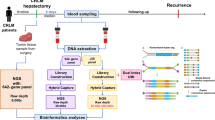
Abstract
Background
For patients with colorectal liver metastases (CRLM) who receive neoadjuvant therapy (NAT), reliable indicators that can early and accurately predict treatment response are lacking. This study was conducted to prospectively investigate the potential of early circulating tumor DNA (ctDNA) dynamics as a precise predictor of NAT response and recurrence in CRLM.
Methods
This study prospectively enrolled 34 patients with CRLM who received NAT, with blood samples collected and subjected to deep targeted panel sequencing at two time points: 1 day before the first and the second cycles of NAT. Correlations of ctDNA mean variant allele frequency (mVAF) dynamics and treatment response were assessed. The performance of early ctDNA dynamics in predicting treatment response was assessed and compared with those of carcinoembryonic antigen (CEA) and cancer antigen 19-9 (CA19-9).
Results
The baseline ctDNA mVAF was significantly associated with pre-NAT tumor diameter (r = 0.65; P < 0.0001). After one cycle of NAT, the ctDNA mVAF declined remarkably (P < 0.0001). The dynamic change in ctDNA mVAF of 50% or more was significantly correlated with better NAT responses. The discriminatory capacity of ctDNA mVAF changes was superior to that of CEA or CA19-9 in predicting radiologic response (area under the curve [AUC], 0.90 vs 0.71 vs 0.61) and pathologic tumor regression grade (AUC, 0.83 vs 0.64 vs 0.67). The early changes in ctDNA mVAF but not CEA or CA19-9 were an independent indicator of recurrence-free survival (RFS) (hazard ratio, 4.0; P = 0.023).
Conclusions
For CRLM patients receiving NAT, an early ctDNA change is a superior predictor of treatment response and recurrence compared with conventional tumor markers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death worldwide.1 The liver remains the most common organ of CRC distal metastasis, and curative-intent resection has long been the standard of care for resectable CRC liver metastases (CRLM).2 However, a high rate of recurrence still constrains the long-term survival for the majority of CRLM patients.3
Although recent studies have demonstrated that neoadjuvant therapy (NAT) might be beneficial, especially for those with a high recurrence risk, a subset of patients remains who respond poorly to NAT and cannot benefit from the treatment.4 In current clinical practice, the treatment response of CRLM patients to NAT is commonly evaluated by imaging tests. However, before a reliable radiologic response assessment is applicable, several cycles of chemotherapy still are needed. This exposes the non-responders to unnecessary toxicity and also may delay their access to alternative, potentially more effective therapeutic regimens. Thus, identification of reliable biomarkers that can predict response to NAT early is urgently needed.5
In recent years, liquid biopsy has shown great application potential as a surrogate biomarker for cancer diagnosis and treatment monitoring.6 Circulating tumor DNA (ctDNA), a subset of plasma cell-free DNA (cfDNA), is found in the peripheral blood of tumor patients due to the continuous release of fragmented DNA from necrotic cancer cells and shows a high concordance with tumor tissue as to genomic alterations.7 Recent evidences have shown that plasma ctDNA could offer a minimally invasive and highly sensitive approach for early screening, disease monitoring, prognostication, and therapeutic response assessment in gastrointestinal cancers.8
In the neoadjuvant setting, existing evidences indicate that the application of ctDNA detection alone or in combination with conventional imaging methods can accurately predict pathologic response and clinical outcome in local advanced rectal cancer and breast cancer.9,10,11,12,13,14 For CRLM patients receiving NAT, however, the role of ctDNA dynamic changes in response and prognosis prediction has not been deeply investigated. Because ctDNA levels are influenced by tumor type, size, and metastatic site, recent studies have shown that CRC patients with liver metastases have a significantly higher level of ctDNA than patients with other metastatic sites.15,16 This indicates that ctDNA has a natural advantage as an ideal surrogate marker to monitor the burden of liver metastases, especially for patients with negative carcinoembryonic antigen (CEA) or cancer antigen 19-9 (CA19-9). On the other hand, the half-life time of ctDNA is about 2 h, which is much shorter than that of clinical tumor markers.17
All the aforementioned evidences indicate that the dynamics of ctDNA may reflect the real-time tumor burden and treatment response earlier and more accurately than conventional tumor markers. However, most previous studies collected blood samples and profiled ctDNA after all cycles of NAT or before surgery, so the role of early dynamic changes of ctDNA in the response prediction remain unclear.
This study investigated the potential of early dynamic changes in ctDNA as a precise predictor of NAT response and CRLM recurrence, especially compared with conventional tumor markers.
Methods
Study Design and Patient Enrollment
This observational, prospective cohort study recruited patients with resectable or borderline resectable CRLM treated with NAT and subsequent surgical resection. The study was designed and conducted in accordance with the Declaration of Helsinki and received approval from the Ethics Committee of Huashan Hospital, Fudan University (approval no. KY2019-606). Written informed consents for sample collection and genetic profiling were obtained from the patients. The study was registered at the Chinese Clinical Trial Registry (ChiCTR2100043990).
The primary end point of the current study was whether early ctDNA dynamics during NAT are predictive of response and recurrence for CRLM.
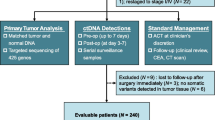
The specific key inclusion criteria specified patients 18 to 75 years of age with an Eastern Cooperative Oncology Group (ECOG) score of 0 or 1, pathologically confirmed colorectal cancer with synchronous or metachronous liver metastases, resectable or borderline resectable liver metastases, and NAT before surgery recommended after an institutional multidisciplinary team (MDT) evaluation. The exclusion criteria ruled out patients with extensive liver metastases, unresectable extrahepatic metastases, NAT already received or refused, refusal to undergo serial blood collection for biomarker analyses, blood samples that did not pass the quality control, no qualification for ctDNA detection, and refusal to undergo subsequent surgery after NAT. Between March 2019 and December 2021, 39 patients were prospectively recruited, and 34 patients were included in the final analysis.
Diagnostic, Treatment, and Follow-up Procedures
The specific information of the diagnostic procedure is summarized in supplementary methods. For the patients with potentially resectable liver metastases, MDT evaluation was performed by hepatobiliary surgeons, gastrointestinal surgeons, oncologists, radiologists, and pathologists. Neoadjuvant therapy was suggested for patients with one or more inferior prognostic factors such as high clinical risk score (CRS), concurrent extrahepatic metastases, RAS/BRAF mutations, right-sided primary tumor, or other factors not suitable for upfront surgery.
The patients then were treated per the recommendations of the MDT, with NAT administered in 2-week cycles. The patients with resectable liver metastases were suggested to receive standard chemotherapy only (mFOLFOX6 or FOLFIRI), whereas those with borderline resectable liver metastases received chemotherapy (mFOLFOX6, FOLFIRI, or FOLFOXIRI) plus anti-VEGF (bevacizumab) or anti-EGFR (cetuximab) targeted therapy based on RAS and BRAF gene mutation status. Adjustment in the dose and timing of chemotherapy and in the number of treatment cycles was decided by MDT discussion based on the radiologic response and chemotherapy-related toxicities. Surgery was scheduled to be performed within 4 to 6 weeks after completion of NAT. Before a resort to surgery, all the patients underwent MDT evaluation. For synchronous CRLM, simultaneous or staged operations were selected for different patients. After the operations, adjuvant therapy was suggested for all the patients.
After surgical resection, all the patients underwent a standardized follow-up procedure to verify their recurrence and survival status. Routine follow‐up evaluation consisted of blood tumor marker levels, physical examination, colonoscopy, and enhanced computed tomography (CT) of the chest/abdomen/pelvic cavity. Scheduled follow-up evaluation was performed every 3 months for the first 3 years, then every 6 months thereafter until 5 years. Recurrence-free survival (RFS) was defined as the time from hepatectomy to tumor relapse or last follow-up visit. Event-free survival (EFS) was defined as the time from diagnosis to tumor relapse or last follow-up visit.
NAT Response Evaluation
The radiologic response was evaluated based on the relative change in the sum of the longest diameters of measurable target lesions according to RECIST v1.1.18 The patients with stable disease (SD) or progressive disease (PD) were classified as radiologic non-responders, whereas those with a partial response (PR) or a complete response (CR) were classified as radiologic responders. Early tumor shrinkage (ETS) was defined as a tumor size decrease of 20% or more at week 8. Investigators were blinded to the results of ctDNA during NAT.
The pathologic response was evaluated using tumor regression grade (TRG) according to Mandard criteria as follows19: TRG1 (absence of residual cancer and a large amount of fibrosis), TRG2 (rare residual cancer cells scattered throughout the fibrosis), TRG3 (more residual tumor cells but predominant fibrosis), TRG4 (residual cancer cells predominating over fibrosis), and TRG5 (no signs of regression). The TRG status was evaluated by two independent investigators blinded to radiologic results. Pathologic responders were defined as patients achieving TRG1/2/3, whereas non-responders were defined as TRG4/5.
ctDNA Measurement and Cutoff Point for ctDNA Change
The peripheral blood samples 1 day before the first and second cycles of NAT were collected. For plasma supernatant collection, 10-ml peripheral blood samples were used. Determination of circulating mutated DNA was performed using a target Next-Generation Sequencing (NGS) panel covering of 61 cancer-related genes (Table S1) at 3D Medicines, Inc., a clinical laboratory accredited by the College of American Pathologists (CAP) and certified by the Clinical Laboratory Improvement Amendments (CLIA). The details are in the Supplementary Data file.
Mean variant allele frequency (mVAF), defined as the average VAF of all variants detected, was chosen as the parameter that reflected the abundance of ctDNA in the plasma. Then the optimal cutoff points for ctDNA mVAF change to predict radiologic response, pathologic response, and recurrence-free survival (RFS) were evaluated. As demonstrated in Tables S2, S3, and S4, ctDNA mVAF changes of 40%, 50%, and 40% showed the best discrimination capacity of clinical (radiologic) response, pathologic response, and RFS benefit, respectively. The point of 50% also performed well (Youden index, 0.57) in predicting clinical response, whereas the point of 40% did not perform as well (Youden index, 0.41) in predicting pathologic response. We therefore chose an mVAF change of 50% as the cutoff point to divide patients into ctDNA responders and non-responders. Consequently, ctDNA responders were defined as those with a ratio of a change in the second to baseline ctDNA mVAF of 50% or more, whereas non-ctDNA responders were defined as those with ctDNA mVAF change of less than 50%.
Statistical Methods
Descriptive statistics were used for patient and tumor characteristics, with continuous variables summarized in terms of medians and ranges. A Pearson correlation test was performed to determine the linear association between continuous values. For comparison of ctDNA responders with non-responders, the chi-square test or Fisher’s exact test was used for categorical factors, whereas the t test or Mann-Whitney test was used for continuous parameters. The median follow-up time, median RFS time, and 1- and 3-year RFS rates were calculated by the Kaplan-Meier method. For the different groups, RFS curves were depicted using Kaplan-Meier plots and compared with the log-rank test.
The effects of individual clinical and mutational variables on RFS were evaluated by the univariable Cox proportional hazards model. Multivariable Cox regression analysis was performed to identify independent prognostic factors and determine their hazard ratios (HRs) and 95% confidence intervals (CIs). Receiver operating characteristic (ROC) curves were depicted for the variables, and the area under the curve (AUC) was calculated to compare their predictive performance.
All data analyses were performed using R (version 3.6.1) or GraphPad Prism software (version 8.0.2). In all analyses, P values lower than 0.05 from a two-sided test were considered statistically significant.
Results
Clinicopathologic Characteristics and Treatment Outcomes
Between March 2019 and December 2021, 39 patients with resectable or borderline resectable CRLM were recruited. After initial screening, 37 patients fulfilled the inclusion criteria and received NAT. During NAT, one patient gave up NAT for personal reasons and was excluded. After NAT, one patient who refused surgery for personal reasons also was excluded from further analysis. Another patient was excluded from analysis due to the lack of qualified blood samples for ctDNA detection. Finally, 34 patients completed NAT and subsequent surgery according to the study protocol (Fig. 1).
Baseline demographics and clinicopathologic and treatment characteristics of all the enrolled patients are presented in Table 1. The median age of the overall cohort was 59.5 years (range, 32.0–74.0 years), and 44.1% were women. The cohort included 22 (64.7%) patients with synchronous metastasis and 12 (35.3%) patients with metachronous disease. In terms of primary tumor sidedness, 23 (67.6%) patients were left-sided, and 11 (32.4%) patients were right-sided. Multiple liver metastases were present in 28 (82.4%) patients, and 20 (58.8%) patients had bilobar involvement. Among the cohort, 18 (52.9%) patients had positive primary nodal status, and 6 (17.6%) patients had extrahepatic metastases. Abnormal CEA levels were present in 27 (79.4%) patients and abnormal CA19-9 levels in 15 (44.1%) patients. Pathology showed poorly differentiated tumors in 23.5% (8/34) of the cases.
The median courses of NAT were 4 (range, 3–8), with 16 (47.1%) patients receiving only standard chemotherapy and 18 (52.9%) patients receiving chemotherapy plus targeted therapy. The objective response rate (ORR) was 79.4%, and the disease control rate (DCR) was 97.1%. All the enrolled patients underwent surgery, with no perioperative mortality. Simultaneous resection was performed for 17 of the 22 synchronous patients, whereas 5 patients underwent staged resection. In terms of pathologic response, 6 (17.6%) patients achieved TRG2, 16 (47.1%) patients achieved TRG3, and 12 (35.3%) patients achieved TRG4. During a median postoperative follow-up time of 18.5 months, the median RFS and EFS times were respectively 13.2 and 16.0 months, and the 1-year RFS rate was 51.3% (Fig. S1). By the end of the follow-up period, 17 of the patients experienced recurrence, and 4 patients died of disease progression.
Baseline Mutational Status of ctDNA and Clinical Relevance
Somatic mutations in ctDNA were detected in all (100%) the patients at baseline. The mutational landscape of the baseline ctDNA is characterized and demonstrated in Fig. S2. A total of 114 mutations in 29 genes were identified, with 1 to 7 (median, 3) mutations detected in each sample. The most commonly detected genes were TP53 (74%), KRAS (29%), and PIK3CA (18%).
The baseline ctDNA mVAF had no correlation with clinicopathologic factors, except for tumor diameter (Fig. 2a; P < 0.0001). Compared with CEA and CA19-9, mVAF demonstrated a stronger correlation with tumor diameter, as evidenced by a correlation coefficient of 0.65 for mVAF compared with 0.31 for CEA and 0.34 for CA19-9 (P < 0.0001). The level of baseline ctDNA mVAF was not significantly associated with RFS (Fig. S3).
Clinical and therapeutic response relevance of baseline and dynamic changes in ctDNA. a Association of baseline ctDNA with tumor diameter. b Dynamic changes in ctDNA between 1 day before the first cycle of NAT and 1 day before the second day of NAT. c Waterfall plot showing the response of patients based on different molecular response status. d Changes in ctDNA mVAF between patients achieving CR/PR and those achieving SD/PD. e The proportion of patients achieving ETS between the molecular responsive group and the non-responsive group. f Receiver operating characteristic (ROC) curves of the fractional ctDNA, CEA, and CA19-9 changes in predicting radiologic response; the area under the curve (AUC) was calculated to compare their predictive performance. g Changes in ctDNA mVAF between patients achieving tumor regression grade (TRG)2/3 and those achieving TRG4. h The proportion of patients achieving pathologic tumor regression (TRG2/3) between the molecular responsive group and the non-responsive group. i ROC curves of the fractional ctDNA, CEA, and CA19-9 changes in predicting pathologic response; the AUC was calculated to compare their predictive performance. ctDNA circulating tumor DNA; NAT neoadjuvant therapy; mVAF median variant allele frequency; CR complete response; PR partial response; SD stable disease; PD progressive disease; CEA carcinoembryonic antigen; CA19-9 cancer antigen 19-9; ETS early tumour shrinkage; TRG tumor regression grade
Similarly, univariable logistic regression was used to investigate the association of baseline gene mutation status with RFS, and only genes with mutation rates greater than 10% were included. The P values of these genes did not achieve statistical significance, with RAS mutations tending to be associated with worse RFS, but the significance of the association was marginal (P = 0.0672; Table S5).
Early ctDNA Dynamics and Radiologic Response
After one cycle of NAT, the ctDNA mVAF declined remarkably (P < 0.0001; Fig. 2b). However, the ctDNA mVAF change had no correlation with clinicopathologic factors (Table 1). We then evaluated the association between mVAF change and radiologic response. According to RECIST criteria, 27 cases were classified as radiologic responders (PR), whereas 7 cases were categorized as non-responders (SD or PD) (Fig. 2c). As demonstrated in Fig. 2d, the change in mVAF was significantly greater in the responsive group than in the non-responsive group (P = 0.0004), and the patients with a molecular response (>50% mVAF decrease) had a greater probability of achieving ETS than the non-response patients (<50% mVAF decrease; 88.0% vs 33.3%; P = 0.0039; Fig. 2e).
We further compared the predictive performance of ctDNA mVAF dynamics with that of tumor markers such as CEA and CA19-9. The ROC curve analysis suggested that the sensitivity and specificity of ctDNA dynamics for radiologic response prediction were significantly higher than those of CEA and CA19-9 (AUC, 0.90 vs 0.71 vs 0.61; Fig. 2f). This indicated that ctDNA dynamics had a better performance than CEA or CA19-9 in predicting radiologic response.
Early ctDNA Dynamics and Pathologic Response
We then evaluated the association between ctDNA mVAF change and pathologic response. According to Mandard TRG grading criteria, 22 patients were classified as pathologic responders (TRG2/3), whereas 12 patients were classified as non-responders (TRG4). As demonstrated in Fig. 2g, the change in mVAF was significantly greater in the pathologic responder group than in the non-responder group (P = 0.0013), and the patients with ctDNA response (>50% mVAF decrease) had a greater probability of achieving a pathologic response than those with a non-response (<50% mVAF decrease; 80.0% vs 22.2%; P = 0.0037; Fig. 2h). In the multivariate analysis, the ctDNA changes remained an independent predictive factor for pathologic response (Table S6).
Next, the performance (predictive accuracy of pathologic response) of ctDNA, CEA, and CA19-9 dynamics also was evaluated and compared. The ROC curve analysis suggested that the sensitivity and specificity of the ctDNA dynamics for pathologic response prediction were much higher than those of CEA and CA19-9 (AUC, 0.83 vs 0.64 vs 0.67; Fig. 2i). This indicated that the ctDNA dynamics had a better performance than CEA or CA19-9 in predicting pathologic response.
Early ctDNA Dynamics and Clinical Outcome
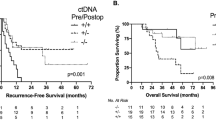
Finally, we evaluated the association between ctDNA dynamics and postoperative recurrence. The specific information about ctDNA dynamic changes, treatment response, and clinical outcomes for each patient was demonstrated in a swimmer plot (Fig. 3a). In the Kaplan-Meier analysis, the molecular response group showed a significantly longer RFS and EFS than the non-response group (Fig. 3b and c; P = 0.0102 for RFS and P = 0.0054 for EFS). In contrast, early changes in CEA and CA19-9 were not significantly associated with RFS (Fig. S4a and S4b). According to the univariate Cox proportional hazard analysis, the variables significantly associated with worse RFS were a ctDNA mVAF change less than 50% (P = 0.015), synchronous metastasis (P = 0.031), right-sided primary tumor (P = 0.043), high CRS score (P = 0.007), and female gender (P = 0.036) (Table 2).
Association of molecular response with clinical outcomes. a Swimmer plot showing ctDNA dynamic changes, treatment response, and clinical outcomes for each patient. b Kaplan-Meier survival plots showing RFS after surgery according to different molecular response groups. c Kaplan-Meier survival plots showing EFS after surgery according to different molecular response groups. ctDNA circulating tumor DNA; RFS recurrence-free survival; EFS event-free survival
In the multivariate analyses, the only remaining independent predictors of RFS were ctDNA mVAF change (HR, 4.0; P = 0.023) and primary tumor sidedness (HR, 3.51; P = 0.018) (Table 2). The aforementioned results indicated that changes in ctDNA mVAF are an early and reliable biomarker to predict recurrence of CRLM.
We then combined the ctDNA alteration with conventional clinicopathologic factors to predict the patients’ survival. Interestingly, the combination of ctDNA alteration and ETS showed significant value in predicting RFS. The patients with a ctDNA response (–) and ETS (–) showed a significantly poorer RFS than those with either a ctDNA response (+) or ETS (+) (Fig. S5). By contrast, neither the combination of ctDNA alteration and tumor marker (CEA and CA19-9) nor a pathologic response (TRG) showed significant value in predicting RFS (Fig. S6).
Predictive Value of ctDNA in CEA- and CA19-9-Negative Cases
We performed a subgroup analysis of the patients with negative baseline CEA or CA19-9. At baseline, 7 (20.6%) patients had a negative CEA (<5 ng/mL), and 19 (55.9%) patients had a negative CA19-9 (<37 U/mL). The CEA-negative subgroup showed no significant correlation between ctDNA response and ETS, pathologic response, or RFS (Fig. S7a). In the CA19-9-negative subgroup, the ctDNA response showed a significant association with ETS (P = 0.046), and a non-significant trend toward an improved pathologic response (P = 0.14) and RFS (P = 0.06) (Fig. S7b).
Representative Cases
All these results suggested that early dynamic changes in ctDNA were a superior predictor of treatment response and recurrence. For example, a 52-year-old woman had a diagnosis of sigmoid colon cancer involving synchronous liver-limited metastases, with wildtype RAS/BRAF. The serial plasma collections showed a remarkable decrease of 98.9% in early ctDNA mVAF change, whereas the decrease in CEA and CA19-9 at this time was not significant. After NAT, the patient achieved a partial response and significant pathologic regression (TRG2) and kept no evidence of disease (NED) status during the follow-up period (Fig. 4a).
Conversely, a 31-year-old woman had a diagnosis of descending colon cancer involving synchronous liver-limited metastases, with wildtype RAS/BRAF. The serial plasma collections showed an increase of 59.7% in ctDNA mVAF change, whereas the levels of CEA and CA19-9 were stable. This patient did not achieve a significant radiologic response or pathologic regression, and finally experienced early recurrence during postoperative adjuvant therapy (Fig. 4b).
Discussion
In recent years, ctDNA has become the focus of research on precision diagnosis and treatment of CRC, ranging from early cancer detection and minimal residual disease (MRD) monitoring to prognosis and treatment response prediction.20 This study focused on the role of ctDNA in the neoadjuvant setting for CRLM patients. The study showed that early ctDNA dynamics was strongly associated with radiologic and pathologic response to NAT in CRLM. Meanwhile, early ctDNA dynamics also were significantly related to postoperative RFS. These findings showed the application potential of ctDNA in CRLM receiving NAT, making it possible to change clinical decisions as early as before the second cycle of NAT and to maximize the effectiveness of NAT.
Neoadjuvant therapy followed by surgery has become a standard of care for selected patients with locally advanced rectal cancer, breast cancer, pancreatic cancer, or CRLM. Because the case of upfront surgery was not favorable, NAT was believed to improve surgery efficacy and outcome, probably by enhancing the chance of achieving a margin-negative (R0) resection. Additionally, NAT also could offer a unique window for real-time monitoring of treatment response. Treatment response to NAT usually is evaluated by imaging tests, but several cycles of chemotherapy are needed until a reliable radiologic response assessment is possible. This exposes non-responders to unnecessary toxicity and also may delay their access to other potentially more effective treatment regimens. Thus, identification of reliable and real-time biomarkers that can early predict response to NAT is urgently needed for personalizing treatment.
Several previous studies have investigated the role of ctDNA detection in locally advanced rectal cancer and breast cancer receiving NAT. Findings have shown that ctDNA detection alone or in combination with conventional imaging methods can accurately predict pathologic response and clinical outcome,9,10,11,12,13,14,15 thus perhaps helping in the selection of the best treatment strategy (i.e., resection or watch-and-wait for rectal cancer) or the choice of the patients who would not benefit from further postoperative therapy, preventing overtreatment. In CRLM, ctDNA detection has shown great advantage in terms of prognosis prediction and MRD surveillance after resection,21,22,23,24 whereas its role in neoadjuvant setting remains unclear. In a previous study, circulating free methylated tumor DNA markers were detected for sensitive assessment of tumor burden and early response monitoring of CRLM patients receiving systemic chemotherapy.25
Most previous studies collected blood samples after all cycles of NAT or before surgery and did not focus on the predictive role of early ctDNA changes in treatment responses to NAT and postoperative outcomes. Findings have shown that CRC patients with liver metastases have significantly higher levels of ctDNA than patients with metastases at other sites.15,16 On the other hand, the findings also have shown that the half-life of plasma ctDNA is as short as less than 2 h,17 making it possible to distinguish the sensitive and non-sensitive CRLM patients early during NAT.
In contrast to previous studies collecting blood samples after NAT, the current study focused on the role of early dynamic changes of ctDNA in response prediction. As demonstrated in our study, after a single cycle of NAT, the mVAF of ctDNA declined early in patients responsive to NAT. These results proved the concept that ctDNA could act as a real-time surrogate marker reflecting the tumor burden and treatment response earlier and more accurately than conventional tumor markers for CRLM. For example, ETS is an early indicator of treatment sensitivity that shows the decrease in tumor burden measured at the time of the first imaging evaluation.26 In most previous studies, ETS was evaluated at 8 weeks,27,28 in some studies, at 6 or 7 weeks.29,30 At the evaluation time point, an ETS of 20% or greater is considered a favorable indicator for CRLM patients. In our study, we showed that ctDNA mVAF reduction measured at 2 weeks was strongly consistent with ETS evaluated at 8 weeks, indicating that ctDNA monitoring is an earlier marker for response assessment than ETS. Besides the aforementioned radiologic and pathologic parameters, ctDNA change also was an independent factor of RFS for CRLM patients in this cohort. Additionally, ctDNA also could provide more genetic mutation information, which reflected tumor biologic behavior and heterogeneity.
Our study had several limitations. First, the sample was relatively small, which may limit the extrapolation of our conclusions. Second, the NAT regimen was not standardized. The patients underwent different approaches based on different tumor burdens, tumor locations, mutation profiles, and therapeutic goals, which made the study closer to real-world investigation, but might also have introduced more confounding factors. Third, although many studies have reported that elevated postoperative ctDNA could possibly help identify high-recurrence risk patients, ctDNA was not measured after curative-intent surgery in our study due to the study design. In the future, additional detection after surgery and during follow-up evaluation should be added to provide more information about the prediction performance of ctDNA for CRLM patients.
Conclusions
This prospective study provided novel evidence that early ctDNA dynamic changes can accurately predict NAT response and recurrence in CRLM. In future clinical practice, selectively prognostic and therapeutic stratification may be suggested early during NAT for CRLM patients with different early ctDNA dynamic changes.
References
Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Tsilimigras DI, Brodt P, Clavien PA, et al. Liver metastases. Nat Rev Dis Primers. 2021;7:27.
Margonis GA, Buettner S, Andreatos N, et al. Prognostic factors change over time after hepatectomy for colorectal liver metastases: a multi-institutional, international analysis of 1099 patients. Ann Surg. 2019;269:1129–37.
Ayez N, van der Stok EP, Grünhagen DJ, et al. The use of neo-adjuvant chemotherapy in patients with resectable colorectal liver metastases: clinical risk score as possible discriminator. Eur J Surg Oncol. 2015;41:859–67.
Yamashita S, Chun YS, Kopetz SE, Vauthey JN. Biomarkers in colorectal liver metastases. Br J Surg. 2018;105:618–27.
Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic: implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297–312.
Cheng ML, Pectasides E, Hanna GJ, Parsons HA, Choudhury AD, Oxnard GR. Circulating tumor DNA in advanced solid tumors: clinical relevance and future directions. CA Cancer J Clin. 2021;71:176–90.
Nordgård O, Tjensvoll K, Gilje B, Søreide K. Circulating tumour cells and DNA as liquid biopsies in gastrointestinal cancer. Br J Surg. 2018;105:e110–20.
Magbanua MJM, Li W, Wolf DM, et al. Circulating tumor DNA and magnetic resonance imaging to predict neoadjuvant chemotherapy response and recurrence risk. NPJ Breast Cancer. 2021;7:32.
Vidal J, Casadevall D, Bellosillo B, et al. Clinical impact of presurgery circulating tumor DNA after total neoadjuvant treatment in locally advanced rectal cancer: a biomarker study from the GEMCAD 1402 trial. Clin Cancer Res. 2021;27:2890–8.
Magbanua MJM, Swigart LB, Wu HT, et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann Oncol. 2021;32:229–39.
Li S, Lai H, Liu J, et al. Circulating tumor DNA predicts the response and prognosis in patients with early breast cancer receiving neoadjuvant chemotherapy. JCO Precis Oncol. 2020;4:244–57.
McDuff SGR, Hardiman KM, Ulintz PJ, et al. Circulating tumor DNA predicts pathologic and clinical outcomes following neoadjuvant chemoradiation and surgery for patients with locally advanced rectal cancer. JCO Precis Oncol. 2021;5:123–32.
Schou JV, Larsen FO, Sørensen BS, et al. Circulating cell-free DNA as predictor of treatment failure after neoadjuvant chemo-radiotherapy before surgery in patients with locally advanced rectal cancer. Ann Oncol. 2018;29:610–5.
Osumi H, Shinozaki E, Takeda Y, et al. Clinical relevance of circulating tumor DNA assessed through deep sequencing in patients with metastatic colorectal cancer. Cancer Med. 2019;8:408–17.
Bando H, Nakamura Y, Taniguchi H, et al. Effects of metastatic sites on circulating tumor DNA in patients with metastatic colorectal cancer. JCO Precis Oncol. 2022;6:e2100535.
Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90.
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228–47.
Rubbia-Brandt L, Giostra E, Brezault C, et al. Importance of histological tumor response assessment in predicting the outcome in patients with colorectal liver metastases treated with neo-adjuvant chemotherapy followed by liver surgery. Ann Oncol. 2007;18:299–304.
Malla M, Loree JM, Kasi PM, Parikh AR. Using circulating tumor DNA in colorectal cancer: current and evolving practices. J Clin Oncol. 2022;40:2846–57.
Newhook TE, Overman MJ, Chun YS, et al. Prospective study of perioperative circulating tumor DNA dynamics in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg. 2022. https://doi.org/10.1097/SLA.0000000000005461.
Wang DS, Yang H, Liu XY, et al. Dynamic monitoring of circulating tumor DNA to predict prognosis and efficacy of adjuvant chemotherapy after resection of colorectal liver metastases. Theranostics. 2021;11:7018–28.
Øgaard N, Reinert T, Henriksen TV, et al. Tumour-agnostic circulating tumour DNA analysis for improved recurrence surveillance after resection of colorectal liver metastases: a prospective cohort study. Eur J Cancer. 2022;163:163–76.
Tie J, Wang Y, Cohen J, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: a prospective cohort study. PloS Med. 2021;18:e1003620.
Bhangu JS, Beer A, Mittlböck M, et al. Circulating free methylated tumor DNA markers for sensitive assessment of tumor burden and early response monitoring in patients receiving systemic chemotherapy for colorectal cancer liver metastasis. Ann Surg. 2018;268:894–902.
Heinemann V, Stintzing S, Modest DP, Giessen-Jung C, Michl M, Mansmann UR. Early tumour shrinkage (ETS) and depth of response (DpR) in the treatment of patients with metastatic colorectal cancer (mCRC). Eur J Cancer. 2015;51:1927–36.
Douillard JY, Siena S, Peeters M, Koukakis R, Terwey JH, Tabernero J. Impact of early tumour shrinkage and resection on outcomes in patients with wild-type RAS metastatic colorectal cancer. Eur J Cancer. 2015;51:1231–42.
Cremolini C, Loupakis F, Antoniotti C, et al. Early tumor shrinkage and depth of response predict long-term outcome in metastatic colorectal cancer patients treated with first-line chemotherapy plus bevacizumab: results from phase III TRIBE trial by the Gruppo Oncologico del Nord Ovest. Ann Oncol. 2015;26:1188–94.
Modest DP, Laubender RP, Stintzing S, et al. Early tumor shrinkage in patients with metastatic colorectal cancer receiving first-line treatment with cetuximab combined with either CAPIRI or CAPOX: an analysis of the German AIO KRK 0104 trial. Acta Oncol. 2013;52:956–62.
Giessen C, Laubender RP, Fischer von Weikersthal L, et al. Early tumor shrinkage in metastatic colorectal cancer: retrospective analysis from an irinotecan-based randomized first-line trial. Cancer Sci. 2013;104:718–24.
Acknowledgment
This study was supported by the Program of Shanghai Outstanding Young Medical Talents Training and the Program of Shanghai Academic/Technology Research Leader (22XD1400300).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Disclosure
There are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, XY., Zhang, R., Han, JH. et al. Early Circulating Tumor DNA Dynamics Predict Neoadjuvant Therapy Response and Recurrence in Colorectal Liver Metastases: A Prospective Study. Ann Surg Oncol 30, 5252–5263 (2023). https://doi.org/10.1245/s10434-023-13604-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-023-13604-2