Abstract
Background
The recurrence rate after hepatic resection of colorectal liver metastases (CRLM) remains high. This study aimed to investigate postoperative circulating tumor DNA (ctDNA) based on ultra-deep next-generation sequencing (NGS) to predict patient recurrence and survival.
Methods
Using the high-throughput NGS method tagged with a dual-indexed unique molecular identifier, named the CRLM-specific 25-gene panel (J25), this study sequenced ctDNA in peripheral blood samples collected from 134 CRLM patients who underwent hepatectomy after postoperative day 6.
Results
Of 134 samples, 42 (31.3%) were shown to be ctDNA-positive, and 37 resulted in recurrence. Kaplan-Meier survival analysis showed that disease-free survival (DFS) in the ctDNA-positive subgroup was significantly shorter than in the ctDNA-negative subgroup (hazard ratio [HR], 2.96; 95% confidence interval [CI], 1.91–4.6; p < 0.05). When the 42 ctDNA-positive samples were further divided by the median of the mean allele frequency (AF, 0.1034%), the subgroup with higher AFs showed a significantly shorter DFS than the subgroup with lower AFs (HR, 1.98; 95% CI, 1.02–3.85; p < 0.05). The ctDNA-positive patients who received adjuvant chemotherapy longer than 2 months showed a significantly longer DFS than those who received treatment for 2 months or less (HR, 0.377; 95% CI, 0.189–0.751; p < 0.05). Uni- and multivariable Cox regression indicated two factors independently correlated with prognosis: ctDNA positivity and no preoperative chemotherapy.
Conclusion
The study demonstrated that ctDNA status 6 days postoperatively could sensitively and accurately predict recurrence for patients with CRLM using the J25 panel.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
What’s New?
Although ctDNA was established as a biomarker for predicting recurrence in patients with CRLM, many issues still needed to be clarified, including the optimal timing of blood collection after surgery sequencing depth, and the sensitivity and specificity of ctDNA for predicting recurrence. This study developed a novel panel containing 25 genes associated with CRLM prognosis. Differences in predicted recurrence were compared between the J25 panel (a panel with fewer genes and greater sequencing depth) and the 642-gene panel (a panel with more genes and relatively smaller sequencing depth).
The ctDNA status values of peripheral blood collection between 6 and 31 days postoperatively were compared. The findings showed that the ctDNA status 6 days postoperatively could sensitively and accurately predict recurrence for patients with CRLM using the J25 panel.
Colorectal cancer (CRC) is the third most common cancer worldwide and the second leading cause of cancer-related deaths.1 Approximately 50–70% of patients will experience metastases, contributing to the high mortality rates reported for CRC.2 Hepatic resection remains the gold standard treatment that can be curative for patients with colorectal liver metastases (CRLM).3
More than half of CRLM patients experience cancer recurrence after hepatic resection, and most of these recurrences occur within 1 year.4 Circulating tumor DNA (ctDNA) could be a useful prognostic marker for minimal residual disease (MRD), intended to describe an occult state of micrometastatic disease undetectable by conventional imaging methods or blood tests after definitive treatment.5 Patients with a high risk of recurrence should receive more intense adjuvant chemotherapy and surveillance, whereas patients with low risk could be managed with de-escalation treatment without compromising survival outcomes.6 The prognostic role of ctDNA-based MRD detection currently is established for many malignant tumors and has been incorporated into standard clinical guidelines.7,8 However, routine MRD evaluation by next-generation sequencing (NGS) has been hindered, mainly because of the difficulties detecting and authenticating low-level mutations, especially single nucleotide variants (SNVs).
Limited studies have established ctDNA as a biomarker for predicting recurrence in CRLM patients.9,10 However, the sample sizes were small, and these studies were designed retrospectively. Furthermore, some issues remain to be clarified, including the optimal timing of postoperative blood collection and the sequencing depth.
The current study developed a novel panel containing 25 genes associated with CRLM prognosis. Differences in predicted recurrence were compared between the J25 panel (a panel with fewer genes and greater sequencing depth) and the 642-gene panel (a panel with more genes and relatively smaller sequencing depth). The study was approved by the Ethics Committee of Beijing Cancer Hospital and performed in accordance with the Declaration of Helsinki.
Methods and Materials
Study Design
Between February 2019 and March 2021, the study enrolled patients with CRLM from Hepatopancreatobiliary Surgery Department I of Beijing Cancer Hospital who underwent hepatic resection. The patients underwent radical resection of the primary tumor and liver metastases. The inclusion criteria specified patients who had resectable CRLM before hepatic resection as determined by a multidisciplinary team (MDT), metastasis identified as liver-limited disease, preserved liver function (indocyanine green [ICG] < 10%), and no other simultaneous malignancies. Patients who underwent only ablation or palliative hepatic resection (R2) were excluded from the study.
Disease Management
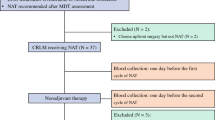
Preoperative staging and evaluation of liver metastasis were performed by computed tomography (CT) scan or MRI of the abdomen and chest for each patient according to standard clinical protocols. Peripheral blood samples were collected a median of 6 days postoperatively, and some patients also had blood collection a median of 31 days after surgery to assess differences in ctDNA at different blood collection times.
All the patients had samples collected after radical resection for both primary and liver metastases. The patients provided liver metastatic tumor tissue samples containing 50% or more tumor cells. After radical resection, patients may have received adjuvant chemotherapy, which consisted mainly of regimens containing 5-fluorouracil (5-FU), leucovorin, and oxaliplatin (mFOLFOX6) or 5-FU, leucovorin, and irinotecan (FOLFIRI) with or without bevacizumab or cetuximab.
All the patients were followed up, with disease-free survival (DFS) and overall survival (OS) as the end points. The patients were followed up every 3 months for the first 2 years with physical examination, carcinoembryonic antigen (CEA), measurement, CA 19-9 serum measurement, and CT scans of the abdominal/thoracic/pelvic area (enhanced MRI could also replace CT).
Sample Processing
Tumor DNA was extracted from 5 to 10 slides (5-mm thick) of formalin-fixed, paraffin-embedded (FFPE) tumor tissue samples using the QIAamp DNA FFPE Tissue Kit (Qiagen, Venlo, Netherlands). Peripheral blood samples were isolated by centrifugation at 1600 g for 10 min, then at 16,000 g for another 10 min to collect plasma and paired white blood cell samples. According to the manufacturer’s instructions, cell-free DNA (cfDNA), containing fractions of ctDNA, was extracted from 3 to 5 ml of plasma using HiPure Circulating DNA Midi Spin Kits (Magen, Guangzhou, China). Genomic DNA (gDNA) was extracted from white blood cell (WBC) samples using the Blood Genomic DNA Mini Kit (CWBIO, Beijing, China). All DNA extracted from peripheral blood samples and tumor tissue samples was estimated by a targeted sequencing strategy capturing all exons of 642 tumor-related genes (Table S1), whereas the ctDNA from peripheral blood samples was analyzed additionally by the J25 panel (described in the following section).
Gene Selection and Performance Verification of the J25 Panel
The J25 panel contained 25 genes that correlated with CRLM prognosis, all of which were confirmed to have relatively high mutation frequencies in the following three independent databases: the 642-gene panel sequencing data accumulated from previous CRLM samples in our department, the Memorial Sloan Kettering (MSK) CRLM database, and The Cancer Genome Atlas CRC database. All the samples could have at least one mutation detected by the J25 panel. The gene list and mutation frequency of each gene are presented in Table S2.
Performance verification also was performed for the J25 panel with standard gradient DNA samples using quality control indicators including effective sequencing depth, probe coverage, and detection sensitivity and accuracy. In standard gradient DNA samples, three selected hotspot mutation loci in CRLM were tested through repeated spike-in measurements of the well-characterized reference material set, GW-OCTM009 (GeneWell, Shenzhen, China), at five dilution allele frequencies (AFs): 0.05, 0.1, 0.3, 0.5, and 1% (Table S3). Furthermore, 145 typical CRLM hotspot mutations in 10 healthy human samples were sequenced with the J25 panel to examine specificity at the mutation level (Table S4).
Hybrid Capture-Based Targeted DNA Sequencing
The target NGS tests of all samples were performed at GloriousMed Clinical Laboratory (Shanghai) Co., Ltd. Two separate custom-designed DNA enrichment panels were used: the NimbleGen SeqCap EZ choice (Roche, Basel, Switzerland) was used to capture the coding regions of the 642 genes, and the dual-index UMI-added xGen Predesigned Gene Capture Pools (IDT, Coralville, IA, USA) were used to capture the coding regions of the J25 panel genes. For each sample, 20–50 ng of cfDNA, 200–500 ng of tumor DNA, or 500 ng of DNA were used for library preparation and quantification, guided by the KAPA Hyper Prep protocols (Roche). Pools of four to six libraries were used to hybridize to the capture panel for 16 h at 47/65 °C. Washing, recovery, and amplification were performed sequentially according to the standard procedures of the NimbleGen SeqCap EZ and IDT panels. The libraries were then purified using KAPA Pure Beads (Roche) and quantified using the Qubit dsDNA HS Assay Kit (Thermo Fisher, Waltham, MA, USA). The final libraries were sequenced on an Illumina Novoseq6000 (Illumina, San Diego, CA, USA) at PE150 mode. The raw sequencing depth was approximately × 6000 for the 642-gene tumor tissue samples, × 20,000 for the 642-gene cfDNA samples, and × 60,000 to × 200,000 for the J25 cfDNA samples.
Identification of Somatic Mutations
Sequencing adapters were trimmed from raw data using Trimmomatic (version 0.36, Babraham Institute, Cambridge, UK). After adapter trimming, the reads then were aligned to the hg19 version of the human reference genome using BWA-MEM in paired-end mode. Polymerase chain reaction (PCR) duplicates were removed using Picard (version 2.8.1) for the 642-gene panel sequencing data or fgbio (version 1.2.0) for the J25 panel sequencing data. For the 642-gene panel sequencing data, base quality scores were further recalibrated by GATK (version 3.7). Somatic and germline mutations were called using Mutect2 (GATK 3.7 for the 642-gene panel, GATK 4.0 for the J25 panel), and VardictJava (Release 1.8.0) with a paired workflow.
Variants then were annotated and filtered using ANNOtate VARiation and an in-house developed script. The details for variant filtering were as follows: (1) for tissue samples using the 642-gene panel, the variant frequency was 4.5% or higher; (2) for blood samples using the 642-gene panel, the variant frequency was 1% or higher, and mutations also were identified in the corresponding tissues; and (3) for blood samples using the J25 panel, the variant read number was 2 or higher, mutations also were identified in the corresponding tissues, and additionally, the variant frequency was 0.001 or higher for samples in which only one variant site was identified. Based on the filtered mutations, the blood sample was classified as ctDNA-positive if it contained mutations. Otherwise it was classified as ctDNA-negative.
Statistical Analysis
Fisher’s exact test was used to compare differences between the two groups. Kaplan-Meier survival curves were generated by the ggsurvplot function in the survminer R package (https://github.com/kassambara/survminer). Uni- and multivariate analyses were performed using the Cox proportional hazards model to identify independent risk factors for DFS. For significant risk factors, the Cox proportional hazards model was reconstructed to calculate the coefficients of these factors. The risk score was calculated based on the coefficients and status of the factors. Time-dependent receiver operating characteristic (ROC) analysis was performed using the survival ROCR package.11 Statistical analysis was conducted using R software (version 3.6.1). All statistical significance thresholds were set at a two-tailed p value of 0.05.
Results
Sensitivity and Specificity of the J25 Panel
The average initial sequencing depth of the UMI library of the J25 panel reached 100,000 ± × 50,000 at initial DNA input of 30 ng or higher. The effective library DNA conversion rate was approximately 70%, with a single-base level error rate of 9.83 × 10−5 (95% confidence interval [CI], 9.3710–5–1.03 × 10–4). In the standard gradient test, the overall compliance was 100% at the sample level for the 15 samples involving three mutation loci and five dilution concentrations. The coefficient of variation of detected AF mutations complied with the requirements (Table S3). In addition, the J25 panel presented no false-positives, and the specificity was 100% in all 145 CRC typical hotspot mutations tested (Table S4).
Patient Characteristics
The study enrolled 134 patients between February 2019 and March 2021. The demographic and clinicopathologic characteristics of the patients are shown in Table 1. The median age was 58 years, and 73% of the patients were male. On the basis of pathologic characteristics, 89.6% had T3–4 diseases and 78.4% were synchronous for liver metastasis timing. The median metastasis lesion number was 3, and the median largest metastasis size was 22 mm. Of the 134 patients 110 (82.1%) received preoperative chemotherapy. During the study period, 84 (62.7%) of the patients experienced recurrences. The median follow-up period after radical resection was 17.3 months (range, 3.3–37.3).
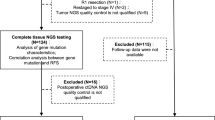
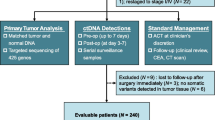
The workflow of sample processing and sequencing experiments is presented briefly in Fig. 1. Surgical tissues from all the patients were sequenced using the 642-gene panel. Using the J25 panel, 134 peripheral blood samples after a median of 6 postoperative days were sequenced, and At the same time, to compare the differences between the two panels, 25 of the 134 patients were sequenced using the 642-gene panel. To assess differences in ctDNA at different blood collection times, peripheral blood samples from 43 of the 134 patients after a median of 31 postoperative days also were sequenced using the J25 panel.
Comparison of the J25 Panel and the 642-Gene Panel
The study tested 25 peripheral blood samples with both J25 panel and 642-gene panel. The J25 panel detected 23 mutations in eight positive samples in a mean of three mutations per sample, and the mean mutation AF was 1.039% (range, 0.019–8.386%). The 642-gene panel detected seven mutations in three positive samples in a mean of two mutations per sample, and the mean mutation AF was 4% (range, 1.3–9.6%) (Fig. 2A). Therefore, mutations with a lower AF could be detected by the J25 panel due to its high depth-sequencing characteristics. Notably, all the mutations identified by the 642-gene panel also were identified by the J25 panel (Fig. 2A). The mutation rate and the concordance between tissue and plasma ctDNA of the J25 panel were significantly better than those of the 642-gene panel, especially for the known prognostic hotspot genes APC and TP53 in CRLM (Fig. 2B).
Comparison of the J25 and 642-gene panels for 25 patients. A The mutation oncoplot included tumor tissue sequenced by the 642-gene panel, plasma ctDNA sequenced by both the J25 and 642-gene panels. B The concordance of APC and TP53 between tissue and plasma ctDNA in the two panels. C, D The Kaplan-Meier survival curves for DFS or OS in ctDNA-positive and ctDNA-negative subgroups with the J25 panel. E, F The Kaplan-Meier survival curves for DFS or OS in ctDNA-positive and ctDNA-negative subgroups with the 642-gene panel. ctDNA, circulating tumor DNA; DFS, disease-free survival; OS, overall survival
Among the 18 patients with recurrence, 44.4% (8/18) were mutation-positive according to the J25 panel, whereas only 16.6% (3/18) had mutations according to the 642-gene panel. The 25 patients were divided into two subgroups by ctDNA-negative or ctDNA-positive status according to the J25 panel or 642-gene panel results. The Kaplan-Meier survival analysis showed that both DFS and OS were significantly shorter in the ctDNA-positive subgroup than in the ctDNA-negative subgroup in both panels (Fig. 2C–F).
A time-dependent ROC curve was used to assess the prognostic value of the biomarkers, including the J25 panel and the 642-gene panel at 3 years. For DFS, the AUC was was 0.72 with the J25 panel and 0.58 with the 642-gene panel (Fig. S1). The two AUCs differed significantly (p = 0.0099). For OS, the AUC was 0.62 with the J25 panel and 0.63 with the 642-gene panel (Fig. S1). The two AUCs showed no difference (p = 0.95). Therefore, the J25 panel could distinguish DFS more significantly between negative and positive groups than the 642-gene panel.
Time Point of Sample Collection for the J25 Panel
The study selected 43 paired peripheral blood samples at two postoperative time points, after a median of 6 days and 31 days, respectively. There were 15 and 13 positive samples on postoperative days 6 and 31, respectively (Fig. S2). The mean mutation AF was 0.165% on day 6 and 3.261% on day 31. Therefore, the sampling on postoperative day 6 did not differ significantly from the sampling on postoperative day 31 days. It is easier to obtain blood samples 6 days after surgery, and some patients have been discharged from the hospital within 31 days, which is not conducive to sample collection. Therefore, we chose to collect peripheral blood 6 days after surgery.
Ability to Predict Recurrence and Survival Using the J25 Panel
In the 134 tumor tissue samples sequenced by the 642-gene panel, the mutation AFs ranged from 5.6 to 67.4%. All the tissue samples had at least one mutation detected in the genes of the J25 panel (Fig. S3). A total of 134 peripheral blood samples were tested by J25 panel. The mutation profile of tissue and blood samples is shown in Fig. 3A. The J25 panel detected 103 mutations in 42 ctDNA-positive samples, for a mean of 2.45 mutations per sample, and the mean mutation AF was 0.824% (range, 0.019–13.308%).
The ability to predict recurrence and survival using the J25 panel for 134 patients. A The mutation oncoplot included tumor tissue sequenced by the 642-gene panel (blue) and plasma ctDNA sequenced by the J25 panel (red). B The mutation oncoplot of 42 ctDNA-positive samples. C, D The Kaplan-Meier survival curves for DFS or OS in ctDNA-positive and ctDNA-negative subgroups with the J25 panel. E The Kaplan-Meier survival curves for DFS grouped by the median allele frequency in the ctDNA-positive group. ctDNA, circulating tumor DNA; DFS, disease-free survival; OS, overall survival
The frequently mutated genes were TP53, APC, KRAS, and PIK3CA in the postoperative blood samples (Fig. 3B). Of the 42 patients who were ctDNA-positive, 37 relapsed. The recurrence rate was 88.1%. Of the 92 patients who were ctDNA-negative, 47 relapsed. The recurrence rate was 51.09%.
The 134 patients were divided into two subgroups by ctDNA-negative or ctDNA-positive status according to the J25 sequencing results. The Kaplan-Meier survival analysis showed that the DFS in the ctDNA-positive subgroup was significantly shorter than in the ctDNA-negative subgroup (hazard ratio [HR], 2.96; 95% CI, 1.91–4.6; Fig. 3C). The OS in the ctDNA-positive subgroup also was significantly shorter than in the ctDNA-negative subgroup (HR, 11.5; 95% CI, 3.28–40.6; Fig. 3D). When the 42 ctDNA-positive samples were further divided by the median of the mean AF (allele frequency), which was 0.1034%, the subgroup with higher AFs showed a significantly shorter DFS than the subgroup with lower AFs (HR, 1.98; 95% CI, 1.02–3.85; p = 0.039; Fig. 3E). The subgroup with higher AFs showed shorter OS than the subgroup with lower AFs, but the difference was not significant (HR, 1.55; 95% CI, 0.488–4.92; p = 0.45; Fig. S4).
The patients were further divided into two groups according to the time of postoperative chemotherapy (> 2 vs. ≤ 2 months). The ctDNA-positive patients differed significantly in DFS between the two groups (p = 0.0042). The subgroup with more than 2 months of chemotherapy showed a significantly longer DFS than those who had chemotherapy for 2 months or less (HR, 0.377; 95% CI, 0.189–0.751; Fig. 4A). The DFS of the ctDNA-negative patients did not differ significantly between the two groups (Fig. 4B). These results had certain guiding significance for postoperative chemotherapy.
Prognostic Factors for DFS
The association of clinical factors, genes, and ctDNA status with recurrence was further analyzed using univariate Cox regression. The following seven factors were significantly associated with DFS: ctDNA status, postoperative CA199 value, bilobar or unilobar localization, metastasis lesion number, preoperative chemotherapy, preoperative CA199 value, and KRAS mutation (p < 0.05; Table S5). Multivariable Cox regression was further performed with these significant factors. The following two factors remained independently correlated with prognosis: ctDNA positivity (HR, 1.89; 95% CI, 1.14–3.11; p = 0.013) and no preoperative chemotherapy (HR, 0.43; 95% CI, 0.19–0.96; p = 0.04; Fig. 5).
Discussion
The recurrence rate of CRLM after radical resection remains high. Currently, imaging examination is the only efficient method for detecting these clues, which can demonstrate residual disease when sufficient tumor size is present.12 Although numerous maneuvers have been designed to predict patients who might experience recurrence with high risk factors, such as clinical risk scores and RAS status, it is difficult to say that these traditional prognostic models reflect precision medicine in modern era.13,14
Several studies have indicated the clinical utility of ctDNA for MRD assessment, monitoring recurrence, and treatment response in many fields of carcinomas.15 However, studies that evaluate ctDNA as a prognostic biomarker in patients with CRLM for MRD detection and prediction of disease progression are limited. The current study was a prospective cohort analysis to investigate the utility of the J25 panel for predicting recurrence in CRLM. The J25 panel is intended to profile cancer somatic mutations comprehensively with a large targeted gene panel for the purpose of reducing economic cost worldwide.
In this study, a J25 panel for CRLM was proposed and constructed by the dual-index, UMI-tagged, high-throughput NGS method. It included the most common 25 gene mutations in colorectal cancer. The J25 panel was superior to the previous 642-gene panel in terms of sensitivity and specificity.
Although the NGS method has been widely applied to monitor MRD in CRLM, it does not explore the time point of blood sampling for postoperative ctDNA detection. Diehl et al.16 demonstrated that the half-life of ctDNA was 114 min after radical resection. Evidence showing the rapid decay of ctDNA after radical tumor resection was presented in a lung cancer study. The authors indicated that cfDNA was metabolized by the third day after R0 resection, and that it could be a baseline value for postoperative surveillance.17
Additionally, Guo et al.18 reported that the ctDNA mutation AF did not differ significantly between 2–4 days and 5–8 days postoperatively. Therefore, a preliminary study was performed to compare sampling times of 6 days and 31 days postoperatively at the beginning of current study, and the findings showed no significant difference. The authors concluded that 6 postoperative days might be a rational sampling time point for ctDNA recurrence monitoring.
The current standard biomarker used for care surveillance is CEA, which has limited sensitivity and specificity and is elevated in only a few patients.19 In recent years, ctDNA has been a promising biomarker receiving significant attention. It is released by dying cancer cells into the bloodstream and theoretically contains the same genetic and epigenetic changes as the cancer cells from which it originated.20 Several studies have declared that detectable ctDNA after surgery in early-stage cancers is associated with a high risk of recurrence.12,21 Several studies have reported that in stages II and III colon and rectal cancer, ctDNA testing can identify the MRD after radical resection, thus identifying patients with the highest risk of recurrence.22
Although limited studies have reported that ctDNA detection could be used to select patients eligible for liver metastasectomy, the sample size for ctDNA testing in these studies was small and available only during and after surgery.23,24 It showed that patients with detectable ctDNA after hepatic resection have markedly shorter recurrence-free survival than those who had no resection (HR, 18; 95% CI, 7.9–40).25
Detection of persistent ctDNA after surgery or adjuvant treatment effectively identifies CRC patients with MRD who ultimately will experience recurrence without additional therapy. Accordingly, several prospective clinical trials of ctDNA-guided adjuvant therapy are currently underway to evaluate whether patients with evidence of MRD through ctDNA detection after surgery, adjuvant therapy, or both may benefit from additional or more intensive systemic therapy to reduce recurrence risk.26 In the current study, CRLM patients with ctDNA positivity who underwent 2 months of adjuvant chemotherapy had improved long-term DFS.
The development of several molecular techniques that allow the high-sensitivity analysis of ctDNA has sparked recent interest in pursuing the clinical role of ctDNA for MRD detection in various tumor types.27 The limited sensitivity of the ctDNA assays is an important concern, especially in the context of resected early-stage colon cancer patients whose ctDNA levels in plasma are quite low. In the series reported by Tie et al.26 and Reinert et al.21 the calculated sensitivity of a single ctDNA measurement in the immediate postoperative period was modest, at approximately 50%. Fragment size analysis, tracking of multiple mutations, serial testing, and adoption of NGS panels that enable testing for a large number of genomic and epigenetic alterations might improve assay sensitivity.28,29 Theoretically, a hybrid capture-based approach is a better choice than an amplicon-based approach for cfDNA-based liquid biopsy applications.30,31
Study Limitation
The current study had potential limitations. First, only one time point was selected (postoperative day 6). Future studies will include regular monitoring of ctDNA dynamics at various time points, including initial diagnosis, before surgery, after surgery on day 6, and after completion of adjuvant chemotherapy. Second, a comparative analysis between the J25 panel and the 624-gene panel was performed on only 25 paired samples. Ideally, the specificity of ctDNA positivity and the mutation AFs in the positive subgroup for predicting recurrence needs to be further validated by expanding the external cohort and extending the follow-up periods.
Conclusion
The current study developed the J25 panel, which has higher sensitivity and accuracy. The ctDNA-positive factor at postoperative day 6 was a sufficient and significant predictor of DFS. The J25 panel outperformed the 642-gene panel in predicting DFS performance and detected signs of recurrence earlier than classical clinical methods. Therefore, this study is expected to translate into an improved recurrence indicator for patients with CRLM.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Adam R, Kitano Y. Multidisciplinary approach of liver metastases from colorectal cancer. Ann Gastroenterol Surg. 2019;3:50–6.
Margonis GA, Buettner S, Andreatos N, et al. Prognostic factors change over time after hepatectomy for colorectal liver metastases: a multi-institutional, international analysis of 1099 patients. Ann Surg. 2019;269:1129–37.
Imai K, Allard MA, Benitez CC, et al. Early recurrence after hepatectomy for colorectal liver metastases: what optimal definition and what predictive factors? Oncologist. 2016;21:887–94.
Naidoo M, Gibbs P, Tie J. ctDNA and adjuvant therapy for colorectal cancer: time to re-invent our treatment paradigm. Cancers. 2021;13:346.
To YH, Lee B, Wong HL, Gibbs P, Tie J. Circulating tumour DNA to guide treatment of gastrointestinal malignancies. Visc Med. 2020;36:388–96.
Alimirzaie S, Bagherzadeh M, Akbari MR. Liquid biopsy in breast cancer: a comprehensive review. Clin Genet. 2019;95:643–60.
Chabon JJ, Hamilton EG, Kurtz DM, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020;580:245–51.
Kobayashi S, Nakamura Y, Taniguchi H, et al. Impact of preoperative circulating tumor DNA status on survival outcomes after hepatectomy for resectable colorectal liver metastases. Ann Surg Oncol. 2021;28:4744–55.
Mason MC, Tzeng CD, Tran Cao HS, et al. Preliminary analysis of liquid biopsy after hepatectomy for colorectal liver metastases. J Am Coll Surg. 2021;233:82–9.
Sing T, Sander O, Beerenwinkel N, Thomas L. ROCR: visualizing classifier performance in R. Bioinformatics. 2005; 21(20):3940–1. https://doi.org/10.1093/bioinformatics/bti623.
Tie J, Cohen JD, Wang Y, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut. 2019;68:663–71.
Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–18 (discussion 318–21).
Tosi F, Magni E, Amatu A, et al. Effect of KRAS and BRAF mutations on survival of metastatic colorectal cancer after liver resection: a systematic review and meta-analysis. Clin Colorectal Cancer. 2017;16:e153–63.
Liu APY, Smith KS, Kumar R, et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell. 2021;39:1519–30.
Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90.
Chen K, Zhao H, Shi Y, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC). Clin Cancer Res. 2019;25:7058–67.
Guo N, Lou F, Ma Y, et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci Rep. 2016;6:33519.
Loupakis F, Sharma S, Derouazi M, et al. Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis Oncol. 2021;5:1166–77.
Warton K, Mahon KL, Samimi G. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr Relat Cancer. 2016;23:R157–71.
Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124–31.
Wang Y, Li L, Cohen JD, et al. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol. 2019;5:1118–23.
Bidard FC, Kiavue N, Ychou M, et al. Circulating tumor cells and circulating tumor DNA detection in potentially resectable metastatic colorectal cancer: a prospective ancillary study to the Unicancer Prodige-14 Trial. Cells. 2019;8:516.
Narayan RR, Goldman DA, Gonen M, et al. Peripheral circulating tumor DNA detection predicts poor outcomes after liver resection for metastatic colorectal cancer. Ann Surg Oncol. 2019;26:1824–32.
Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346–92.
Tie J, Cohen JD, Wang Y, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5:1710–7.
Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJA. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018;18:1174–96.
Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10:eaat4921.
Chakrabarti S, Xie H, Urrutia R, Mahipal A. The promise of circulating tumor DNA (ctDNA) in the management of early-stage colon cancer: a critical review. Cancers. 2020;12:2808.
Samorodnitsky E, Jewell BM, Hagopian R, et al. Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum Mutat. 2015;36:903–14.
Garcia-Garcia G, Baux D, Faugere V, et al. Assessment of the latest NGS enrichment capture methods in clinical context. Sci Rep. 2016;6:20948.
Acknowledgment
We thank Hong-Wei Wang and Juan Li for their helpful suggestion in data analyzing. This study was supported by grants (nos. 81874143 and 31971192) from the National Nature Science Foundation of China and the Beijing Hospitals Authority Youth Program (code: QMS20201105).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
There are no conflicts of interest.
Ethical Approval
The study was approved by the Ethics Committee of Beijing Cancer Hospital. The study was performed in accordance with the Declaration of Helsinki
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10434_2023_13362_MOESM4_ESM.pdf
The mutation oncoplot of tumor tissue sequenced by 642-gene panel in 134 patients. Genes in the J25 panel were highlighted in red. All tissue samples had at least one mutation detected in the genes of the J25 panel (PDF 26 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, W., Jin, KM., Zhang, MH. et al. Recurrence Prediction by Circulating Tumor DNA in the Patient with Colorectal Liver Metastases After Hepatectomy: A Prospective Biomarker Study. Ann Surg Oncol 30, 4916–4926 (2023). https://doi.org/10.1245/s10434-023-13362-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-023-13362-1