Abstract
The intricate process of protein binding orchestrates crucial drug interactions within the bloodstream, facilitating the formation of soluble complexes. This research endeavours to improve the dissolution and oral bioavailability of Rifampicin (RMP) by strategically manipulating drug-protein binding dynamics and the hydrophobic characteristics of human serum albumin (HSA). Various precipitation techniques leveraging methanol, ammonium sulfate, and heat treatment were meticulously employed to tailor the properties of colloidal albumin (HSA NPs). The resultant complexes underwent comprehensive characterization encompassing evaluations of hydrophobicity, size distribution, surface charge, and structural analyses through FTIR, TG-DSC, XRD, and morphological examinations. The findings revealed a significant binding affinity of 78.07 ± 6.6% with native albumin, aligning with prior research. Notably, the complex RMP-HSA NPs-M13, synthesized via the methanolic precipitation method, exhibited the most substantial complexation, achieving a remarkable 3.5-fold increase, followed by the ammonium sulfate (twofold) and heat treatment (1.07-fold) methods in comparison to native albumin binding. The gastric simulated media exhibited accelerated drug release kinetics, with maximal dissolution achieved within two hours, contrasting with the prolonged release observed under intestinal pH conditions. These findings translated into significant improvements in drug permeation, as evidenced by pharmacokinetic profiles demonstrating elevated Cmax, AUC, t1/2, and MRT values for RMP-HSA NPs-M13 compared to free RMP. In summary, this innovative approach underscores the potential of precipitation methods in engineering stable colloidal carrier systems tailored to enhance the oral bioavailability of poorly soluble drugs, offering a pragmatic and scalable alternative to conventional surfactants, polymers, or high-energy methods for complex formation and production.
Highlights
-
Colloidal albumin produced by methanol exhibits superior physiochemical and pharmaceutical performance.
-
Albumin-Rifampicin conjugates are thought to be a potential method for improving drug release and oral bioavailability.
-
Albumin-Rifampicin complex shows better pharmacokinetic characteristics compared to free Rifampicin.

Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
A significant hurdle in drug development is addressing poor bioavailability, with over 40% of marketed drugs estimated to have low water solubility (lipophilic). In the United State pharmacopoeia, this figure exceeds 30% [1]. The inadequate solubility and extremely low dissolution rate of these drugs in gastrointestinal fluids often lead to insufficient bioavailability, particularly for Biopharmaceutics Classification System II class drugs [2]. Rifampicin is an antibiotic that is effective against infections like tuberculosis. Due to its low aqueous solubility (slightly soluble), high logP value (3 to 4), and high plasma protein binding (nearly 80%), designing its parenteral, transmucosal, and oral liquid formulations for rifampicin proves challenging. Furthermore, efforts to enhance rifampicin’s solubility through cyclodextrin complexation have shown limited improvement in its permeability [3].
Various formulation strategies have been devised to overcome low solubility and bioavailability issues, including micronization, solubilization via co-solvents, permeation enhancers, surfactant dispersions, salt formation, and precipitation techniques [4]. However, these techniques have limitations, constraining their utility in enhancing solubility. Alternative methods such as liposomes [5], microemulsions [6], and solid dispersions [7] They have shown restrained success and failed to address regulatory compliance, limiting their application across a broad range of biopharmaceuticals. Therefore, there is a pressing need for a novel approach to tackle formulation challenges, enhancing efficacy and optimising therapy in pharmaco-economics.
In this study, we leverage the natural affinity of hydrophobic drugs for plasma proteins to improve their aqueous distribution. Drug-protein binding is a reversible process involving weak bonds like hydrogen, hydrophobic, and ionic bonds. Human serum albumin, with a plasma concentration of 35 to 50 mg/mL and a high drug binding capacity, is known to bind with various drugs such as warfarin, diazepam, ketoprofen, and rifampicin. Albumin is known to enhance the solubility and dissolution rate of poorly aqueous soluble drugs owing to its amphiphilic and hydrocolloidal nature, high binding affinity for large biopharmaceuticals, and biosurfactants-like properties [8].
We prepared RMP-albumin complexes with both native and colloidal albumin. The techniques employed to produce colloidal albumin included the antisolvent, heat precipitation, and ammonium sulphate precipitation methods. The surface hydrophobicity of colloidal albumin and its impact on drug complexation were also evaluated. The resulting complexes were characterised for size, zeta potential, drug-albumin interaction, surface hydrophobicity, solubility, dissolution rate and ability to improve bioavailability.
Materials and Methods
Materials
Rifampicin was obtained from BV Patel Perd Centre, Ahmedabad. At the same time, human serum albumin was sourced from Central Drug House (P) Ltd. Dichloromethane, n-Octanol, and methanol of analytical grade was acquired from SRL Chemicals, Mumbai, India. All remaining chemicals and reagents were of analytical grade and were utilised without additional purification. Purified Milli-Q water from Millipore, Billerica, MA, USA, which had been degassed and filtered through 0.45 mm hydrophilic PVDF filters (Millipore Millex-HV), was employed in all experimental procedures.
Methods
Assay Method Development
The RMP estimation procedure was modified based on the report of Liu et al. (2021), with some modifications. RMP solutions were prepared in methanol and PBS pH 7.4, in the concentration ranging from 10 to 50 µg/mL. Absorbance readings were taken at 475 nm, the maximum absorption wavelength, and correlated with their respective concentrations. By establishing a linear regression equation with an r-squared value surpassing 0.99, the concentration of RMP could be determined in future assays [9,10,11].
Investigating Solubility, logP Value and Protein Binding Affinity of RMP with Native Albumin
Solubility determination of RMP was carried out by adding an excess of RMP to 10 mL of distilled water in sealed glass vials at 37 ºC. The liquids were shaken for 24 h, and then 1 mL of solution was withdrawn and centrifuged at 1200 rpm for 5 min to remove excessive RMP. A 20 µL aliquot of the resulting solution was injected into a High-Performance Liquid Chromatography (HPLC), and drug content was measured. For logP determination, A known amount of RMP was added to 10 mL water and shaken for 1 h. Finally, 10 mL n-octanol was added to this solution and shaken for another 1 h to ensure complete mixing in a separating funnel. Finally, the mixture solution was kept at a tripod stand for stabilization for 2 h, and the two immiscible layers were separated for analyzing RMP in each phase. Finally, the logP value was calculated using a standard equation. About 5%w/v albumin solution was made in phosphate buffer pH 7.4, and the excess drug was mixed into 10 mL of it to make a supersaturated solution RMP. The dispersion was kept at 37 ºC under mild agitation in a thermostat orbital shaker for 6 h. The dispersion was then subjected to dialysis for 12 h in phosphate buffer at pH 7.4 to separate the free drug from the complex. The drug concentration in the complex was determined by UV spectrophotometer at λ max 475 nm.
Development of Colloidal Dispersion of Albumin and Its Complex with RMP
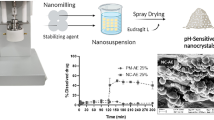
Colloidal albumin (HSA NPs) was synthesized using different physical and chemical precipitants, namely methanol (HSA NPs-M), ammonium sulfate (HSA NPs-AS), and heat (HSA NPs-H). In essence, a solution containing 5% w/v albumin in phosphate buffer at pH 7.4 underwent precipitation by exposure to aqueous solutions of methanol (0.2–0.8%v/v) and ammonium sulfate (0.2–0.7% w/v), as well as varying temperatures from 15–35 ºC. Following a brief 5-min exposure to the respective precipitants, the resulting colloidal dispersions were centrifuged at 20,000 rpm using an ultra-centrifuge. Subsequently, the pellets were collected and re-dispersed in phosphate buffer at pH 7.4. Optimization was carried out to determine the optimal levels of physical and chemical precipitants, ensuring colloidal albumin possessed desired physicochemical characteristics without interfering with the subsequent complexation process [12, 13].
The experimental conditions were optimized based on the physical properties of the developed colloidal system. The batch exhibiting favorable particle size distribution was complexed with RMP (RMP-HSA NPs). A 50 mL optimized colloidal dispersion in phosphate buffer (pH 7.4) with a concentration of 5%w/v was mixed with a 5 mL aqueous solution of RMP (0.25%w/v) for 2 h. Additionally, a super-saturated aqueous RMP solution was enclosed in a dialysis tube (molecular weight cutoff 2 kDa) at both ends and introduced into the dispersion to release free drugs into the medium continuously. As the complexation progressed, the free drug concentration in the medium decreased, compensated by the drug from the dialysis bag. This allowed exploration of the maximum affinity of colloidal albumin for complexation. The dispersion was then incubated at 37 ºC in phosphate buffer at pH 7.4, using an orbital thermostat shaker with gentle agitation (50 revolutions per minute (rpm)). The drug concentration in the complex is at various intervals. was determined by UV spectroscopy.
Characterization of RMP-HSA NPs
Drug Content
The RMP-HSA NPs underwent dialysis in a phosphate buffer (pH 7.4) for ample duration to prevent any additional drug from escaping into the external environment. Subsequently, the resulting complex was blended with an equal-volume solution of methanol and water (5% v/v). This methanolic dispersion of the RMP-HSA NPs underwent sonication for 2 min, then subjected to centrifugation at 15,000 rpm for 10 min. The drug concentration in the supernatant was then assessed via UV–Vis spectroscopy at 475 nm.
Particle Size Determination
The particle size and size distribution of the HSA NPs obtained through various precipitation methods (HSA NPs-M, HSA NPs-AS, and HSA NPs-H) were assessed using the Beckman Coulter Delsa TM Nano C zeta sizer. Approximately 2 mL of the dispersion was loaded into cuvettes of the Beckman Coulter instrument, and measurements of size and polydispersity index (PDI) were taken. Similarly, the particle size and size distribution of RMP-HSA NPs-M, RMP-HSA NPs-AS, and RMP-HSA NPs-H were also evaluated using the same procedure [14].
Surface Charge
The physical stability of colloidal albumin in a dispersion is dictated by its surface charge. This charge is assessed by measuring zeta potential, which quantifies the electrophoretic mobility of particles in an electric field. Therefore, the zeta potential was determined utilizing the Beckman Coulter DelsaTM Nano C Zeta sizer [15].
Surface Hydrophobicity
This experiment assessed the surface hydrophobicity of colloidal albumin complexes formed through various techniques. The assessment employed the Rose Bengal method, following the established procedure outlined in previous literature [16, 17]. Various albumin concentrations were dissolved in water (2% w/v, 4% w/v, and 6% w/v) and subjected to precipitation using distinct methods (methanol, ammonium sulfate, and heat-sensitive precipitation) according to predefined experimental parameters. Subsequently, 10 mg of Rose Bengal dye was introduced into each solution [18]. Aliquots were incubated for 24 h for dye binding with a hydrophobic surface. After 24 h, the aliquots were centrifuged, and absorbance was measured at 549 nm.
Fourier-Transform Infrared Spectroscopy
The Fourier Transform Infrared (FTIR) spectra of RMP, HSA, HSA NPs, and RMP-HSA NPs were examined using a Perkin-Elmer Spectrum RX1 FT-IR spectrometer. Before FT-IR analysis, all samples were thoroughly dried to ensure a moisture content of less than 3% and powdered for mixing with potassium bromide to form pellets. The measurements were conducted within the range of 4000 to 800 cm − 1 with a resolution of 2 cm−1.
Thermogravimetric Analysis
A differential scanning calorimetry (DSC) coupled with thermogravimetric (TG) analysis was conducted to verify the integration of RMP into the HSA NPs. DSC is a method used for thermal analysis, examining crystallinity, polymorphic transitions, and energy changes during formulation. The phase transitions of RMP, HSA NPs, and RMP-HSA NPs were analysed using a DSC-TG instrument (DSC 60, Shimadzu Co.) with perforated aluminum-sealed 50 µL pans. The analysis was conducted under nitrogen gas (20 ml/min) with a heating rate of 10 ºC/min, ranging from 50 to 250°C [19].
X-ray Diffraction Study
X-ray diffraction is a crucial technique in examining alterations in the crystalline arrangement of a drug throughout formulation development. For the analysis of RMP, HSA NPs, and RMP-HSA NPs, X-ray Diffraction (XRD) was conducted using the XPERT-PRO instrument, employing Cu Kα radiation with a wavelength of 1.542 Å, operated at 40 mA and 45 kV. The scanning rate was set at 10°/min, spanning from 5° to 50° of 2θ, with a wavelength increment of 0.017° [20].
Morphological Investigation
The surface morphology of the RMP-HSA NPs was investigated using a Scanning Electron Microscope (SEM) EVO M-10 (Zeiss), which employs a beam of electrons in a raster scan pattern to examine the sample surface. Before imaging, the sample was mounted on a holder and coated with platinum using a sputter coater. An accelerating voltage of 15 kV was applied during imaging. The SEM images were subsequently utilized to determine the average diameter of the colloidal albumin and the drug-colloidal albumin complex.
Evaluation of RMP-HSA NPs
Solubility Determination
Solubility assessments of RMP, RMP-HSA complex, and RMP-HSA NPs (prepared via various methods) were conducted by introducing an excessive amount of these samples into phosphate buffer at pH 7.4 to achieve saturation. Sealed tubes containing the samples were placed on a vortex shaker for 24 h at 37°C, followed by a 2-day equilibration period. Subsequently, the solution underwent centrifugation and filtration through a 0.1 µm membrane filter (Millipore Corporation). The resulting filtrate was appropriately diluted and subjected to analysis using a UV spectrophotometer at 475 nm [10].
Determination of Partition Coefficient
The logP of RMP and RMP-HSA NPs were determined according to the method reported by Yue et al. (2010) [21] with slight modification. In summary, samples containing 500 mg of RMP were divided into three separate vials and added to 5 mL of n-octanol. These mixtures were then agitated at 100 rpm for 24 h in a shaker water bath set at 25°C. Subsequently, aqueous phases consisting of solutions with varying pH levels (HCl pH 1.2, PBS pH 6.8, and PBS pH 7.4) were introduced to the n-octanol solutions and agitated for 24 h under the same conditions. Afterwards, the aqueous and n-octanol phases were separated and centrifuged at 10,000 rpm for 15 min. The resulting solutions were filtered using a 0.45 μm PVDF syringe filter (Millipore Millex-HV) and analysed for drug content using a UV–Vis spectrophotometer at 475 nm. The logarithm of the n-octanol/water partition coefficient (log P) was calculated using the formula logP = log10(P), where P represents the partition coefficient at different pH values ranging from 1.2 to 7.4.
Stability Studies
RMP has been noted to exhibit instability under specific pH conditions when exposed to the biological environment along with isoniazid at gastric pH. Maintaining its stability in biological environments is crucial for maximizing its efficacy. Hence, the stability of both RMP and RMP-HSA NPs was assessed under gastric pH 1.2 conditions in the presence of isoniazid [22].
In Vitro Dissolution Study
The dissolution behavior of RMP, RMP-HSA complex, and RMP-HSA NPs-M13 was investigated using a USP type I automated dissolution test apparatus (ElectrolabTDT-08L, India) in phosphate buffer at pH 1.2 and pH 6.8. Samples equivalent to 500 mg of RMP in 3 mL water were prepared and immediately placed into a dialysis bag (2 kDa molecular weight cutoff), which was then placed in baskets and immersed in dissolution vessels containing 900 mL buffer at pH 1.2 for 2 h, followed by 900 mL PBS at pH 6.8 for 4 h. The dissolution media were maintained at 37 ± 0.5 ºC and stirred at 50 rpm. Periodically, 1 mL aliquots of the samples were collected over 2 h initially and then over 4 h. Collected volumes were replenished with fresh dissolution medium as needed. All samples were filtered using a 0.45 µ PVDF syringe filter, specifically the Millipore Millex-HV, and the concentration of RMP was determined using a UV–Vis spectrophotometer at 475 nm [23].
Ex Vivo Permeation Studies
This study was performed according to the procedure mentioned by Sharma et al., [24] with some modifications. The gut mucosa of goats was procured from a nearby slaughterhouse in Ringer's solution. These mucosal samples were affixed onto enhancer cells, ensuring the mucosal side faced the formulation, while the inner surface exposed an area of 1.77 cm2 to the dissolution medium (pH 7.4; 150 mL) maintained at 37 ± 0.5°C and 50 rpm. Samples (both RMP and RMP-HSA NPs-M13), equivalent to 500 mg of RMP, were dispersed in 3 mL buffer at pH 1.2 and promptly introduced into the enhancer cell, which was then placed inside a glass jar for studying drug permeation through the mucosal membrane over time. At predetermined intervals over 3 h, 3 mL samples were withdrawn, filtered through a 0.45 μm membrane filter, and subjected to analysis using a UV–Vis spectrophotometer at 475 nm. The cumulative amount of drug permeated across the mucosa per unit of time was graphed to determine the rate of RMP permeation [25].
In Vivo Pharmacokinetic Study
Quantification of RMP in Plasma Samples
The levels of Rifampicin in the plasma were assessed using HPLC (Thermo Scientific, LTQ XL, Germany). A Phenomenex C18 column (250 mm × 4.6 mm, 5 µm) was the stationary phase, maintained at ambient temperature. The mobile phase consisted of a mixture of ammonium acetate and acetonitrile in a ratio of 30:70. A flow rate of 1.0 mL/min was maintained, and samples were subjected to analysis via HPLC [26].
Rifampicin Extraction and Sample Preparation
To extract RMP, 200 µL of plasma was placed into a micro-centrifuge vial, followed by the addition of 500 µL of methanol. The mixture was vigorously vortexed and then centrifuged at 10,000 rpm for 10 min. After centrifugation, the supernatant was isolated, and 20 µL samples were injected for analysis using HPLC [27].
Pharmacokinetic Studies of RMP and RMP-HSA NPs-M13
Male albino, Sprague Dawley rats, weighing between 150 and 200 g were allocated into two groups, each consisting of six rats per time point. One group received a dose of 15 mg/kg of RMP, while the other received an equivalent dose of the RMP complex. Blood samples were collected from the retro-orbital plexus of the rats at various time intervals and transferred into heparin-containing micro-centrifuge vials. Subsequently, the samples were centrifuged at 10,000 rpm for 10 min to separate the plasma, which was then stored at -20°C until analysis. Quantification of RMP was conducted using HPLC with Noscapine employed as an internal standard. Parameters such as maximum concentration (Cmax) and time to achieve maximum concentration (Tmax) were directly determined from the plasma concentration–time profile. Additionally, the area under the concentration–time curve (AUC0–48 h or AUC0–∞), elimination half-life (t1/2), and mean residence time (MRT) were calculated following the procedure outlined by Singh et al. in 2014. The relative bioavailability (F) of RMP-HSA NPs compared to RMP alone was assessed using the following equation.
Statistical Analysis
All the data are presented in means ± standard deviation (SD). Statistical analysis was performed using a paired ‘t’ test using GraphPad Prism software version 5.0. A significant difference was regarded as p < 0.05.
Results and Discussion
Method of Estimation
The UV absorption peak of Rifampicin in methanol and phosphate buffer was determined to be 475 nm, aligning with the reported λ max for Rifampicin, thereby confirming the purity of the provided drug. Figure 1 illustrates the UV absorbance spectrum of the drug across the 200–800 nm range, along with the standard curves for Rifampicin in methanol and PBS at pH 7.4. This figure displays the absorption maxima of Rifampicin and its corresponding calibration curves in methanol and PBS at pH 7.4. Linear responses were observed within the 10–50 µg/ml concentration range, with correlation coefficients of r2 = 0.9986 and 0.9979, respectively, indicating adherence to Beer’s law within these concentration ranges.
Aqueous Solubility, logP and Protein Binding Affinity of RMP with Native Albumin
The saturation solubility of Rifampicin in water was measured to be 1.03 mg/mL, indicating its restricted solubility in water. Solubility studies further demonstrated Rifampicin's pronounced affinity for organic solvents and limited solubility in aqueous environments, affirming its inherently lipophilic character. Table I delineates Rifampicin's solubility profile alongside its protein binding affinity with native albumin. Specifically, the drug content within native albumin was determined as 40 mg in a 10 mL solution of 5% albumin, likely attributed to the exposure of hydrophilic albumin groups within a phosphate buffer at pH 7.4. Rifampicin's calculated log P value stood at 1.83, affirming its significant lipophilic properties. Consequently, minimal interaction between the drug and native albumin was anticipated. However, despite this expectation, the literature indicates over 80% plasma protein binding of Rifampicin. Notably, the observed plasma protein binding closely aligned with previously reported findings in the literature [28].
Colloidal Albumin and Its Complex with RMP
Colloidal albumin was obtained per the methodology outlined in "Development of Colloidal Dispersion of Albumin and Its Complex with RMP" Section and subsequently fine-tuned to optimize its particle size and zeta potential. Various chemical and physical parameters were adjusted throughout the development and optimization process. Table II illustrates the optimization results of colloidal albumin achieved through precipitation induced by methanol, ammonium sulfate, and heat treatments. No viable batch was obtained with methanol concentrations up to 0.50% v/v in aqueous solution. The particle size of colloidal albumin ranged from HSA-NPs-M8 (21.5 nm) to HSA-NPs-M14 (149.2 nm), indicating a significant precipitation effect on the albumin solution. In each case, a negative surface charge exceeding 20 mV was observed. Higher methanol concentration leads to complete denaturation of proteins; hence, a phase separation can be obtained. A high concentration of the organic solvent may facilitate the complete exposure of the hydrophobic surface of the protein; however, a controlled increase in chemical denaturant concentration poses significant utility in managing colloidal dispersions. Here, the methanolic treatment in the tested concentration range posed tremendous utility in preparing albumin NPs. Similarly, batches treated with ammonium sulfate increased particle size, ranging from 30.5 nm to 148.2 nm at concentrations of 0.55% w/v and 0.7% w/v, respectively. The negative surface charge exceeded 20 mV in all cases. The particles with tiny appearance and size are a virtue of controlled process parameters and material attributes. Treating HAS solution with ammonium sulfate led to the formation of a colloidal solution in the tested ranges; however, using higher concentrations led to larger particles, which could be of limited utility. Heat treatment also yielded satisfactory batches, albeit with larger particle sizes, reaching up to 220.7 nm for HSA NPs-H5. Gradual temperature increases lead to tiny particles forming at 25 ºC and above. HSA molecules, i.e., solubilized in water, are stabilized by water exposure over the albumin surface if the system is stored in fixed condition. Stimuli that may trigger the exposure of the hydrophobic surface of the albumin molecule to some extent help to form smaller precipitates. Little change in system temperature from 20 to 25 ºC led to the formation of smaller tiny precipitates after sufficient incubation. However, these particles cannot be observed from the naked eye unless they are sufficiently large. Further, an increase in temperature led to an increase in size and complete precipitation (phase separation) was observed above 45 ºC. This could be due to the complete denaturation of the protein structure. In blood, HSA be in a colloidal form that is stabilized by various components present in blood. The proteins, peptides, hormones, enzymes, and other components help prevent the unfolding of the proteinaceous structure and, hence, are more stable. However, HSA in water is in solution form and is more prone to destabilization under small changes in stimuli conditions, and unfolding the proteinaceous structure is comparatively easy. The most optimal particle size was achieved through chemical treatment.
Characteristics of RMP-HSA colloidal complex
Particle Size and Size Distribution
The particle sizes of HSA NPs-M13, HSA NPs-AS10, and HSA NPs-H3 were initially measured at 66.9 ± 4.7 nm, 78.4 ± 7.9 nm, and 98.1 ± 9.1 nm, respectively. Upon complexing the drug with these particles, a significant increase in size was observed, measuring 185.6 ± 24.4 nm, 248.6 ± 18.83 nm, and 256.5 ± 18.6 nm for HSA NPs-M13, HSA NPs-AS10, and HSA NPs-H3, respectively. This indicates successful drug adsorption onto the surface of colloidal albumin (see Fig. 2). The findings indicate that methanol-treated precipitates exhibited a higher propensity for complexation, followed by ammonium sulfate and heat treatments. This preference could be attributed to the systemic effects induced by methanol. With its low surface tension, methanol rapidly displaces water from the hydration layer, approaching the albumin surface. This reduces polarity within the microenvironment, prompting structural rearrangements in the protein and increased surface hydrophobicity. Conversely, heat precipitation batches led to an overall increase in enthalpy-induced flash precipitation of albumin, resulting in larger particle sizes and PDI. Ammonium sulfate, with its capacity for charge neutralization, induced protein precipitations without significant structural modifications, ultimately limiting their surface hydrophobicity. These findings are corroborated by reports emphasizing the usefulness of methanol-based precipitation techniques in producing more sustainable hydrocolloids [30].
Size distribution and zeta potential of placebo and RMP-loaded albumin NPs by different methods, respectively. a Particle size distribution and b Zeta potential values. Pictures c, d, and e depict the particle size distribution curves of the drug-loaded particles prepared by methanol, ammonium sulphate, and heat treatments, respectively
Surface Charge
The examination of zeta potential revealed that both colloidal albumin and rifampicin, when in phosphate buffer at pH 7.4, carry a negative charge, indicating a lack of likelihood for ionic interaction between colloidal albumin and the drug. Additionally, zeta potential was found to have minimal influence on the complexation process. Upon the introduction of the drug to the colloidal albumin, there was a slight decrease in zeta potential, likely due to the slightly increased size of the drug-albumin complex. Figure 2 illustrates the diverse physicochemical characteristics of the optimized formulations.
RMP Assay in Colloidal Albumin Complex
The drug concentration within the albumin complex of the optimized batch was assessed, revealing the highest drug content in the methanol-treated batches, followed by those treated with ammonium sulfate and heat precipitation. This discrepancy may be attributed to the elevated surface hydrophobicity and larger surface area observed in the methanol-treated batch. Furthermore, the polydispersity index (PDI) of the methanol-treated batch was notably lower compared to the batches treated with ammonium sulfate and heat precipitation, indicating a more uniform distribution of colloidal albumin particles concerning their size distribution and, consequently, an increased overall surface area. Conversely, the lower drug content observed in the heat precipitation batch could be attributed to its higher PDI, larger particle size, and reduced surface hydrophobicity. The findings are summarized in Table III.
Surface Hydrophobicity
Surface hydrophobicity analysis revealed that colloidal albumin precipitates obtained through methanol treatment exhibited the highest surface hydrophobicity, followed by precipitation with ammonium sulfate and heat treatments. Methanol treatment creates conducive conditions for the self-precipitation of albumin by displacing water from the hydration layer. This water displacement increases polarity and prompts protein folding, increasing surface hydrophobicity. Additionally, the volume of methanol used plays a crucial role in determining the precipitates' size and PDI. It was observed that both size distribution and PDI increased proportionally with the concentration of methanol. In contrast, ammonium sulfate-induced protein precipitation through charge neutralization did not significantly alter the structural configuration of the proteins. Consequently, the surface hydrophobicity remained relatively low regardless of particle size. On the other hand, heat-induced precipitation resulted in flash precipitation due to the system's increased enthalpy, leading to a broader size distribution and lower surface hydrophobicity. Our findings align with those reported by He et al. (2013), corroborating the impact of different precipitation methods on the surface properties of albumin precipitates [31] (Fig. 3).
Infrared Spectrum
The IR spectra were acquired and scrutinized to detect interactions between RMP and albumin functional groups. The results of the IR spectroscopy refute the possibility of covalent bonding between the drug and albumin. However, the obtained peaks indicate some physical or weak interactions with albumin, likely attributed to the bonding of certain hydrophilic functional groups of RMP with albumin. All RMP peaks below 500 cm-1 vanished in the HSA, HSA NPs, and RMP-HSA NPs groups, while a peak at 480 cm-1 was consistently present across all groups. Additionally, a peak at 770 cm−1 showed modification (reduced intensity and shifted position) when complexed with albumin. Furthermore, the peak at 820 cm−1 (Ar − H) shifted to 799 cm−1, and another peak at 1033 cm−1 shifted to 1050 cm−1, possibly due to the influence of albumin. The RMP peak at 1240 cm−1 (C − O − C) shifted to 1259 cm−1 in the HSA, HSA NPs, and RMP-HSA NPs groups [32]. Figure 4i illustrates the IR spectra of both the drug and the colloidal albumin complex following methanol induction. While the fingerprint region remained consistent, the peak at 2634 cm−1 shifted to 2660 cm−1, attributed to the characteristic peaks of albumin. Notably, a significant peak at 3470 cm-1 (− OH) was observed to shift to 3380 cm−1. The presence of peaks at 2812 cm−1 (− CH3) was consistent across all tested groups. These peak shifts, although minimal, suggest the potential for a physical interaction between the drug and albumin.
Thermogravimetric-Differential Scanning Calorimetry
A comprehensive analysis using TG-DSC was conducted to validate the incorporation of RMP within the HSA NPs. Figure 4ii illustrates the TG-DSC curves for RMP-HSA NPs, RMP alone, and HSA NPs. In the case of RMP, a distinctive endothermic peak was detected at 117 ºC, indicative of a change in enthalpy (as depicted in Fig. 4b). This peak, not accompanied by any mass loss, characterized the phase transition of RMP, closely aligning with its melting point. Furthermore, an exothermic peak emerged at 162 ºC, accompanied by a mass loss of up to 48% within the range of 120–175 ºC, likely corresponding to the decomposition temperature of RMP. HSA-NPs displayed an endothermic peak at 150 ºC, likely corresponding to their melting phase. However, a notable shift in temperature towards higher values was observed in the DSC curves of pure albumin, possibly due to the increased structuring of albumin within the nanoparticles. After heating the RMP-HSA-NPs samples, another endothermic peak was evident in the DSC analysis at 195 ºC, precisely aligning with the temperature corresponding to unstructured albumin outside nanoparticles. Subsequently, the subsequent peak in the DSC curve corresponded to the decomposition of the protein within the nanoparticles, occurring within a temperature range of 285–338 ºC, accompanied by a weight loss exceeding 45%. The DSC curve for RMP immobilized within nanoparticles exhibited peaks characteristic of RMP but with reduced intensity. This thermal behaviour suggests that the drug did not undergo significant chemical alteration upon immobilization within the albumin nanoparticles, a finding consistent with the FT-IR results [33, 34].
X-Ray Powder Diffraction
The XRD analysis revealed distinct findings for the pure and the drug-albumin complex. In the case of the plain drug, a sharp peak was observed at 2ɵ 17.6, indicative of its crystalline structure, while for the RMP-HSA NPs complex, this peak shifted slightly to 18.9 (Fig. 4iii). Notably, a broad peak at a 2ɵ value of 12.3, characteristic of albumin alone, was absent in the RMP and its NP complex. Additionally, the peak at a 2ɵ value of 36 experienced a shift to 37.5 due to the presence of RMP in the complex. Another distinctive peak corresponding to RMP at a 2ɵ value 43.5 was also evident in the RMP-HSA NPs complex. While albumin exhibited some degree of surface modification of RMP, the complete surface alteration was not fully apparent in the graphs of HSA NPs, RMP, and RMP-HSA NPs. These findings underscore the potential of this physical complex to enhance solubility without altering the active moiety extensively. Moreover, the XRD results align well with those obtained from FTIR analysis and TG-DSC curves, further corroborating the insights gained from this investigation [35].
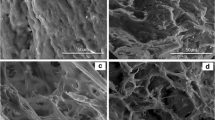
Morphology of the Prepared NPs
The surface morphology of the optimized RMP-HSA NPs-M13 formulation is depicted in Fig. 5. These images illustrate the formation of diminutive nanoparticles, with approximately five or more particles observed within a length of 1 µm, thus corroborating the particle size findings obtained through zeta-sizer analysis. Examination of drug content reaffirms robust drug loading post a six-hour incubation period, ensuring maximal drug deposition onto the surface of the nanoparticles [36]. The comparatively smaller size observed in the methanolic batch is ascribed to its increased surface hydrophobicity, which fosters a greater attraction to non-polar drugs. Conversely, the larger particle size observed in the ammonium sulfate and heat-treated groups is attributed to their relatively lower degree of surface hydrophobicity.
Effectiveness of RMP-HSA NPs
Solubility Determination
RMP-HSA NPs-M13 demonstrated the highest aqueous solubility at 12.29 ± 0.96 mg/mL, followed by RMP-HSA NPs-AS10 at 9.48 ± 0.47 mg/mL, and RMP-HSA NPs-H3 at 8.50 ± 0.73 mg/mL, in contrast to the solubility of RMP alone at 1.03 mg/mL. The colloidal complex based on albumin increased RMP's solubility by 5 (Fig. 6A). The superior solubility of RMP-HSA NPs-M13 can be attributed to its heightened surface hydrophobicity, smaller particle size, and increased surface area available for drug adsorption. Furthermore, the higher drug content in RMP-HSA NPs-M13 resulted in a lower albumin content per unit mass of the drug-albumin complex. Conversely, RMP-HSA NPs-AS10 and RMP-HSA NPs-H3 exhibited lower drug content, leading to higher albumin content per unit mass of the complex. Additionally, the lower surface hydrophobicity in RMP-HSA NPs-AS10 and RMP-HSA NPs-H3 led to a higher affinity for water, further increasing medium viscosity and diminishing drug solubility. Kudva et al. (2010) suggest that the presence of albumin enhances the solubility of curcumin, a hydrophobic compound, in the developed complex [37]. Figure 6 compares the solubility of RMP in RMP-HSA NPs-M13, RMP-HSA NPs-AS10 and RMP-HSA NPs-H3.
Partition coefficient
Table IV illustrates the logP values of RMP and RMP-HSA NPs-M13 across various media. RMP-HSA NPs-M13 demonstrated a reduced log P value compared to RMP, indicating a shift towards higher partitioning into the aqueous phase and decreased lipophilic character. This enhancement in aqueous solubility observed in the RMP-HSA NPs-M13 complex can be attributed to the effective binding capacity of albumin, the ample surface area facilitating physical interaction between RMP and albumin, and the resultant solubilizing effect.
Stability Studies
Stability assessments were conducted for both RMP and the RMP-HSA NPs-M13 complex, individually and in the presence of isoniazid (INH). Equal concentrations of RMP (20 mg) were subjected to pH 1.2 and 37 ºC conditions, with samples extracted after 4 h for analysis. The results revealed notable stability of RMP, both in its standalone form and when encapsulated within the RMP-HSA NPs-M13 complex, even in the presence of INH (Fig. 6B).
Dissolution Profile of RMP-HSA NPs-M13 Against RMP
Figure 7A illustrates the dissolution profiles of both RMP and RMP-HSA NPs-M13 in HCl (pH 1.2) and phosphate buffer (pH 6.8). In the gastric media, RMP-HSA NPs-M13 demonstrated significantly improved dissolution (76.8%) compared to RMP (40.46%) after two hours. Conversely, at pH 6.8, RMP and RMP-HSA NPs-M13 exhibited limited drug release after 4 h. Notably, most RMP-HSA NPs-M13 dissolution occurred within 2 h at gastric pH. RMP-HSA NPs-M13 showed a 1.89-fold increase in dissolution compared to RMP alone. The dissolution behaviour of RMP is notably influenced by the pH of the dissolution medium, aligning with differences in solubility [29]. Given RMP's basic nature, it exhibits enhanced solubility in acidic pH conditions. The complex effectively enhances solubility and dissolution in both acidic and basic environments.
Permeation Profile of of RMP-HSA NPs Against RMP Through Gastric Mucosa
Most RMP release occurred within a 2-h under gastric pH conditions, aligning with its heightened solubility at pH 1.2. Consequently, the permeation study was conducted solely at gastric pH for 3 h. When administered alone, RMP exhibited a 37.8% drug permeation through the gastric mucosa within 2 h (considering this period as the gastric emptying point), closely matching the 40.46% release observed during the same timeframe under gastric pH conditions. The formation of RMP complexes with HSA NPs resulted in an augmented dissolution profile, influencing its permeation behaviour (Fig. 7B). Notably, more than 74% drug permeation was achieved within 2 h with RMP-HSA NPs-M13, underscoring the remarkable partitioning effect of this complex compared to the drug alone. Corresponding to the dissolution profile findings, RMP-HSA NPs-M13 demonstrated a twofold increase in drug permeation, highlighting its significant potential in enhancing drug absorption through the gastric mucosa and time to maximum concentration (Tmax). While the permeation profile provides insights into its kinetic behavior, further validation through in vivo pharmacokinetic studies in small animals was conducted to confirm the improved kinetics [38]. Animal experiments were carried out as per the ARRIVE guidelines, the EU Directive 2010/63/EU for animal experiments and in accordance with the current guidelines of CCSEA India. The proposed activity was duly approved by CCSEA through the Institutional Animal Ethics Committee prior to the commencement of the experimental work.
Oral Pharmacokinetic Profile of of RMP-HSA NPs-M13 Against RMP in Animals
The pharmacokinetic study aimed to compare the oral pharmacokinetic patterns of RMP with RMP-HSA NPs-M13, assessing drug concentrations in rat plasma over 96 h post-oral administration. Plasma underwent liquid–liquid extraction. Figure 8 illustrates the mean plasma concentration–time curves of RMP and RMP-HSA NPs-M13. These plots depict the plasma concentration against time following oral administration of RMP-HSA NPs-M13 (at approximately 15 mg/kg) and RMP (15 mg/kg) to six rats.
The data collected is summarized in Table V. RMP-HSA NPs-M13 demonstrated a higher Cmax of 77.2 µg/mL with a Tmax of 3 h, whereas RMP alone exhibited a 29.9 µg/mL Cmax within 2 h. The mean AUC0–∞ values for RMP-HSA NPs-M13 and free RMP were 573.93 and 194.26 µg h/mL, respectively. The relative bioavailability of RMP-HSA NPs-M13 (AUC0–∞) compared to free RMP (AUC0–∞) was 295.44%. The observed increase in relative bioavailability of RMP-HSA NPs-M13 post-oral administration can be attributed to several factors: firstly, albumin, a significant constituent of blood, contributes to biocompatibility, and secondly, the enhancement of RMP's aqueous solubility within the RMP-HSA NPs-M13 complex. This improvement in bioavailability may also be ascribed to the increased rate and/or extent of RMP absorption into the intestinal mucosa facilitated by albumin, thus playing a pivotal role in enhancing drug absorption through the intestinal mucosa. It is documented that RMP is a potent enzyme inducer, causing its metabolism, which could potentially decrease its bioavailability and efficacy. Nevertheless, RMP-HSA NPs-M13, by augmenting RMP absorption, may mitigate RMP's metabolic processes in vivo, leading to increased bioavailability. However, the precise mechanism underlying the reduction in rifampicin metabolism through this approach warrants further investigation.
Conclusion
The study offers novel insights into enhancing RMP's dissolution rate and oral bioavailability through biomolecular intervention by forming its colloidal complex with HSA. We successfully synthesized RMP–HSA NPs using a physicochemical stimuli-sensitive precipitation technique. Characterization via FTIR, TG-DSC, and PXRD techniques confirmed complex formation through methods such as methanol, ammonium sulfate, and heat treatment, indicating a robust physical interaction between the drug and albumin with varying strengths, as corroborated by hydrophobicity, solubility and dissolution studies. The surface properties of albumin, influenced by the concentration and nature of precipitants and process parameters, play a critical role in determining drug-binding affinity. Studies underscored the significant utility of albumin complexes in enhancing RMP’s aqueous solubility, dissolution and oral bioavailability. Complex revealed excellent stability at pH 1.2 even in the presence of INH, compared to free RMP. Complex showed enhanced dissolution rate and efficient permeation through mucosal membrane. Pharmacokinetic analyses demonstrated notable improvements in parameters such as Cmax, AUC, t1/2, and MRT compared to RMP alone, indicative of enhanced rifampicin bioavailability in rats. In conclusion, the physicochemical stimuli-sensitive precipitation approach holds promise for generating stable colloidal carrier systems using biomolecules with high affinity for hydrophobic moieties. This method offers a viable alternative for enhancing the aqueous solubility of poorly soluble drugs, particularly in cases where synthetic surfactants, polymers, or high-energy methods present challenges in simplicity and scalability.
Data Availability
The authors confirm the data supporting the findings of this study are available within the manuscript.
References
Giliyar C, Fikstad DT, Tyavanagimatt S. Challenges and opportunities in oral delivery of poorly water-soluble drugs. Drug Deliv Technol. 2006;6:57–63.
Urbanetz NA. Stabilization of solid dispersions of nimodipine and polyethylene glycol 2000. Eur J Pharm Sci. 2006;28(1–2):67–76.
Tewes F, Brillault J, Couet W, Olivier JC. Formulation of rifampicin-cyclodextrin complexes for lung nebulization. J Control Release. 2008;129(2):93–9.
Rogers TL, Gillespie IB, Hitt JE, Fransen KL, Crowl CA, Tucker CJ, et al. Development and characterization of a scalable controlled precipitation process to enhance the dissolution of poorly water-soluble drugs. Pharm Res. 2004;21(11):2048–57.
Riaz M. Review article: stability and uses of liposomes. Pak J Pharm Sci. 1995;8(2):69–79.
Jadhav KR, Shaikh IM, Ambade KW, Kadam VJ. Applications of microemulsion based drug delivery system. Curr Drug Deliv. 2006;3(3):267–73.
Leuner C, Dressman J. Improving drug solubility for oral delivery using solid dispersions. Eur J Pharm Biopharm. 2000;50(1):47–60.
Khoder M, Abdelkader H, ElShaer A, Karam A, Najlah M, Alany RG. Efficient approach to enhance drug solubility by particle engineering of bovine serum albumin. Int J Pharm. 2016;515(1–2):740–8.
Dennis MS, Zhang M, Meng YG, Kadkhodayan M, Kirchhofer D, Combs D, et al. Albumin binding as a general strategy for improving the pharmacokinetics of proteins*. J Biol Chem. 2002;277(38):35035–43.
Rai VK, Dwivedi H, Yadav NP, Chanotiya CS, Saraf SA. Solubility enhancement of miconazole nitrate: binary and ternary mixture approach. Drug Dev Ind Pharm. 2014;40(8):1021–9.
Jena SK, Singh C, Dora CP, Suresh S. Development of tamoxifen-phospholipid complex: novel approach for improving solubility and bioavailability. Int J Pharm. 2014;473(1–2):1–9.
Ming Y, Li B, Fu R, Xing H, Liu Y, Duan D, et al. Bovine serum albumin nanoparticle-mediated delivery of sorafenib for improving hepatocellular carcinoma therapy. J Nanosci Nanotechnol. 2021;21(10):5075–82.
Gao Y, Nai J, Yang Z, Zhang J, Ma S, Zhao Y, et al. A novel preparative method for nanoparticle albumin-bound paclitaxel with high drug loading and its evaluation both in vitro and in vivo. PLoS ONE. 2021;16(4):e0250670.
Rai VK, Roy A, Sharma A, Rath G, Kar B, Ghosh G, et al. Development and pharmaceutical evaluation of azelaic acid and Vitamin E oil-based nanoemulgel of tacrolimus for topical application in plaque psoriasis. J Pharm Innov. 2023;18(4):1616–25.
Alam M, Mishra A, Yadav KS, Pradhan D, Kar B, Ghosh G, et al. Development and Evaluation of Dutasteride Nanoemulgel for the Topical Delivery against Androgenic Alopecia. Pharm Nanotechnol. 2024. https://doi.org/10.2174/0122117385269151231031161411.
Pustulka SM, Ling K, Pish SL, Champion JA. Protein nanoparticle charge and hydrophobicity govern protein corona and macrophage uptake. ACS Appl Mater Interfaces. 2020;12(43):48284–95.
Beloqui A, Solinís M, Gascón AR, del Pozo-Rodríguez A, des Rieux A, Préat V. Mechanism of transport of saquinavir-loaded nanostructured lipid carriers across the intestinal barrier. J Control Release. 2013;166(2):115–23.
Xiao Y, Wiesner MR. Characterization of surface hydrophobicity of engineered nanoparticles. J Hazard Mater. 2012;215–216:146–51.
Farhan A, Jadah A. Study of the kinetics of thermal degradation of unsaturated polyester and polyester/ silica nanoparticles composites by TGA and DSC analysis techniques. Periodico Tche Quimica. 2020;17:437.
Yadav P, Yadav A. Preparation and characterization of BSA as a model protein loaded chitosan nanoparticles for the development of protein-/peptide-based drug delivery system. Futur J Pharm Sci. 2021;7:200.
Yu JN, Zhu Y, Wang L, Peng M, Tong SS, Cao X, et al. Enhancement of oral bioavailability of the poorly water-soluble drug silybin by sodium cholate/phospholipid-mixed micelles. Acta Pharmacol Sin. 2010;31(6):759–64.
Singh S, Mariappan TT, Sharda N, Kumar S, Chakraborti A. The reason for an increase in decomposition of rifampicin in the presence of isoniazid under acid conditions. Pharm Pharmacol Commun. 2010;6:405–10.
Kang RK, Mishr N, Rai VK. Guar Gum micro-particles for targeted co-delivery of doxorubicin and metformin HCL for improved specificity and efficacy against colon cancer: in vitro and in vivo studies. AAPS PharmSciTech. 2020;21(2):48.
Sharma A, Upadhyay DK, Sarma GS, Kaur N, Gupta GD, Narang RK, et al. Squalene integrated NLC based gel of tamoxifen citrate for efficient treatment of psoriasis: A preclinical investigation. J Drug Deliv Sci Technol. 2020;56:101568.
Elhabak M, Ibrahim S, Abouelatta SM. Topical delivery of l-ascorbic acid spanlastics for stability enhancement and treatment of UVB induced damaged skin. Drug Deliv. 2021;28(1):445–53.
Singh C, Bhatt TD, Gill MS, Suresh S. Novel rifampicin–phospholipid complex for tubercular therapy: Synthesis, physicochemical characterization and in-vivo evaluation. Int J Pharm. 2014;460(1):220–7.
Patil JS, Suresh S. Physicochemical characterization, in vitro release and permeation studies of respirable rifampicin-cyclodextrin inclusion complexes. Indian J Pharm Sci. 2009;71(6):638–43.
Alghamdi WA, Al-Shaer MH, Peloquin CA. Protein binding of first-line antituberculosis drugs. Antimicrob Agents Chemother. 2018;62(7):e00641-18.
Arca HÇ, Mosquera-Giraldo LI, Pereira JM, Sriranganathan N, Taylor LS, Edgar KJ. Rifampin stability and solution concentration enhancement through amorphous solid dispersion in cellulose ω-Carboxyalkanoate matrices. J Pharm Sci. 2018;107(1):127–38.
Storp B, Engel A, Boeker A, Ploeger M, Langer K. Albumin nanoparticles with predictable size by desolvation procedure. J Microencapsul. 2012;29(2):138–46.
He W, Lu Y, Qi J, Chen L, Hu F, Wu W. Food proteins as novel nanosuspension stabilizers for poorly water-soluble drugs. Int J Pharm. 2013;441(1–2):269–78.
Naik KM, Nandibewoor ST. Spectral characterization of the binding and conformational changes of bovine serum albumin upon interaction with an anti-fungal drug, methylparaben. Spectrochim Acta A Mol Biomol Spectrosc. 2013;105:418–23.
Alves E, Valadão Cardoso A. Effect of hydroxyurea (HU) on gelatinization mechanism of type I collagen suspensions. Matéria (Rio de Janeiro). 2018;23. https://doi.org/10.1590/S1517-707620180004.0545.
Bronze-Uhle ES, Costa BC, Ximenes VF, Lisboa-Filho PN. Synthetic nanoparticles of bovine serum albumin with entrapped salicylic acid. Nanotechnol Sci Appl. 2017;10:11–21.
Schianti JN, Cerize NN, Oliveira AM, Derenzo S, Gongora-Rubio MR. Rifampicin nanosuspension production using microfluidic device. InNanotech Conf Expo. 2011;1:229–232.
Mishra N, Yadav KS, Rai VK, Yadav NP. Polysaccharide encrusted multilayered nano-colloidal system of andrographolide for improved hepatoprotection. AAPS PharmSciTech. 2017;18(2):381–92.
Kudva AK, Manoj MN, Swamy BM, Ramadoss CS. Complexation of amphotericin B and curcumin with serum albumins: solubility and effect on erythrocyte membrane damage. J Exp Pharmacol. 2011;3:1–6.
Sinha P, Srivastava N, Rai VK, Mishra R, Ajayakumar PV, Yadav NP. A novel approach for dermal controlled release of salicylic acid for improved anti-inflammatory action: Combination of hydrophilic-lipophilic balance and response surface methodology. J Drug Deliv Sci Technol. 2019;52:870–84.
Acknowledgements
The authors would also like to thank the Researchers Supporting Project Number (RSPD2024R1082), King Saud University, Riyadh, Saudi Arabia. King Saud University, Riyadh, Saudi Arabia for funding this research work. The authors also acknowledge the financial support of the Department of Biotechnology (DBT), Govt. India.
Funding
The DBT Builder project funded the research with order no (BT/INF/22/SP45078/2022).
Author information
Authors and Affiliations
Contributions
Vineet Kumar Rai: Data collection, experimental work, compilation, and manuscript writing. Deepak Pradhan: Proofreading, software and language editing. Jitu Halder: Data analysis, language editing and critical analysis. Tushar Kanti Rajwar: Compilation, manuscript writing. Ritu Mahanty: Data collection and experimental work. Ivy Saha: Data collection, experimentation and statistics. Priyanka Dash: Animal experimentation and proof reading. Chandan Dash: Software analysis, data collection and proof reading. Saroj Kumar Rout: Proof Reading. Jameel Al-Tamimi: Data Analysis, language editing. Hossan Ebaid: data collection and proof reading. Salim Manoharadas: Data Analysis, language editing and funding support. Biswakanth Kar: Animal experiment and data interpretation. Goutam Ghosh: Conceptualization of the topics and critical analysis. Goutam Rath: Conceptualization of the topic and design of the table of content, supervision, data analysis and paper writing, editing and proof reading.
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rai, V.K., Pradhan, D., Halder, J. et al. Physicochemical Stimuli-Mediated Precipitation Approach for the Modulation of Rifampicin’s Dissolution and Oral Bioavailability. AAPS PharmSciTech 25, 189 (2024). https://doi.org/10.1208/s12249-024-02915-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12249-024-02915-6