Abstract
Purpose
Despite curative-intent treatment, recurrence is common for patients with colorectal cancer (CRC). Currently, prediction of disease recurrence and prognostication following surgery is based upon vague clinical factors and more precise and dynamic biomarkers for risk stratification and treatment decisions are urgently needed. Circulating tumor DNA (ctDNA) is a promising biomarker for patients undergoing treatment for resectable CRC.
Methods
In this review, we provide an overview of the data supporting current uses of ctDNA for CRC, including localized CRC and resectable colorectal liver metastases (CLM), as well as descriptions of important ongoing clinical trials using ctDNA in the care of patients with CRC.
Results
The detection of ctDNA following curative-intent therapy is associated with disease recurrence, and multiple trials are investigating its role in determining need and duration for adjuvant therapy for localized CRC. In addition, ctDNA reliably predicts prognosis for patients with CLM, with trials underway studying ctDNA-guided treatment sequencing and intensity.
Conclusion
The detection of ctDNA is a sensitive and dynamic biomarker for disease recurrence in CRC. Many investigations are underway into ctDNA’s potential role in surveillance and treatment algorithms, and it has the potential to become a critical biomarker to determine individualized strategies for treatment sequencing, choice, and duration of therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Circulating tumor DNA (ctDNA) is a promising biomarker for tumor monitoring throughout disease management in many cancers [1]. Multiple existing serum-based biomarkers are used to guide treatment, but these have significant limitations, such as the presence in patients without cancer, and absence in a significant fraction of patients with advanced malignancy [2,3,4]. For colorectal cancer (CRC) specifically, the only recommended blood-based biomarker is a carcinoembryonic antigen (CEA), which lacks both sensitivity and specificity [5, 6]. Moreover, even when elevated in the setting of CRC, CEA does not accurately predict disease recurrence or direct treatment strategies. Currently, prognostication and surveillance following treatment for CRC are often based on vague, imprecise clinical characteristics. Thus, there is an urgent unmet need for sensitive and dynamic biomarkers for patients with CRC.
Presence and levels of ctDNA can be assessed by non-invasive measurements of tumor-specific DNA fragments within the patient’s blood. These fragments are shed by the tumor into the peripheral bloodstream and can be assessed as part of a patient’s total circulating free DNA (cfDNA) [7]. Serial measurement of ctDNA can be used as a sensitive, dynamic indicator of disease burden [8]. In addition, ctDNA can be used as a “liquid biopsy,” facilitating low-risk, minimally invasive genetic analysis of cancers that are difficult to sample, or only accessible by fine-needle aspirate, which may not provide enough sample for genotyping [9]. These liquid biopsies may also offer a more comprehensive picture of tumor heterogeneity and evolution compared to a single-site tissue biopsy [10]. Beyond demonstrating the genomics of a tumor at a single time point, serial measurement of specific targetable mutations, such as KRAS, offers an understanding of the genomic evolution of a patients tumor(s) throughout treatment, which can have significant treatment implications, particularly as more targeted treatment options become available [1, 11].
There is a wide range of methodology for measuring ctDNA. Assays are typically performed on plasma samples instead of serum samples due to the higher yield of ctDNA during immune-cell lysis [12]. A variety of assay platforms are available, but assay techniques can generally be divided into polymerase chain reaction- (PCR-) or next-generation sequencing- (NGS-) based techniques. PCR-based techniques are built upon primers complementary to a panel of known mutation sequences, whereas NGS-based techniques theoretically sequence the whole genome, though they still ultimately focus on a specific panel of genes or “hotspots.” While there are many commercially available assays, the Colon and Rectal-Anal Task Force of the United States National Cancer Institute developed general guidelines for the standardization of assay methodology and analytical validity for ctDNA use in CRC [10].
This novel biomarker shows promise in multiple aspects of CRC care including detection of clinically actionable mutations, risk stratification for recurrence after surgery and/or adjuvant therapy, measurement of the effectiveness of adjuvant therapy, and early detection of recurrence. With an explosion of trials investigating its role, ctDNA is rapidly evolving into an essential biomarker for the precision treatment of multiple malignancies, to include CRC (Table 1). In the following review, we will summarize the current data and on-going trials examining the role of ctDNA in the treatment of CRC and discuss future directions for this novel biomarker.
ctDNA for Localized CRC
Minimal Residual Disease Detection for Stage I–III Disease
Patients with stage I–III CRC are generally able to undergo curative-intent resection. After such a resection, however, decisions on who is at high enough risk of recurrence to justify adjuvant therapy can be difficult and are based mostly on clinical factors. As a result, there is a wide spectrum of disease recurrence within a given stage and many patients who are rendered ostensibly disease-free by surgery may have a microscopic disease that cannot be seen either grossly or radiographically. Detection of such occult disease can be accomplished through the detection of ctDNA in the postoperative setting, termed minimal residual disease (MRD), which has been shown to be a reliable predictor of disease recurrence.
In a prospective multicenter cohort of 130 patients with stages I to III CRC, ctDNA levels were measured pre-operatively, post-operatively at day 30, and every third month for 3 years to determine its association with disease recurrence. Patients who were found to be ctDNA positive at postoperative day 30 were 7 more times likely to have the recurrence within 3 years than ctDNA-negative patients. Also, patients found to be ctDNA positive after adjuvant therapy were 17 times more likely to recur [13]. Wang and colleagues also demonstrated that serial ctDNA measurement was a sensitive marker of disease recurrence in stage I, II, and III resected CRC. At a median follow-up of 49 months, they found a recurrence rate of 77% in patients with a positive ctDNA level and a recurrence rate of 0% in patients with a negative level [6]. These associations persisted even when controlled for clinicopathologic features such as age, T and N staging, and pathologic response to therapy in both studies [6, 13]. This powerful predictor of recurrence may serve as a better determinant than traditional staging when deciding which patients really need adjuvant therapies.
While detecting a positive ctDNA has a clear role in prognostication, the specific quantity of ctDNA found may also be used to further stratify recurrence risk. Tie et al. demonstrated that recurrence risk after surgical resection in stage II/III CRC cancer increases exponentially with increasing ctDNA mutant allele frequency (MAF), with hazard ratios of 1.2, 2.5, and 5.8 for MAF of 0.1%, 0.5%, and 1% respectively [14]. Therefore, integration not only of a binary ctDNA analysis (present or not present), but a quantitative analysis, may further enhance clinicians’ ability to stratify recurrence risk for patients in this adjuvant setting.
ctDNA-Guided Therapy
Because of its association with a high risk of recurrence, postoperative ctDNA may identify patients with resected stage II–III CRC in whom there is a clear benefit from adjuvant chemotherapy. In particular, the use of adjuvant chemotherapy for patients with stage II CRC remains controversial and decisions for its application are based upon high-risk clinical factors [15,16,17,18]. ctDNA offers an opportunity not only to give adjuvant therapy when the recurrence risk is high, but also to omit chemotherapy when the risk of recurrence is low. In a study of patients with resected stage II CRC, Tie and colleagues found that ctDNA was detected in 7.9% of patients who did not receive adjuvant chemotherapy and 79% of these patients recurred during study follow-up [19]. Interestingly, only 9.8% of patients recurred who were ctDNA-negative following surgery, highlighting ctDNA as a potential marker for residual disease and risk of recurrence. Most recently in the DYNAMIC trial, a ctDNA-guided approach was compared to standard clinicopathologic factors for the selection of patients for adjuvant therapy for stage II CRC and resulted in a reduced use of adjuvant chemotherapy without any reduction in 2-year RFS [20]. Moreover, this trial supported the use of ctDNA-guided treatment decisions and confirmed the low risk of recurrent disease in those patients with stage II CRC without detectable ctDNA after surgery.
In Japan, a large prospectively adaptive platform trial termed CIRCULATE-Japan (NCT04457297) is examining the utility of ctDNA levels in determining the need for adjuvant chemotherapy for resectable stage II–IV CRC, with a goal of enrolling 2500 patients. This adaptive platform utilizes the over-arching GALAXY data registry, which will include all patients who undergo complete surgical resection for stage II–IV CRC and will include the collection of serial ctDNA levels before surgery, and throughout the postoperative period, as well CT scans every 6 months for 7 years. This registry will be used to screen patients into adjuvant “de-escalation” and “escalation” trials. Patients with negative post-operative ctDNA levels at 4 weeks will be randomized into the VEGA trial, comparing no adjuvant chemotherapy to 3 months of CAPOX with a seven-year follow-up. Patients with negative ctDNA at 4 weeks post op who then go on to develop a positive ctDNA at any time between 4 weeks and 2 years will be enrolled in the ALTAIR trial. In this trial, both arms will receive four cycles of adjuvant CAPOX followed by randomization to receive 6 months of trifluridine/tipiracil (FTD/TPI) or placebo and followed for 5 years [21]. This large adaptive trial may elucidate the role of ctDNA in patient selection for adjuvant therapy.
An important component within the CIRCULATE-Japan trial is the DENEB study, which will prospectively enroll 200 patients from the GALAXY registry with pT1 CRC with high-risk features after complete local resection scheduled for additional colorectal resection with lymphadenectomy. The study will compare the ability of pre-operative ctDNA levels to predict lymph node metastases (LNM) versus pathologic features such as depth of submucosal invasion > 1000 μm, lymphovascular invasion, high grade, and poor differentiation following completion resection [22]. Under current guidelines, patients with these high-risk features undergo additional resection but only 6–16% of these patients have identified LNM after resection [23]. This study aims to determine if ctDNA can be used to better predict LNM in these patients compared to these classic pathologic criteria, with implications of reducing overtreatment in certain low-risk populations that may be sufficiently treated with local resection alone [22].
Further exciting studies are investigating the role of ctDNA in guiding adjuvant therapy for stage II and stage III CRC, whether chemotherapy is needed for all or only for those with detectable ctDNA, escalation, or de-escalation. For patients with low-risk stage II CRC typically not offered chemotherapy (stage IIA), COBRA is a prospective phase II/III trial comparing ctDNA-guided treatment decisions. Patients are randomized to either standard-of-care surveillance or ctDNA-informed arm where those with detectable ctDNA receive 6 months of adjuvant FOLFOX/CAPOX compared to surveillance for those without detectable ctDNA. This study aims to evaluate if ctDNA can indicate which patients may benefit from chemotherapy in this cohort and conversely who may remain without chemotherapy on surveillance, as well as if chemotherapy can increase survival in those with detectable ctDNA in patients with stage IIA CRC [24]. Within stage III and high-risk stage II CRC where adjuvant chemotherapy is standard of care, NRG-GI008 (CIRCULATE-US) aims to evaluate whether escalation may benefit those with detectable ctDNA or whether those without detectable ctDNA postoperatively may forego chemotherapy and enter surveillance. Patients with resected stage III CRC without postoperative ctDNA detection will enter cohort A and randomize to either standard of care FOLFOX/CAPOX or active surveillance with ctDNA monitoring. Those patients with resected stage II or III CRC with detectable ctDNA postoperatively, or patients under surveillance in cohort A that become ctDNA positive, may enter cohort B and randomize to either standard of care FOLFOX/CAPOX or escalation to FOLFOXIRI. This trial aims to investigate an overall ctDNA-guided approach to both escalation and de-escalation for patients with resected stage II or III CRC [25]. Lastly, the DYNAMIC-III trial randomizes patients to ctDNA-informed chemotherapy strategies vs. standard of care (i.e., ctDNA-blinded) strategies, and is separated into two cohorts of patients with stage III CRC based on post-operative ctDNA positivity [26]. In the ctDNA-negative cohort, a de-escalation strategy is compared to standard-of-care treatment, based on 3-year recurrence-free survival (RFS). In the ctDNA-positive patients, an escalation treatment strategy is compared to the standard of care as measured by 24-month RFS. Primarily, the DYNAMIC-III trial aims to demonstrate the non-inferiority of ctDNA-guided therapy in terms of RFS [27]. These studies offer further opportunities for individualized care and determination of adjuvant chemotherapy efficacy.
Implications for Surveillance
Even in the setting of an initial negative ctDNA, clinical recurrence may be predicted earlier with serial ctDNA monitoring than traditional strategies of post-operative surveillance. Compared to standard-of-care radiologic imaging, ctDNA monitoring has offered an impressive 8.7 months of mean lead time to detection of recurrence (p < 0.001), with an additional fivefold increase of ctDNA levels prior to radiologic detection [13]. In another longitudinal cohort from Denmark, elevated ctDNA levels predicted recurrence with a lead time of 9.4 months compared to radiologic imaging, and ctDNA quantitative dynamics demonstrated a correlation with tumor volume [28]. Another study found that a positive ctDNA level preceded radiologic detection of recurrence by a median of 3 months, which may impact decision-making on imaging intervals and perhaps the modality of imaging at later visits [6].
Ultimately, the goal of surveillance strategies is to improve survival via early detection, but it is not clear that diagnosing recurrence sooner will translate to better survival outcomes. Historically, more intensive surveillance strategies have resulted in improved survival [29], but this has become a point of contention more recently [30, 31]. Further, it is unclear as of yet if early detection of ctDNA without a radiologic correlate will improve patient outcomes. A randomized trial IMPROVE-IT2 is currently underway in Denmark to compare standard-of-care CT scan surveillance and ctDNA-guided surveillance in stage III or high-risk stage II CRC patients who underwent surgical resection. In the experimental arm, patients undergo serial ctDNA measurements every 4 months, with a positive result triggering a whole-body FDG-PET/CT scan assessment. The goals of this study are to determine if ctDNA-guided surveillance increases the proportion of patients receiving curative intent resection or local treatment of metastatic disease, as well as outcomes including overall survival, quality of life, cost-effectiveness, and time to recurrence detection [32]. Beyond this, multiple additional studies are currently underway investigating the integration of ctDNA into surveillance for patients with stage I–III CRC as ctDNA has been repeatedly shown to be a sensitive marker for recurrence detection (Table 1).
ctDNA for Resectable Colorectal Liver Metastases
The liver is the most common site of metastasis from CRC, and unfortunately, the 5-year OS for patients with stage IV disease is approximately 14% [33]. Despite this, some patients with CLM may achieve long-term survival, and these patients should be considered for curative-intent therapy [34]. Indeed, the 5-year OS for patients who are able to undergo curative-intent hepatectomy for CLM is between 40 and 60% [35, 36]. While long-term survival can be achieved, rates of recurrence after curative-intent hepatectomy for CLM are between 50 and 70%, and the application of systemic chemotherapy has shown inconsistent benefit [37,38,39,40]. To date, the addition of chemotherapy, whether via perioperative or adjuvant approach, has only been shown to improve RFS, not overall survival (OS). One may hypothesize that the use of a more refined and dynamic biomarker may better inform selection for chemotherapy based upon the risk of recurrence, which may lead to improved outcomes.
Pre-Hepatectomy ctDNA Detection for Patients with CLM
Preoperative detection of ctDNA in patients undergoing hepatectomy for CLM has been shown to predict worse outcomes. Narayan and colleagues drew intraoperative blood samples in 59 patients undergoing hepatectomy for CLM and found that 67% of these patients had detectable ctDNA [41]. The authors found an association with the detection of ctDNA, specifically circulating mutations in TP53, was associated with decreased 2-year disease-specific survival. In a similar study, Kobayashi and colleagues detected ctDNA preoperatively in 80% of patients undergoing initial hepatectomy in a limited cohort of patients with solitary resectable CLM in Japan. At a median follow-up of 39 months, the median RFS of patients preoperatively ctDNA positive was 12.5 months, while the median RFS was not reached in those without ctDNA detection [42]. Although the difference was not statistically significant, median OS for patients with detectable ctDNA preoperatively tended to be shorter compared to those ctDNA-negative (78.1 months vs. NR). The majority of patients did not undergo preoperative chemotherapy in this study, thus reflecting the natural history of preoperative ctDNA detection. In a report from MD Anderson Cancer Center, preoperative ctDNA was detected in 71% of patients undergoing curative-intent hepatectomy for CLM, but preoperative ctDNA status was not associated with either RFS or OS [43]. Interestingly, in this cohort, patients were treated with perioperative chemotherapy, so their preoperative ctDNA sampling was following preoperative chemotherapy, and it is possible some patients experienced ctDNA clearance following preoperative chemotherapy. It is likely that, while the proportion of patients with CLM with a negative preoperative ctDNA assay is low, this group will have improved outcomes, and might benefit from a de-escalation of therapy.
Minimal Residual Disease Detection Following Hepatectomy for CLM
Similar to MRD for patients with resected localized colorectal cancer, the detection of ctDNA following curative-intent resection of CLM is associated with worse oncologic outcomes. In a seminal study in 2008, Diehl and colleagues showed in a small cohort of patients that the detection of ctDNA following curative-intent surgery was associated with recurrence [8]. In this cohort of 18 patients, 16 of whom had stage IV disease, the authors found that postoperative ctDNA detection was associated with recurrence and most recurred within 1 year [8]. Furthermore, in a preliminary analysis of 63 patients who underwent hepatectomy for CLM at MD Anderson Cancer Center, and had ctDNA analysis throughout their treatment course, patients with detectable ctDNA at any time postoperatively had significantly worse 2-year OS from surgery (70% vs. 100%, p = 0.005). In a subsequent prospective cohort of 48 patients who underwent curative-intent hepatectomy for CLM at our institution, 38% of patients had detectable ctDNA postoperatively and those ctDNA-positive patients had a median RFS of 7.5 months, compared to 33 months for those ctDNA-negative [43]. Even further, in a larger study of 105 patients, 30% of patients had detectable ctDNA within 180 days of curative-intent hepatectomy for CLM, and ctDNA-positive patients had significantly worse median RFS at 6.3 months compared to 12.2 months for those ctDNA-negative [44]. Similarly, Reinert and colleagues performed a longitudinal study of 115 patients with perioperative and serial postoperative surveillance plasma samples up to 3 years in patients who underwent curative-intent resection of CLM [45]. Among the 40 patients with plasma samples drawn within 30 days of surgery and prior to initiation of adjuvant therapy in the study, 67.5% were ctDNA-negative and 32.5% were ctDNA-positive. All patients with detectable ctDNA within 30 days of surgery recurred compared to 55.6% of those ctDNA-negative after a median follow-up of 19.7 months in those without recurrence.
Detection of ctDNA postoperatively not only identifies patients at high risk for recurrence, but the serial sampling of ctDNA postoperatively continues to provide critical information for patients and providers. For example, in a prospective study of 54 patients who underwent hepatectomy for resectable CLM from 2011–2014, Tie and colleagues found that 24% of patients were ctDNA-positive postoperatively and these patients had an 83% risk of recurrence [46]. Patients in this study received chemotherapy in either a perioperative or adjuvant therapy approach and underwent serial ctDNA analyses longitudinally throughout care. Although a small cohort of patients, 8 of 11 patients with persistently detectable ctDNA following completion of adjuvant therapy recurred, while 2 of the 3 patients who cleared their ctDNA following adjuvant chemotherapy remained disease free at 60 and 82 months after hepatectomy, supporting the application of chemotherapy for patients with MRD [46]. Thus, serial ctDNA analyses throughout the continuum of care may afford a window into treatment efficacy for patients undergoing curative-intent surgery for CLM.
Patients without detectable ctDNA following curative-intent hepatectomy for CLM are at lower risk for recurrence, however, do still recur. In our series of 105 patients with ctDNA analysis within 180 days of hepatectomy, 70% of patients were ctDNA-negative, yet almost half of these patients still recurred within 1 year [44]. All of these patients who experienced early recurrence had extrahepatic disease resected at the time of hepatectomy, indeterminate lung nodules, had multiple CLM, or underwent margin-positive resection. Indeed, Tie and colleagues reported a Kaplan–Meier estimate of a 5-year RFS of 69.3% for those ctDNA-negative postoperatively and Reinert and colleagues reported a 55.6% rate of recurrence in ctDNA-negative patients in their study [45, 46]. Interestingly, Reinert and colleagues found that in 87.5% of postoperative ctDNA-negative patients who recurred, the initial site of recurrence was the lungs and lung recurrences were over 15 times more likely to be in ctDNA-negative patients [45]. Although some of these negative results may be due to platform sensitivity, careful consideration of ctDNA-negativity following hepatectomy is important when discussing ctDNA-guided postoperative treatment and surveillance decisions.
Perioperative ctDNA Dynamics and CLM
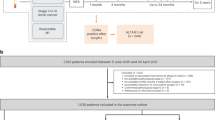
Perioperative measurement of ctDNA around curative-intent surgery for CLM may offer more refined prognostication than a single time point. Patients with ctDNA detected preoperatively may experience molecular clearance after preoperative systemic therapy or surgery, and this may offer more information about the prognosis. In the aforementioned prospective study of patients undergoing curative-intent hepatectomy for CLM at our institution, 23% did not have ctDNA detected either before or after surgery (ctDNA-/-), 29% were positive preoperatively and remained positive postoperatively (ctDNA + / +), and 40% experienced molecular clearance of preoperatively detected ctDNA (ctDNA ±) [43]. Patients ctDNA ± had outcomes similar to those without perioperative ctDNA detection (ctDNA-/-), supporting the idea that curative-intent surgery can render a patient disease free even when ctDNA positive prior to surgery (Fig. 1). Moreover, this supports efforts to identify patients who may be at risk for MRD on the basis of ctDNA dynamics.
Perioperative ctDNA dynamics for patients undergoing curative-intent hepatectomy for CLM. Kaplan–Meier analysis for A recurrence-free and B overall survival based upon dynamic perioperative ctDNA detection. Log-Rank p values. Adapted with permission from Newhook et al. Ann Surg 2022 [43]
In the study discussed above, Tie and colleagues reported the association of ctDNA dynamics and risk for recurrence following curative-intent surgery for CLM [46]. Overall, they again reported the strong association with postoperative ctDNA detection with both RFS and OS compared to those ctDNA negative. For those patients who continued onto adjuvant chemotherapy with serial samples available (n = 11), 3 patients experienced clearance of ctDNA with chemotherapy and 2 of these patients remained without recurrence after a median follow-up of 50.5 months. Those patients without clearance on adjuvant chemotherapy recurred. The detection of ctDNA following completion of oncologic therapy was associated with a 0% 5-year RFS; however, perhaps, most importantly clearance of ctDNA (ctDNA-) was associated with a 75.6% 5-year RFS [46].
In this same study, 23 patients (43%) underwent neoadjuvant chemotherapy and 21 (91%) of these patients had detectable ctDNA prior to treatment, compared to 80% reported by Kobayashi and 71% preoperative detection reported by our group at MD Anderson Cancer Center [42, 43, 46]. Interestingly, the rate of ctDNA detection decreased with each cycle of neoadjuvant chemotherapy in the Tie et al. study, and overall a 40-fold decrease in variant allele frequency detected over the course of neoadjuvant therapy in this cohort [46]. Most patients (59%) had undetectable ctDNA following neoadjuvant therapy; however, ctDNA clearance with neoadjuvant therapy was not associated with improved RFS, which may be due to the small sample size [46]. It remains to be elucidated whether ctDNA dynamics within the neoadjuvant therapy phase of care is associated with oncologic outcomes or can predict which patients are at the highest risk for MRD.
Predictors of MRD and ctDNA-Informed Treatment Strategies for CLM
As MRD is a surrogate for recurrence, the detection of ctDNA following hepatectomy identifies patients with occult metastatic disease and is a potential endpoint for future research and clinical trials. Furthermore, preoperative identification of patients at high risk for MRD may allow improved stratification of patients for surgery and/or novel treatment strategies. Nishioka and colleagues from MD Anderson Cancer Center sought to identify patient and tumor characteristics associated with postoperative ctDNA detection within 180 days of curative-intent hepatectomy for CLM [44]. In this study of 105 patients, having multiple tumors and a comutation in RAS + TP53 was associated with postoperative ctDNA detection within 180 days [44]. Similarly, Tie and colleagues found that having more than 1 CLM was associated with postoperative ctDNA detection; however, also noted that patients with left-sided, node-positive primary tumors and elevated postoperative CEA levels were associated with MRD [46]. This is important because preoperative identification of patients at the highest risk for MRD may inform treatment intensity or sequencing. Moreover, these patients may be candidates for novel treatment strategies in the preoperative setting to decrease the proportion of patients with MRD following hepatectomy for CLM.
Future Directions
The association of ctDNA detection with outcomes for patients with CRC makes it a promising biomarker that may augment treatment decisions, sequencing, and surveillance, and has the potential to directly inform the use of the standard of care and/or novel therapies for these patients. Multiple on-going prospective clinical trials are evaluating the impact of ctDNA detection and ctDNA-guided therapy on outcomes of patients across the spectrum of the disease and soon will provide data to potentially solidify the use of ctDNA for patients with CRC.
Much of the impact of ctDNA detection has been focused on those who are ctDNA positive, but there is much potential to impact care by identifying those who are ctDNA-negative following curative-intent surgery for CRC. Under current treatment paradigms, some patients are undergoing intensive therapies that may not provide benefits. Even in the setting of metastatic disease, the use and sequencing of systemic chemotherapy for patients with resectable CLM remain an area of debate. The use of ctDNA will hopefully allow clinicians to identify patients that may avoid potentially morbid therapies, impacting quality of life, and reserve lines of chemotherapy for future use in case of recurrence. Indeed, that is the question being asked in a prospective clinical trial of ctDNA-guided adjuvant chemotherapy at The University of Texas MD Anderson Cancer Center, the REACT-CLM trial (Risk-stratifiEd Adjuvant ChemoTherapy for Colorectal Liver Metastases; NCT05062317). Within a perioperative chemotherapy treatment model, patients undergoing curative-intent hepatectomy following preoperative chemotherapy undergo ctDNA analysis postoperatively and those without ctDNA detection will de-escalate or forego postoperative chemotherapy at the discretion of their provider. Those ctDNA-positive patients will either continue with intensive systemic chemotherapy or additional treatment options according to the recommendations of their oncology team. With a primary endpoint of 12-month RFS among ctDNA-negative patients, the aim of this trial is to evaluate ctDNA-guided treatment decisions for patients with CLM with a focus on de-escalation of chemotherapy for low-risk, ctDNA-negative patients following hepatectomy (Fig. 2).
Although dynamic changes in ctDNA detection in the preoperative setting has yet to be associated with outcomes, this may be due to the small sample sizes in published studies. Dynamic changes in variant allele frequencies have been associated with outcomes for patients with unresectable stage IV disease undergoing palliative intent treatment and thus may be an efficacious biomarker for response to chemotherapy [47]. Therefore, studies with larger sample sizes may elucidate if these findings are translatable to patients undergoing preoperative therapy for CRC. Furthermore, tumor tissue somatic mutational profiling may complement ctDNA dynamics in identifying patients who may benefit from novel treatment strategies in the preoperative setting that may be aimed at decreasing MRD following curative intent surgery for CRC.
Conclusion
The detection of ctDNA is an exciting, sensitive, and dynamic biomarker for CRC with the potential to evaluate unique mutational data for each patient. As a liquid biopsy or a cancer detection assay, ctDNA identifies patients with MRD following curative-intent therapy and is associated with recurrence. Many investigations are underway into ctDNA’s role in surveillance and treatment algorithms, and it may become a pivotal data point in personalized decision-making including treatment sequencing, choice, intensity, and duration of therapies. The use of ctDNA assays may lead to increasingly nuanced, personalized treatment strategies for patients throughout the spectrum of disease for colorectal cancer.
References
Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6(224):224ra24.
Ruibal Morell A. CEA serum levels in non-neoplastic disease. Int J Biol Markers. 1992;7(3):160–6.
Ballehaninna UK, Chamberlain RS. Serum CA 19–9 as a biomarker for pancreatic cancer-a comprehensive review. Indian J Surg Oncol. 2011;2(2):88–100.
Wanebo HJ, et al. Preoperative carcinoembryonic antigen level as a prognostic indicator in colorectal cancer. N Engl J Med. 1978;299(9):448–51.
Chao M, Gibbs P. Caution is required before recommending routine carcinoembryonic antigen and imaging follow-up for patients with early-stage colon cancer. J Clin Oncol. 2009;27(36):e279–80; author reply e281.a.
Wang Y, et al. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol. 2019;5(8):1118–23.
Diehl F, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci. 2005;102(45):16368–73.
Diehl F, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14(9):985–90.
Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med. 2010;16(9):398–406.
Dasari A, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat Rev Clin Oncol. 2020;17(12):757–70.
Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497(7447):108–12.
Merker JD, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. Arch Pathol Lab Med. 2018;142(10):1242–53.
Reinert T, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5(8):1124–31.
Tie J, et al. Prognostic significance of postsurgery circulating tumor DNA in nonmetastatic colorectal cancer. individual patient pooled analysis of three cohort studies. Int J Cancer. 2020;148(4):1014–26.
Rebuzzi SE, et al. Adjuvant chemotherapy for stage II colon cancer. Cancers. 2020;12(9).
André T, et al. Adjuvant fluorouracil, leucovorin, and oxaliplatin in stage II to III colon cancer: updated 10-year survival and outcomes according to BRAF mutation and mismatch repair status of the MOSAIC study. J Clin Oncol. 2015;33(35):4176–87.
Argilés G, et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2020;31(10):1291–305.
Benson AB, et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19(3):329–359.
Tie J, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8(346).
Tie J, et al. Circulating Tumor DNA Analysis Guiding Adjuvant Therapy in Stage II Colon Cancer. N Engl J Med. 2022;386(24):2261–72.
Taniguchi H, et al. CIRCULATE-Japan: circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021;112(7):2915–20.
Miyo M, et al. DENEB: Development of new criteria for curability after local excision of pathological T1 colorectal cancer using liquid biopsy. Cancer Sci. 2022;113(4):1531–4.
Ichimasa K, et al. Risk stratification of T1 colorectal cancer metastasis to lymph nodes: current status and perspective. Gut Liver. 2021;15(6):818–26.
Verbus EA, et al. Circulating tumor DNA as a predictive biomarker in adjuvant chemotherapy for patients with stage 2A colon cancer (COBRA). Ann Surg Oncol. 2021;28(8):4095–7.
Sahin IH, et al. Minimal residual disease-directed adjuvant therapy for patients with early-stage colon cancer: CIRCULATE-US. Oncology (Williston Park). 2022;36(10):604–8.
Anandappa G, et al. TRACC: tracking mutations in cell-free DNA to predict relapse in early colorectal cancer—a randomized study of circulating tumour DNA (ctDNA) guided adjuvant chemotherapy versus standard of care chemotherapy after curative surgery in patients with high risk stage II or stage III colorectal cancer (CRC). J Clin Oncol. 2020;38(15_suppl):TPS4120-TPS4120.
Tie J. Circulating tumour DNA analysis informing adjuvant chemotherapy in stage III colon cancer: a multicentre phase II/III randomised controlled study (DYNAMIC-III). 2017, Australian New Zealand Clinical Trials Registry: ANZCTR.
Schøler LV, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res. 2017;23(18):5437–45.
Renehan AG, et al. Impact on survival of intensive follow up after curative resection for colorectal cancer: systematic review and meta-analysis of randomised trials. BMJ. 2002;324(7341):813.
Jeffery M, et al. Follow-up strategies for patients treated for non-metastatic colorectal cancer. Cochrane Database Syst Rev. 2016;11:CD002200.
Snyder RA, et al. Association between intensity of posttreatment surveillance testing and detection of recurrence in patients with colorectal cancer. JAMA. 2018;319(20):2104–15.
Nors J, et al. IMPROVE-IT2: implementing noninvasive circulating tumor DNA analysis to optimize the operative and postoperative treatment for patients with colorectal cancer – intervention trial 2 Study protocol. Acta Oncol. 2020;59(3):336–41.
Siegel RL, et al. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
Kawaguchi Y, et al. Contour prognostic model for predicting survival after resection of colorectal liver metastases: development and multicentre validation study using largest diameter and number of metastases with RAS mutation status. Br J Surg. 2021;108(8):968–75.
Kanas GP, et al. Survival after liver resection in metastatic colorectal cancer: review and meta-analysis of prognostic factors. Clin Epidemiol. 2012;4:283–301.
Kawaguchi Y, et al. Conditional recurrence-free survival after resection of colorectal liver metastases: persistent deleterious association with RAS and TP53 Co-Mutation. J Am Coll Surg. 2019;229(3):286–294 e1.
Portier G, et al. Multicenter randomized trial of adjuvant fluorouracil and folinic acid compared with surgery alone after resection of colorectal liver metastases: FFCD ACHBTH AURC 9002 trial. J Clin Oncol. 2006;24(31):4976–82.
Ychou M, et al. A randomized phase III study comparing adjuvant 5-fluorouracil/folinic acid with FOLFIRI in patients following complete resection of liver metastases from colorectal cancer. Ann Oncol. 2009;20(12):1964–70.
Nordlinger B, et al. Perioperative FOLFOX4 chemotherapy and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC 40983): long-term results of a randomised, controlled, phase 3 trial. Lancet Oncol. 2013;14(12):1208–15.
Bridgewater JA, et al. Systemic chemotherapy with or without cetuximab in patients with resectable colorectal liver metastasis (New EPOC): long-term results of a multicentre, randomised, controlled, phase 3 trial. Lancet Oncol. 2020;21(3):398–411.
Narayan RR, et al. Peripheral circulating tumor DNA detection predicts poor outcomes after liver resection for metastatic colorectal cancer. Ann Surg Oncol. 2019;26(6):1824–32.
Kobayashi S, et al. Impact of preoperative circulating tumor DNA status on survival outcomes after hepatectomy for resectable colorectal liver metastases. Ann Surg Oncol. 2021;28(8):4744–55.
Newhook TE, et al. Prospective Study of perioperative circulating tumor DNA dynamics in patients undergoing hepatectomy for colorectal liver metastases. Ann Surg. 2022.
Nishioka Y, et al. Effect of co-mutation of RAS and TP53 on postoperative ctDNA detection and early recurrence after hepatectomy for colorectal liver metastases. J Am Coll Surg. 2022;234(4):474–83.
Reinert T, et al. Circulating tumor DNA for prognosis assessment and postoperative management after curative-intent resection of colorectal liver metastases. Int J Cancer. 2022;150(9):1537–48.
Tie J, et al. Circulating tumor DNA dynamics and recurrence risk in patients undergoing curative intent resection of colorectal cancer liver metastases: A prospective cohort study. PLoS Med. 2021;18(5)–e1003620.
Tie J, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26(8):1715–22.
Author information
Authors and Affiliations
Contributions
TEN was responsible for the initial conception and outline of the manuscript, and for Figs. 1 and 2. AMA created Table 1. All authors contributed significantly to each draft of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Adams, A.M., Vreeland, T.J. & Newhook, T.E. Circulating Tumor DNA: Towards More Individualized Treatment for Patients with Resectable Colorectal Cancer. J Gastrointest Canc 54, 1071–1081 (2023). https://doi.org/10.1007/s12029-022-00888-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-022-00888-y