85(10), 1424–1432.
Cassinelli, G., Dal Bo, L., Favini, E., et al. (2018). Supersulfated low-molecular-weight heparin synergizes with IGF1R/IR inhibitor to suppress synovial sarcoma growth and metastases. Cancer Letters, 415, 187–197.
Kazarin, O., Ilan, N., Naroditzky, I., et al. (2014). Expression of heparanase in soft tissue sarcomas of adults. Journal of Experimental & Clinical Cancer Research, 33, 39.
Ramani, V. C., Purushothaman, A., Stewart, M. D., et al. (2013). The heparanase/syndecan-1 axis in cancer: Mechanisms and therapies. The FEBS Journal, 280(10), 2294–2306.
Cassinelli, G., Zaffaroni, N., & Lanzi, C. (2016). The heparanase/heparan sulfate proteoglycan axis: A potential new therapeutic target in sarcomas. Cancer Letter, 382(2), 245–254.
Gallagher, J. (2015). Fell-Muir lecture: Heparan sulphate and the art of cell regulation: A polymer chain conducts the protein orchestra. International Journal of Experimental Pathology, 96(4), 203–231.
Karamanos, N. K., Piperigkou, Z., Theocharis, A. D., et al. (2018). Proteoglycan chemical diversity drives multifunctional cell regulation and therapeutics. Chemical Reviews, 118(18), 9152–9232.
Billings, P. C., & Pacifici, M. (2015). Interactions of signaling proteins, growth factors and other proteins with heparan sulfate: Mechanisms and mysteries. Connective Tissue Research, 56(4), 272–280.
Knelson, E. H., Nee, J. C., & Blobe, G. C. (2014). Heparan sulfate signaling in cancer. Trends in Biochemical Sciences, 39(6), 277–288.
Lanzi, C., Zaffaroni, N., & Cassinelli, G. (2017). Targeting heparan sulfate proteoglycans and their modifying enzymes to enhance anticancer chemotherapy efficacy and overcome drug resistance. Current Medicinal Chemistry, 24(26), 2860–2886.
Nagarajan, A., Malvi, P., & Wajapeyee, N. (2018). Heparan sulfate and heparan sulfate proteoglycans in cancer initiation and progression. Front Endocrinol (Lausanne), 9, 483.
Theocharis, A. D., & Karamanos, N. K. (2019). Proteoglycans remodeling in cancer: Underlying molecular mechanisms. Matrix Biology, 75-76, 220–259.
Williamson, D., Selfe, J., Gordon, T., et al. (2007). Role for amplification and expression of glypican-5 in rhabdomyosarcoma. Cancer Research, 67(1), 57–65.
Li, F., Shi, W., Capurro, M., et al. (2011). Glypican-5 stimulates rhabdomyosarcoma cell proliferation by activating hedgehog signaling. The Journal of Cell Biology, 192(4), 691–704.
Filmus, J., & Capurro, M. (2014). The role of glypicans in hedgehog signaling. Matrix Biology, 35, 248–252.
Almazán-Moga, A., Zarzosa, P., Molist, C., et al. (2017). Ligand-dependent hedgehog pathway activation in Rhabdomyosarcoma: The oncogenic role of the ligands. British Journal of Cancer, 117(9), 1314–1325.
Thway, K., Selfe, J., Missiaglia, E., et al. (2011). Glypican-3 is expressed in rhabdomyosarcomas but not adult spindle cell and pleomorphic sarcomas. Journal of Clinical Pathology, 64(7), 587–591.
Capurro, M. I., Xu, P., Shi, W., et al. (2008). Glypican-3 inhibits hedgehog signaling during development by competing with patched for hedgehog binding. Developmental Cell, 14(5), 700–711.
Takeuchi, M., Higashino, A., Takeuchi, K., et al. (2015). Transcriptional dynamics of immortalized human mesenchymal stem cells during transformation. PLoS One, 10(5), e0126562.
Tang, W. Q., Hei, Y., Kang, L., et al. (2014). Heparanase-1 and components of the hedgehog signalling pathway are increased in untreated alveolar orbital rhabdomyosarcoma. Clinical & Experimental Ophthalmology, 42(2), 182–189.
Ridgway, L. D., Wetzel, M. D., & Marchetti, D. (2011). Heparanase modulates Shh and Wnt3a signaling in human medulloblastoma cells. Experimental and Therapeutic Medicine, 2(2), 229–238.
Zhang, Y., & Rosenberg, A. E. (2017). Bone-forming tumors. Surgical Pathology Clinics, 10(3), 513–535.
Sivaraj, K. K., & Adams, R. H. (2016). Blood vessel formation and function in bone. Development, 143(15), 2706–2715.
Kansara, M., Teng, M. W., Smyth, M. J., et al. (2014). Translational biology of osteosarcoma. Nature Reviews. Cancer, 14(11), 722–735.
Karsenty, G., Kronenberg, H. M., & Settembre, C. (2009). Genetic control of bone formation. Annual Review of Cell and Developmental Biology, 25, 629–648.
Dwivedi, P. P., Lam, N., & Powell, B. C. (2013). Boning up on glypicans--opportunities for new insights into bone biology. Cell Biochemistry and Function, 31(2), 91–114.
Cool, S. M., & Nurcombe, V. (2005). The osteoblast-heparan sulfate axis: control of the bone cell lineage. The International Journal of Biochemistry & Cell Biology, 37(9), 1739–1745.
Lieberman, J. R., Daluiski, A., & Einhorn, T. A. (2002). The role of growth factors in the repair of bone. Biology and clinical applications. The Journal of Bone and Joint Surgery. American Volume, 84-A(6), 1032–1044.
Jochmann, K., Bachvarova, V., & Vortkamp, A. (2014). Heparan sulfate as a regulator of endochondral ossification and osteochondroma development. Matrix Biology, 34, 55–63.
Nurcombe, V., Goh, F. J., Haupt, L. M., et al. (2007). Temporal and functional changes in glycosaminoglycan expression during osteogenesis. Journal of Molecular Histology, 38(5), 469–481.
Guimond, S. E., & Turnbull, J. E. (1999). Fibroblast growth factor receptor signalling is dictated by specific heparan sulphate saccharides. Current Biology, 9(22), 1343–1346.
Saijo, M., Kitazawa, R., Nakajima, M., et al. (2003). Heparanase mRNA expression during fracture repair in mice. Histochemistry and Cell Biology, 120(6), 493–503.
Kram, V., Zcharia, E., Yacoby-Zeevi, O., et al. (2006). Heparanase is expressed in osteoblastic cells and stimulates bone formation and bone mass. Journal of Cellular Physiology, 207(3), 784–792.
Aldi, S., Eriksson, L., Kronqvist, M., et al. (2019). Dual roles of heparanase in human carotid plaque calcification. Atherosclerosis. https://doi.org/10.1016/j.atherosclerosis.2018.12.027.
Haupt, L. M., Murali, S., Mun, F. K., et al. (2009). The heparan sulfate proteoglycan (HSPG) glypican-3 mediates commitment of MC3T3-E1 cells toward osteogenesis. Journal of Cellular Physiology, 220(3), 780–791.
Mansouri, R., Haÿ, E., Marie, P. J., et al. (2015). Role of syndecan-2 in osteoblast biology and pathology. Bonekey Reports, 4, 666.
Teplyuk, N. M., Haupt, L. M., Ling, L., et al. (2009). The osteogenic transcription factor Runx2 regulates components of the fibroblast growth factor/proteoglycan signaling axis in osteoblasts. Journal of Cellular Biochemistry, 107(1), 144–154.
Jackson, R. A., Nurcombe, V., & Cool, S. M. (2006). Coordinated fibroblast growth factor and heparan sulfate regulation of osteogenesis. Gene, 379, 79–91.
Montero, A., Okada, Y., Tomita, M., et al. (2000). Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. The Journal of Clinical Investigation, 105(8), 1085–1093.
Ornitz, D. M., & Marie, P. J. (2015). Fibroblast growth factor signaling in skeletal development and disease. Genes & Development, 29(14), 1463–1486.
Pomin, V. H. (2016). Paradigms in the structural biology of the mitogenic ternary complex FGF:FGFR:Heparin. Biochimie, 127, 214–226.
Jackson, R. A., Murali, S., van Wijnen, A. J., et al. (2007). Heparan sulfate regulates the anabolic activity of MC3T3-E1 preosteoblast cells by induction of Runx2. Journal of Cellular Physiology, 210(1), 38–50.
Brown, A. J., Alicknavitch, M., D’Souza, S. S., et al. (2008). Heparanase expression and activity influences chondrogenic and osteogenic processes during endochondral bone formation. Bone, 43(4), 689–699.
Han, Q., Liu, F., & Zhou, Y. (2013). Increased expression of heparanase in osteogenic differentiation of rat marrow stromal cells. Experimental and Therapeutic Medicine, 5(6), 1697–1700.
Smith, P. N., Freeman, C., Yu, D., et al. (2010). Heparanase in primary human osteoblasts. Journal of Orthopaedic Research, 28(10), 1315–1322.
Drobic, B., Pérez-Cadahía, B., Yu, J., et al. (2010). Promoter chromatin remodeling of immediate-early genes is mediated through H3 phosphorylation at either serine 28 or 10 by the MSK1 multi-protein complex. Nucleic Acids Research, 38(10), 3196–3208.
Gibor, G., Ilan, N., Journo, S., et al. Heparanase is expressed in adult human osteoarthritic cartilage and drives catabolic responses in primary chondrocytes. Osteoarthritis and Cartilage, 26(8), 1110–1117.
Yang, Y., Ren, Y., Ramani, V. C., et al. (2010). Heparanase enhances local and systemic osteolysis in multiple myeloma by upregulating the expression and secretion of RANKL. Cancer Research, 70(21), 8329–8338.
Chanalaris, A., Clarke, H., Guimond, S. E., et al. (2019). Heparan sulfate proteoglycan synthesis is dysregulated in human osteoarthritic cartilage. The American Journal of Pathology, 189(3), 632–647.
Chow, W. A. (2018). Chondrosarcoma: Biology, genetics, and epigenetics. F1000Res. https://doi.org/10.12688/f1000research.15953.1.
Bovée, J. V., Hogendoorn, P. C., Wunder, J. S., et al. (2010). Cartilage tumours and bone development: molecular pathology and possible therapeutic targets. Nature Reviews. Cancer, 10(7), 481–488.
Polychronidou, G., Karavasilis, V., Pollack, S. M., et al. Novel therapeutic approaches in chondrosarcoma. Future Oncology, 13(7), 637–648.
Sonne-Holm E, Wong C, Sonne-Holm S et al. Multiple cartilaginous exostoses and development of chondrosarcomas–a systematic review. Danish Medical Journal 61(9):A4895.
Cuellar, A., & Reddi, A. H. (2013). Cell biology of osteochondromas: bone morphogenic protein signalling and heparan sulphates. International Orthopaedics, 37(8), 1591–1596.
Jennes, I., Pedrini, E., & eta l, Z. M. (2009). Multiple osteochondromas: Mutation update and description of the multiple osteochondromas mutation database (MOdb). Human Mutation, 30(12), 1620–1627.
Pacifici, M. (2017). Hereditary Multiple Exostoses: New insights into pathogenesis, clinical complications, and potential treatments. Current Osteoporosis Reports, 15(3), 142–152.
McCormick, C., Leduc, Y., Martindale, D., et al. (1998). The putative tumour suppressor EXT1 alters the expression of cell-surface heparan sulfate. Nature Genetics, 19(2), 158–161.
Hameetman, L., Bovée, J. V., Taminiau, A. H., et al. (2004). Multiple osteochondromas: Clinicopathological and genetic spectrum and suggestions for clinical management. Hereditary Cancer in Clinical Practice, 2(4), 161–173.
Koziel, L., Kunath, M., Kelly, O. G., et al. (2004). Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Developmental Cell, 6(6), 801–813.
Pacifici, M. (2018). The pathogenic roles of heparan sulfate deficiency in hereditary multiple exostoses. Matrix Biology, 71-72, 28–39.
Jones, K. B., Piombo, V., Searby, C., et al. (2010). A mouse model of osteochondromagenesis from clonal inactivation of Ext1 in chondrocytes. Proceedings of the National Academy of Sciences of the United States of America, 107(5), 2054–2059.
de Andrea, C. E., Reijnders, C. M., Kroon, H. M., et al. (2012). Secondary peripheral chondrosarcoma evolving from osteochondroma as a result of outgrowth of cells with functional EXT. Oncogene, 31(9), 1095–1104.
de Andrea, C. E., & Hogendoorn, P. C. (2012). Epiphyseal growth plate and secondary peripheral chondrosarcoma: the neighbours matter. The Journal of Pathology, 226(2), 219–228.
Trebicz-Geffen, M., Robinson, D., Evron, Z., et al. (2008). The molecular and cellular basis of exostosis formation in hereditary multiple exostoses. International Journal of Experimental Pathology, 89(5), 321–331.
Yang, L., Hui, W. S., Chan, W. C., et al. (2010). A splice-site mutation leads to haploinsufficiency of EXT2 mRNA for a dominant trait in a large family with multiple osteochondromas. Journal of Orthopaedic Research, 28(11), 1522–1530.
Huegel, J., Enomoto-Iwamoto, M., Sgariglia, F., et al. (2015). Heparanase stimulates chondrogenesis and is up-regulated in human ectopic cartilage: A mechanism possibly involved in hereditary multiple exostoses. The American Journal of Pathology, 185(6), 1676–1685.
Presto, J., Thuveson, M., Carlsson, P., et al. (2008). Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proceedings of the National Academy of Sciences of the United States of America, 105(12), 4751–4756.
Veraldi, N., Parra, A., Urso, E., et al. (2018). Structural Features of Heparan Sulfate from Multiple Osteochondromas and Chondrosarcomas. Molecules, 23, 3277.
Lamanna, W. C., Kalus, I., Padva, M., et al. (2007). The heparanome--the enigma of encoding and decoding heparan sulfate sulfation. Journal of Biotechnology, 129(2), 290–307.
Waaijer, C. J., de Andrea, C. E., Hamilton, A., et al. (2012). Cartilage tumour progression is characterized by an increased expression of heparan sulphate 6O-sulphation-modifying enzymes. Virchows Archiv, 461(4), 475–481.
Schrage, Y. M., Hameetman, L., Szuhai, K., et al. (2009). Aberrant heparan sulfate proteoglycan localization, despite normal exostosin, in central chondrosarcoma. The American Journal of Pathology, 174(3), 979–988.
van Oosterwijk, J. G., Meijer, D., van Ruler, M. A., et al. (2013). Screening for potential targets for therapy in mesenchymal, clear cell, and dedifferentiated chondrosarcoma reveals Bcl-2 family members and TGFβ as potential targets. The American Journal of Pathology, 182(4), 1347–1356.
Rickel, K., Fang, F., & Tao, J. (2017). Molecular genetics of osteosarcoma. Bone, 102, 69–79.
Lin, Y. H., Jewell, B. E., Gingold, J., et al. (2017). Osteosarcoma: Molecular pathogenesis and iPSC modeling. Trends in Molecular Medicine, 23(8), 737–755.
Tataria, M., Quarto, N., Longaker, M. T., et al. (2006). Absence of the p53 tumor suppressor gene promotes osteogenesis in mesenchymal stem cells. Journal of Pediatric Surgery, 41, 624–632.
Lee, D. F., Su, J., Kim, H. S., et al. (2015). Modeling familial cancer with induced pluripotent stem cells. Cell, 161(2), 240–254.
Sottnik, J. L., Campbell, B., Mehra, R., et al. (2014). Osteocytes serve as a progenitor cell of osteosarcoma. Journal of Cellular Biochemistry, 115(8), 1420–1429.
Orosco, A., Fromigué, O., Bazille, C., et al. (2007). Syndecan-2 affects the basal and chemotherapy-induced apoptosis in osteosarcoma. Cancer Research, 67(8), 3708–3715.
Na, K. Y., Bacchini, P., Bertoni, F., et al. (2012). Syndecan-4 and fibronectin in osteosarcoma. Pathology, 44(4), 325–330.
Chau, B. N., Diaz, R. L., Saunders, M. A., et al. (2009). Identification of SULF2 as a novel transcriptional target of p53 by use of integrated genomic analyses. Cancer Research, 69(4), 1368–1374.
Baraz, L., Haupt, Y., Elkin, M., et al. (2006). Tumor suppressor p53 regulates heparanase gene expression. Oncogene, 25(28), 3939–3947.
Kim, H. S., Kim, J. H., Oh, J. H., et al. (2006). Expression and clinical significance of heparanase in osteosarcoma. Journal of the Korean Orthopaedic Association, 41(1), 43–51.
Zeng, C., Ke, Z., Luo, C., et al. (2010). Heparanase participates in the growth and invasion of human U-2OS osteosarcoma cells and its close relationship with hypoxia-inducible factor-1α in osteosarcoma. Neoplasms, 57(6).
Fan, L., Wu, Q., Xing, X., et al. (2011). Targeted silencing of heparanase gene by small interfering RNA inhibits invasiveness and metastasis of osteosarcoma cells. Journal of Huazhong University of Science and Technology, 31(3), 348–352.
Wang, Y., Yang, X. Y., Yamagata, S., et al. (2013). Involvement of Ext1 and heparanase in migration of mouse FBJ osteosarcoma cells. Molecular and Cellular Biochemistry, 373, 63–72.
Rivara, S., Milazzo, F. M., & Giannini, G. (2016). Heparanase: A rainbow pharmacological target associated to multiple pathologies including rare diseases. Future Medicinal Chemistry, 8(6), 647–680.
Yang, Y., MacLeod, V., & al, D. Y. (2007). The syndecan-1 heparan sulfate proteoglycan is a viable target for myeloma therapy. Blood, 110(6), 2041–2048.
Ritchie, J. P., Ramani, V. C., Ren, Y., et al. (2011). SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 axis. Clinical Cancer Research, 17(6), 1382–1393.
Manier, S., Sacco, A., Leleu, X., et al. (2012). Bone marrow microenvironment in multiple myeloma progression. Journal of Biomedicine & Biotechnology, 2012, 157496.
Redini, F., & Heymann, D. (2015). Bone tumor environment as a potential therapeutic target in Ewing sarcoma. Frontiers in Oncology, 5, 279.
Erkizan, H. V., Uversky, V. N., & Toretsky, J. A. (2010). Oncogenic partnerships: EWS-FLI1 protein interactions initiate key pathways of Ewing’s sarcoma. Clinical Cancer Research, 16(16), 4077–4083.
Kamura, S., Matsumoto, Y., Fukushi, J. I., et al. (2010). Basic fibroblast growth factor in the bone microenvironment enhances cell motility and invasion of Ewing’s sarcoma family of tumours by activating the FGFR1-PI3K-Rac1 pathway. British Journal of Cancer, 103(3), 370–381.
Yang, Y., Macleod, V., Miao, H. Q., et al. (2007). Heparanase enhances syndecan-1 shedding: A novel mechanism for stimulation of tumor growth and metastasis. The Journal of Biological Chemistry, 282(18), 13326–13333.
Cassinelli, G., Favini, E., Dal Bo, L., et al. (2016). Antitumor efficacy of the heparan sulfate mimic roneparstat (SST0001) against sarcoma models involves multi-target inhibition of receptor tyrosine kinases. Oncotarget, 7(30), 47848–47863.
Hostettler, N., Naggi, A., Torri, G., et al. (2007). P-selectin- and heparanase-dependent antimetastatic activity of non-anticoagulant heparins. The FASEB Journal, 21(13), 3562–3572.
Simon, M. P., Pedeutour, F., Sirvent, N., et al. (1997). Deregulation of the platelet-derived growth factor B-chain gene via fusion with collagen gene COL1A1 in dermatofibrosarcoma protuberans and giant-cell fibroblastoma. Nature Genetics, 15(1), 95–98.
Xie, Y., Skytting, B., Nilsson, G., et al. (1999). Expression of insulin-like growth factor-1 receptor in synovial sarcoma: Association with an aggressive phenotype. Cancer Research, 59(15), 3588–3591.
Sun, Y., Gao, D., Liu, Y., et al. (2006). IGF2 is critical for tumorigenesis by synovial sarcoma oncoprotein SYT-SSX1. Oncogene, 25(7), 1042–1052.
Basappa, M. S., Kavitha, C. V., et al. (2010). A small oxazine compound as an anti-tumor agent: A novel pyranoside mimetic that binds to VEGF, HB-EGF, and TNF-α. Cancer Letters, 297(2), 231–243.
Lanzi, C., & Cassinelli, G. (2018). Heparan sulfate mimetics in cancer therapy: The challenge to define structural determinants and the relevance of targets for optimal activity. Molecules, 23(11), 2915.
Madia, V. N., Messore, A., Pescatori, L., et al. (2018). Novel Benzazole derivatives endowed with potent Antiheparanase activity. Journal of Medicinal Chemistry, 61(15), 6918–6936.
Messore, A., Madia, V. N., Pescatori, L., et al. (2018). Novel symmetrical Benzazolyl derivatives endowed with potent anti-Heparanase activity. Journal of Medicinal Chemistry, 61(23), 10834–10859.
Acknowledgments
We want to acknowledge Enrica Favini, Laura Dal Bo and all the colleagues that collaborated in studies on heparanase in sarcomas, and Dr. Nadia Zaffaroni, head of the Molecular Pharmacology Unit, for supporting this research. For the preparation of this Chapter, we thank Alessandro Noseda (Leadiant Biosciences) for providing roneparstat, Monica Tortoreto for in vivo experiments with A204 rhabdoid tumor, and Eugenio Scanziani and Vittoria Castiglioni (Mouse & Animal Pathology Lab (MAPLab), Fondazione Filarete) for immunohistochemical analyses. Finally, we are grateful to Israel Vlodavsky for his encouragement to explore the heparanase world.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Cassinelli, G., Lanzi, C. (2020). Heparanase: A Potential Therapeutic Target in Sarcomas. In: Vlodavsky, I., Sanderson, R., Ilan, N. (eds) Heparanase. Advances in Experimental Medicine and Biology, vol 1221. Springer, Cham. https://doi.org/10.1007/978-3-030-34521-1_15
Download citation
DOI: https://doi.org/10.1007/978-3-030-34521-1_15
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-34520-4
Online ISBN: 978-3-030-34521-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)