Abstract
Anaerobic fungus–methanogen co-cultures from rumen liquids and faeces can degrade lignocellulose efficiently. In this study, 31 fungus–methanogen co-cultures were first obtained from the rumen of yaks grazing in Qinghai Province, China, using the Hungate roll-tube technique. The fungi were identified according to morphological characteristics and internal transcribed spacer (ITS) sequences. The methanogens associated with each fungus were identified by polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) and 16S rRNA gene sequencing. They were five co-culture types: Neocallimastix frontalis + Methanobrevibacter ruminantium, Neocallimastix frontalis + Methanobrevibacter gottschalkii, Orpinomyces joyonii + Methanobrevibacter ruminantium, Caecomyces communis + Methanobrevibacter ruminantium, and Caecomyces communis + Methanobrevibacter millerae. Among the 31 co-cultures, during the 5-day incubation, the N. frontalis + M. gottschalkii co-culture YakQH5 degraded 59.0%–68.1% of the dry matter (DM) and 49.5%–59.7% of the neutral detergent fiber (NDF) of wheat straw, corn stalk, rice straw, oat straw and sorghum straw to produce CH4 (3.0–4.6 mmol/g DM) and acetate (7.3–8.6 mmol/g DM) as end-products. Ferulic acid (FA) released at 4.8 mg/g DM on corn stalk and p-coumaric acid (PCA) released at 11.7 mg/g DM on sorghum straw showed the highest values, with the following peak values of enzyme activities: xylanase at 12,910 mU/mL on wheat straw, ferulic acid esterase (FAE) at 10.5 mU/mL on corn stalk, and p-coumaric acid esterase (CAE) at 20.5 mU/mL on sorghum straw. The N. frontalis + M. gottschalkii co-culture YakQH5 from Qinghai yaks represents a new efficient combination for lignocellulose biodegradation, performing better than previously reported fungus–methanogen co-cultures from the digestive tract of ruminants.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Abundant plant biomass is an underused feedstock for bioenergy production. Recently, anaerobic fungi have been reported to efficiently break down lignocellulosic biomass (Young et al. 2018). Studies have shown that anaerobic fungi can produce different extracellular cell–wall-degrading enzymes, including cellulases, hemicellulases, esterases, and multienzyme complexes called cellulosomes, to decompose lignocellulose materials to produce H2, formate, acetate, ethanol and CO2 (Gilmore et al. 2020; Mi et al. 2009, 2016). The activity of plant cell-wall-degrading enzymes secreted by the anaerobic fungi Neocallimastix patricciarum and Neocallimastix frontalis is higher than that secreted by commercial Trichoderma reesei, Aspergillus oryzae, Aspergillus nidulans and Penicillium pinophilum used in industry (Dijkerman et al. 1997; Yang and Xie 2010; Cao et al. 2013). Therefore, these fungi have wide application prospects in bioenergy, the feed industry, biogas fermentation and other related fields. Anaerobic fungi are classified in the phylum Neocallimastigomycota, which contains one class, Neocallimastigomycetes, one order, Neocallimastigales, one family, Neocallimasticaceae and 18 genera based on flagellum numbers, growth patterns and rhizoid forms (Chang and Park 2020). The whole genome sequences of Anaeromyces robustus, Neocallimastix californiae, Pecoramyces ruminantium, Piromyces finnis and Piromyces sp. E2 are available in the Joint Genome Institute Database (Chang and Park 2020).
Anaerobic fungi use different carbon sources as substrates for mixed acid fermentation, and the metabolites are mainly formate, acetate, lactate, ethanol, H2 and CO2 (Li et al. 2016, 2021). Methanogens can use the metabolites of anaerobic fungi, thus eliminating the feedback inhibition effect of the metabolites on growth to accelerate anaerobic fungus reproduction, promote the production of fungal ATP, and improve the activity and yield of enzymes (Wei et al. 2016). Co-cultures of an anaerobic fungus with a methanogen significantly improved their ability to degrade lignocellulosic substrates and produced a large amount of CH4 and acetate as fermentation end-products (Jin et al. 2011), which provided a theoretical basis for applying anaerobic fungi in biogas fermentation engineering, along with high activity xylanase and acetate production.
Yak (Bos grunniens) is a rare herbivorous ruminant resource in the bovine genus that can adapt to high altitudes, cold, and anoxia (Long et al. 2008; Song et al. 2021). Yaks thrive in harsh environments and graze on wild grasses as their main source of nutrition. A large number of unique, complex, and diverse microbial communities in the yak rumen can synergistically degrade low-quality wild herbage and dry, withered cold-season grass to provide yaks with energy and nutrients, making the yak rumen a natural anaerobic fermentation system for efficiently degrading lignocellulose (Huang et al. 2021a, b; Huang et al. 2021a, b). Globally, there are more than 14 million yaks, but China is the centre of origin, with the largest numbers mainly grazing on the Qinghai–Tibetan Plateau at a 3000–6000 m elevation year round. There are approximately 4.9 million heads of yak in Qinghai Province, accounting for the largest share (38%) of the total number of yaks in China (Fan et al. 2021; Sun et al. 2011).
China is known as the yak capital of the world. The number of yaks in Qinghai Province ranks first in China. To date, no reports have been published about fungus–methanogen co-cultures from yaks grazing in Qinghai Province of China. There are a large number of different types of yaks and vegetation in the six main production areas of Qinghai Province. Different kinds of natural herbage eaten by grazing yaks lead to different microbial floras in their rumens of yaks among different areas. Therefore, there are different combinations of natural anaerobic fungus–methanogen co-cultures present in the rumen of grazing yaks in different areas. These fungus–methanogen co-cultures can efficiently degrade lignocelluloses. As there is an urgent need to effectively improve the utilization rate of many types of straws, studies focusing on the yak rumen microbiota are needed. Previous studies have reported some fungus–methanogen co-cultures from the rumen of yaks grazing in Tianzhu Tibetan Autonomous Prefecture in Gansu Province of China, with high fibrolytic enzyme activities (Wei et al. 2015, 2016, 2017). It is necessary to further systematically study fungus–methanogen co-cultures from the rumen of yaks in different areas. These microbial resources have not been fully exploited. We hypothesized that fungus–methanogen co-cultures from the rumen of yaks grazing in Qinghai Province can efficiently degrade lignocelluloses with high fibrolytic enzyme activities. This study unprecedentedly focused on isolating fungus–methanogen co-cultures from the rumen of Qinghai yaks, and further explored their capacity to degrade wheat straw, oat straw, corn stalk, rice straw and sorghum straw.
Materials and methods
Animal diet and fungus–methanogen co-cultures isolation
Plateau yak is one of the most valued yak breeds in Qinghai Province, China, and has the genes and characteristics of wild yaks. Xinghai County in Hainan Tibetan Autonomous Prefecture in Qinghai Province of China, at an altitude of 4100 m (34°48′–36°14′ N, 99°01′–100°21′ E), was chosen as the experimental site. In the spring (March), plateau yaks (n = 20, male, 4–5 years) grazing in a pasture in Xinghai County were randomly chosen to isolate single anaerobic fungi. The pasture type was alpine meadow with Festuca ovina L. as the main species. The use of animals and the procedure for rumen sample collection were approved by local farms in Hainan Tibetan Autonomous Prefecture of Qinghai Province and the Animal Ethics Committees of the Gansu Academy of Sciences (Gansu, China).
Fresh rumen liquid was collected from the rumen of each yak through a dedicated rumen content collector comprising a stainless steel stomach tube and a tiny vacuum pump. The rumen liquid samples were quickly inoculated into anaerobic tubes containing 9.0 mL basal anaerobic fungus medium and 100 mg air-dried chopped wheat straw, which were autoclaved at 121 °C for 20 min. After inoculation, 1600 IU/mL penicillin and 2000 IU/mL streptomycin were added to kill bacteria, and then the culture was placed at 39 °C. The basal anaerobic fungus medium comprised (per litre): yeast extract, 1.0 g; tryptone, 1.0 g; NaHCO3, 7.0 g; resazurin (1.0 g/L), 1 mL; yak rumen fluid without cells centrifuged at 10,000×g for 15 min at 4 °C, 170 mL; Salt solution I, 165 mL; Salt solution II, 165 mL; l-cysteine hydrochloride, 1.7 g; and distilled water to 1000 mL. Salt solution I contained NaCl, 6.0 g/L; (NH4)2SO4, 3.0 g/L; KH2PO4, 3.0 g/L; CaCl2·2H2O, 0.4 g/L and MgSO4·2H2O, 0.6 g/L. Salt solution II contained 4.0 g/L K2HPO4.
After several subcultures, the single anaerobic fungi were isolated using the Hungate roll-tube technology. Bacterial contamination in each culture was checked by PCR with the primers 968f/1401r (Su et al. 2008). The formation of CH4 was examined using headspace gas chromatography to ensure that methanogens were present in each co-culture. By this method, we isolated single anaerobic fungi and obtained fungus–methanogen co-cultures. The cultures were transferred every 4 days to anaerobic fungus medium with 1% (w/v) wheat straw at 39 °C.
Identification of fungus–methanogen co-cultures and phylogenetic analysis
The cultures incubated for 2 to 3 days in Hungate agar tubes were observed under a light microscope. The cultures incubated in anaerobic fungus liquid medium with or without straw were observed under a phase contrast microscope (Leica, DMIL-PH1, Germany). The anaerobic fungi were first identified according to their morphological features.
The cultures incubated in 0.1% glucose (w/v) liquid medium without straw at 39 °C for 4 days were collected by centrifugation at 10,000×g at 4 °C to extract total genomic DNA. The collected pellets were finely ground using liquid nitrogen. DNA extraction was performed using the Fast DNA SPIN Kit for soil (MP Biomedical, Solon, OH, United States) according to the manufacturer’s instructions. Cell lysis in this kit was performed with sodium phosphate buffer and MT buffer in Lysing Matrix E tubes using a Precellys 24 bead beater for 40 s at a speed of 6.0 m/s.
The ITS sequence of the anaerobic fungi was amplified with the forward primer GM1 (5′-TGTACACACCGCCCGTC-3′) and reverse primer GM2 (5′-CTGCGTTCTTCATCGAT-3′) as reported by Li and Heath. The diversity of methanogens in each co-culture was analysed by the polymerase chain reaction–denaturing gradient gel electrophoresis (PCR–DGGE) method, with the primers 519f/915r GC according to Cheng et al. (2009). The 16S rDNA of methanogens was amplified to identify them using the primers Met86F (5′-GCTCAGTAACACGTGG-3′) and Met1340R (5′-CGGTGTGTGCAAGGAG-3′), and the amplification conditions of the Met primers were modified according to Wei et al. (Wei et al. 2017). All sequences of PCR products were determined by Beijing Genomics Institute (BGI) and all sequences were submitted to the GenBank database.
The amplified products were all visualized and purified by 1% agarose gel electrophoresis. The sequences obtained were aligned by using ClustalX V1.83. A Basic Local Alignment Search Tool (BLAST) search was performed with the obtained sequences to determine the homology with sequences already available in the GenBank database. The evolutionary relationships of the methanogens were plotted using the neighbour-joining method. Phylogenetic analyses and phylogenetic trees construction were conducted in MEGA 7.0.
Scanning electron microscopy
The fungus cultures grown in anaerobic medium containing 100 mg chopped straw at 1% (w/v) at 39 °C for 72 h were centrifuged at 1000×g for 5 min. The precipitate was rinsed three times with phosphate-buffered saline (PBS, pH 7.2). The samples for scanning electron microscopy (SEM) observation were prepared according to Wei et al. (2016).
Screening of co-cultures
According to a number of previous studies, the lignocellulose degradation capability of the fungus–methanogen co-cultures was positively correlated with gas production (Jin et al. 2011; Wei et al. 2015; Getachew et al. 2004; Gasmi-Boubaker et al. 2005; Sebata et al. 2011). Thus, the fungus–methanogen co-culture with wheat straw as the substrate with the highest gas production during the 5-day culture period was selected as the optimum co-culture, and its capacity for degrading five straws and their respective fermentation end-products were determined.
Experimental design and sampling
Wheat straw, oat straw, corn stalk, rice straw and sorghum straw were used as substrates, respectively. All substrates were sun-dried, chopped and ground to pass through a 2 mm screen. The inocula were the fungus–methanogen co-culture and its fungus pure culture obtained by adding chloramphenicol (50 μg/mL final concentration) to inhibit methanogens. Medium without the inoculum was used as the control. Each substrate (1 g) was added to 123 anaerobic bottles, each containing 90 mL basal medium. The 10-mL inoculum was added to basal medium sparged with highly purified CO2 (Li et al. 2020a, b; Solomona et al. 2016; Henske et al. 2017; Ferraro et al. 2018; Hanafy et al. 2018; Joshi et al. 2018; Peng et al. 2018) and supplemented with 1600 IU/mL penicillin and 2000 IU/mL streptomycin. The cultures were incubated at 39 °C for 5 days. During the 5-day incubation, 3 bottles of the fungus–methanogen co-culture were taken out daily to determine end-products, such as CH4 and acetate, as well as xylanase, carboxymethyl cellulase (CMCase), filter paper ase (FPase), ferulic acid esterase (FAE), acetyl esterase (AE), and p-coumaric acid esterase (CAE) activities, ferulic acid (FA), p-coumaric acid (PCA), vanillic acid (VA) and protocatechuic acid (PA) releases. And 3 bottles of the fungus pure culture were taken out daily to determine end-products. The pellets of the centrifuged cultures were collected for the analysis of in vitro dry matter digestibility (IVDMD) and neutral detergent fibre digestibility (NDFD) by the fungus–methanogen co-culture on the five straws.
Enzyme profile assays
The cultures were centrifuged at 5000×g for 10 min at 4 °C to obtain the supernatants to determine xylanase, CMCase, FPase, FAE, AE and CAE activity according to Wei (2016), Cao (2011) and the state standard of the People's Republic of China (GB/T 23874-2009). One unit of enzyme activity was defined as the amount of enzyme that released 1.0 μmol xylose, glucose, FA, p-nitrophenol, or PCA per minute per millilitre at pH6.8 and 39 °C.
Fibre digestibility determination
According to AOAC (2005) Official Methods of Analysis (18th Association of Analytical Chemists, Washington DC, USA), the samples in each bottle were centrifuged and dried at 105 °C for 24 h to determine IVDMD. NDF contents were determined as described by Van Soest et al. Alpha amylase was not used, but sodium sulfite was added to each sample for the NDF assay. The calculation was as follows: IVDMD or NDFD (%) = [(initial DM or NDF of the feed taken for incubation − DM or NDF of the residue)/(initial DM or NDF of the feed taken for incubation)] × 100. The dried residue and NDF content of the dried residue in the uninoculated control were taken as the initial DM and NDF values, respectively.
Phenolic acid release measurement
The extraction methods of FA, PCA, VA and PA from the supernatants of cultures and high-performance liquid chromatography (HPLC) analyses were performed according to the newly improved HPLC method reported by Wang et al. (2013).
Total gas production and CH4 measurement
The cumulative gas production during the 5-day incubation was measured by an AGRS-III automated trace gas recording system for real-time detection of anaerobic microbial growth. CH4 was measured by gas chromatography (GC), with an HP-Innowax (19091N-133) capillary column, high purity nitrogen as the carrier gas, and a hydrogen flame ionization detector. The determination conditions of CH4 were established as follows: total pressure 130 kPa, total flow rate 30.2 mL/min, column flow rate 1.7 mL/min, linear velocity 39.8 cm/s, column temperature 80 °C, gasification chamber temperature 100 °C, and detection chamber temperature 120 °C.
End-product analysis
The cultures were centrifuged at 10,000×g for 10 min at 4 °C to obtain the supernatants. The formate and acetate concentrations in the supernatants were determined by HPLC (Water 2489, USA) on an instrument equipped with an Agilent SB-Aq chromatography column (26 mm × 250 mm, 5 μm, Agilent, USA), a 2489UA detector, a 7725i manual injector (Rheodyne, USA), and a UV2450 ultraviolet–visible spectrometer (Hitachi, Japan). The determination conditions were as follows: mobile phase 5 mmol/L KH2PO4-H3PO4 buffer solution (pH = 2.4), flow velocity 0.5 mL/min, detection wavelength 214 nm, column temperature 25 °C, and injection volume 20 μL. The l-lactate and d-lactate concentrations in the supernatants were analysed using an l,d-lactate Assay Kit (Nanjing Jiancheng Institute of Biological Engineering, Nanjing, China). The ethanol concentration was determined using gas chromatography according to the modified method of Boonchuay et al. (2021).
Statistical analysis
Data are shown as the mean ± standard deviation (SD) and were analysed using one-way analysis of variance (ANOVA) follwed by Tukey’s test with SPSS 18.0 software (Microsoft). p < 0.05 was considered to indicate a statistically significant difference.
Results
Isolation and identification of fungus–methanogen co-cultures from Qinghai yaks
The gas chromatography analysis showed that all isolated fungus–methanogen co-cultures produced CH4, which confirmed the presence of methanogens in each co-culture. Bacterial specific-PCR amplification showed no detectable bacteria in any co-culture. In this study, 31 fungus–methanogen co-cultures were obtained from the rumen of plateau yaks grazing in Hainan Tibetan Autonomous Prefecture in Qinghai Province, China (Table 1). Among the 31 co-cultures, there were 5 types: N. frontalis + M. ruminantium, N. frontalis + M. gottschalkii, O. joyonii + M. ruminantium, C. communis + M. ruminantium, and C. communis + M. millerae. They were identified and named N. frontalis + M. ruminantium co-culture YakQH1-YakQH4, N. frontalis + M. gottschalkii co-culture YakQH5-YakQH16, O. joyonii + M. ruminantium co-culture YakQH17-YakQH23, C. communis + M. ruminantium co-culture YakQH24-YakQH26, and C. communis + M. millerae co-culture YakQH27-YakQH31. Each anaerobic fungal strain was symbiotic with a methanogen species. The fungal isolates were named N. frontalis YakQH1-YakQH16, O. joyonii YakQH17-YakQH23, and C. communis YakQH24-YakQH31, and the methanogen isolates were named M. ruminantium YakQH1-YakQH4, M. gottschalkii YakQH5-YakQH16, M. ruminantium YakQH17-YakQH23, M. ruminantium YakQH24-YakQH26, and M. millerae YakQH27-YakQH31 (Table 1).
The 31 fungi in the co-cultures were observed and identified using light microscopy, phase contrast microscopy, and SEM according to morphological characteristics and the number of zoospore flagella (Figs. 1, 2, 3). The fungal isolates N. frontalis YakQH1-YakQH16, O. joyonii YakQH17-YakQH23, and C. communis YakQH24-YakQH31 showed the morphological features of N. frontalis, O. joyonii, and C. communis, respectively. The ITS sequences of the 31 fungi in the co-cultures were deposited in GenBank under accession numbers MH482796-MH482826 (Table 1). Through sequence homology comparison with the NCBI database, among the 31 co-cultures, all fungal isolates had a similarity of 99%–100% with N. frontalis strain Yak16, O. joyonii strain Yak1, and Caecomyces sp. AGRL-11 registered in GenBank.
Morphological diversity of anaerobic fungi isolates in Hungate agar roll-tubes. A The fungus N. frontalis YakQH5 with an endogenous sporangium and a network of rhizoids. B The fungus O. joyonii YakQH17 produced a rhizomycellium complex with an extensive network of hyphae, single or branched sporangiophores developed from the hyphae, and several globose sporangia full of zoospores were produced from the sporangiophores. C The fungus C. communis YakQH24 colony. Bars = 100 μm
Growth stages of the fungus C. communis YakQH24 in liquid glucose culture medium under phase -contrast microscopy. A A live uniflagellate zoospore. Bars = 10 μm. B The fungus C. communis YakQH24 showed bulbous rhizoids after 24 h incubation. Bars = 10 μm. C The C. communis YakQH24 produced more bulbous rhizoids after 96 h incubation. Bars = 50 μm
SEM of the anaerobic fungus N. frontalis YakQH5. A The mycelium of the N. frontalis YakQH5 penetrated into the thick-walled tissue of wheat straw after 24 h incubation. B The N. frontalis YakQH5 grew with an endogenous sporangium growth after 48 h incubation. C The dense hyphae of the N. frontalis YakQH5 fully penetrated into the wheat straw tissue to physically degrade lignocellulose after 96 h incubation. Bars = 20 μm
The DGGE results confirmed that one fungus was natively associated with only one methanogen in each co-culture. The 16S rRNA gene sequences of the 31 methanogens in the co-cultures were deposited in GenBank under accession numbers MH443285-MH443315 (Table 1). All methanogen isolates (YakQH1-YakQH31) in the 31 co-cultures belonged to Methanobrevibacter sp. based on the 16S rRNA gene sequences. Through sequence homology comparison with the NCBI database, among the 31 co-cultures, all methanogen isolates were found to have a similarity of 99%-100% with M. ruminantium strain YakM2, M. gottschalkii strain PG, and M. millerae strain ZA-10 registered in GenBank.
Phylogenetic tree construction of methanogens
A phylogenetic tree of the 16S rRNA gene sequences of methanogen isolates YakQH1-YakQH31 was constructed. Methanomicrobium mobile BP was used as an outgroup. The results showed that the 31 strains of methanogens included 3 species: M. ruminantium, M. gottschalkii, and M. millerae (Fig. 4).
Phylogenetic tree of the 16S rRNA gene sequences of the methanogens isolates YakQH1–YakQH31 in the fungus–methanogen co-cultures. The topology of the tree was estimated by bootstraps based on 1000 replications. Bootstrap values more than 70% were shown on the major branch points. The scale bar corresponds to five changes per 100 positions. The 16S rRNA sequences determined in the present study and the closest relatives were marked in bold type. Methanomicrobium mobile BP (NR_044726) was used as an out group. GenBank accession numbers were given in brackets
Screening of co-cultures
Among the 31 fungus–methanogen co-cultures, the 12 co-cultures of N. frontalis + M. gottschalkii grew most stably and rapidly and degraded wheat straw in anaerobic tubes to produce the highest total gas yields of 181–230 mL/g DM during the 5-day incubation, obviously more than other co-cultures. Particularly, the N. frontalis + M. gottschalkii co-culture YakQH5 was the most remarkable and was thus selected to determine its ability to biodegrade five types of roughage as substrates. The preservation number of the N. frontalis + M. gottschalkii co-culture YakQH5 was No. 19299 in the China General Microbiological Culture Collection Center (CGMCC).
Enzyme profiles
During the 5-day incubation, in anaerobic bottles, the N. frontalis + M. gottschalkii co-culture YakQH5 degraded wheat straw, corn stalk, rice straw, oat straw and sorghum straw and showed the highest activity values of 12,910 mU/mL xylanase on wheat straw, 929 mU/mL CMCase and 1187 mU/mL FPase on rice straw, 10.5 mU/mL FAE and 245 mU/mL AE on corn stalk, and 20.5 mU/mL CAE on sorghum straw (Table 2).
Fibre digestibility
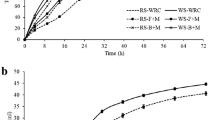
During the 5-day incubation, in anaerobic bottles, the N. frontalis + M. gottschalkii co-culture YakQH5 degraded 60.5% of wheat straw, 67.2% of corn stalk, 68.1% of rice straw, 59.0% of oat straw, and 63.1% of sorghum straw (Fig. 5). Additionally, the co-culture YakQH5 had NDFD values of 49.5% on wheat straw, 59.7% on corn stalk, 55.8% on rice straw, 51.0% on oat straw, and 53.1% on sorghum straw (Fig. 5). The composition of the forages used as substrates before incubation was shown in Table 3.
Phenolic acid release
During the 5-day incubation, in anaerobic bottles, the N. frontalis + M. gottschalkii co-culture YakQH5 degraded wheat straw, corn stalk, rice straw, oat straw, and sorghum straw to release FA 3.4–4.8 mg/g DM, PCA 3.4–11.7 mg/g DM, VA 0.4–0.7 mg/g DM and PA 0.5–0.9 mg/g DM, respectively. The peak values were as follows: FA 4.8 mg/g DM on corn stalk, PCA 11.7 mg/g DM on sorghum straw, VA 0.7 mg/g DM on rice straw, and PA 0.9 mg/g DM on wheat straw (Fig. 6).
Fermentation end-products
During the 5-day incubation, in anaerobic bottles, the N. frontalis + M. gottschalkii co-culture YakQH5 degraded wheat straw, corn stalk, rice straw, oat straw and sorghum straw to produce high total gas yields (the gas was mainly composed of CH4) of 315 mL/g DM, 261 mL/g DM, 300 mL/g DM, 290 mL/g DM, and 284 mL/g DM, respectively (Fig. 7). Meanwhile, the N. frontalis + M. gottschalkii co-culture YakQH5 degraded these lignocellulosic materials to produce the highest end-product yields as follows: CH4 4.6 mmol/g DM on wheat straw and acetate 8.6 mmol/g DM on rice straw (Table 4). The fungus N. frontalis YakQH5 degraded these lignocellulosic materials to produce maxima of H2 3.9 mmol/g DM on wheat straw, formate 2.5 mmol/g DM on sorghum straw, acetate 5.5 mmol/g DM on rice straw, lactate 2.5 mmol/g DM on sorghum straw, and ethanol 45.8 mmol/g DM on wheat straw (Table 4).
Discussion
Diversity of the fungus–methanogen co-cultures from the rumen of grazing yaks
In this study, 31 natural fungus–methanogen co-cultures were first obtained from the rumen fluid of grazing yaks in spring in Qinghai Province, China, comprising 5 combination types: N. frontalis + M. ruminantium, N. frontalis + M. gottschalkii, O. joyonii + M. ruminantium, C. communis + M. ruminantium, and C. communis + M. millerae. In 2015, we isolated 20 natural fungus–methanogen co-cultures from the rumen fluid of grazing yaks in spring in a Wushaoling pasture of Tianzhu Tibetan Autonomous Prefecture in Gansu Province, China, including 4 combination types: N. frontalis + M. ruminantium, O. joyonii + M. ruminantium, O. joyonii + M. millerae, and Piromyces + M. ruminantium (Wei et al. 2015, 2016). Thus, there were many types of natural fungus–methanogen co-cultures in the rumen of grazing yaks. Furthermore, when compared to the reported natural fungus-methanogen co-cultures isolated from the rumen or faeces of ruminants and non-ruminants by Bauchop et al. (1981), Jin et al. (2011), Leis et al. (2014), Sun et al. (2014), Li et al. (2016) and Li et al. (2021) and grazing yaks by Wei et al. (2015, 2017), in this study, 3 new types of natural anaerobic fungus–methanogen co-culture combinations were first obtained from the rumen of yaks, namely: N. frontalis + M. gottschalkii, C. communis + M. ruminantium, and C. communis + M. millerae. These three types of fungus-methanogen co-cultures all included one fungus and one methanogen, and each methanogen coexisting with each fungus belonged to Methanobrevibacter sp., consistent with the natural fungus–methanogen co-cultures isolated from the rumen or faeces of ruminants and non-ruminants previously reported by Jin et al. (2011), Leis et al. (2014), Sun et al. (2014), Li et al. (2016), Li et al. (2021) and Wei et al. (2015, 2017).
Our study revealed that different combinations of natural fungus–methanogen co-cultures in the rumen of grazing yaks in different regions, probably because of the different types of wild herbages eaten by the grazing yaks in different areas, and these different types of natural fungus–methanogen co-cultures differed in their ability to degrade lignocelluloses. This suggests that there are new and abundant microbial resources for efficiently degrading lignocelluloses in the rumen of yaks grazing on the Qinghai-Tibet Plateau, which have not yet been fully explored.
Roughage degradation by fungus–methanogen co-cultures from the rumen of grazing yaks
During the 5-day incubation, the N. frontalis + M. gottschalkii co-culture YakQH5 degraded the 5 kinds of roughages and showed degradation potential, including high lignocellulose-degrading enzyme activities, IVDMD 59.0%-68.1% (from oat straw to rice straw), NDFD 49.5%-59.7% (from wheat straw to corn stalk) and large amounts of FA and PCA releases, which are described in Sect. 3. Accordingly, we found that the degradation degrees of roughages were different. Ranked in terms of highest to lowest decomposition, the substrates were rice straw, corn stalk, sorghum straw, wheat straw, and oat straw, while in terms of highest to lowest degradation, they were corn stalk, rice straw, sorghum straw, oat straw, and wheat straw. The N. frontalis + M. gottschalkii co-culture YakQH5 degraded lignocelluloses by secreting main-chain degrading polysaccharide hydrolases (CMCase, FPase and xylanase) and side-chain degrading esterases (FAE, AE and CAE) with high activities, which could be key lignin-degrading enzymes in enhancing plant cell wall degradation. All these enzymes acted synergistically to effectively decompose lignocelluloses. The N. frontalis + M. gottschalkii YakQH5 degraded sorghum straw to release PCA 11.7 mg/g DM (70.1 μg/mL) as a result of the high PCA content in sorghum straw, consistent with the high activity of CAE when using sorghum stalk as a substrate, implying that the N. frontalis + M. gottschalkii YakQH5 from the rumen of Qinghai yaks can decompose lignin efficiently. Meanwhile, the N. frontalis + M. gottschalkii co-culture YakQH5 degraded wheat straw, corn stalk, rice straw, oat straw and sorghum straw to release very small amounts of VA and PA. The yields of VA and PA releases appeared unrelated to their contents in the roughages. Further study is needed to clarify this finding.
Among the 31 fungus–methanogen co-cultures, the N. frontalis + M. gottschalkii co-culture YakQH5 was screened out with only wheat straw as substrate by measuring gas production. In this case, the lignocellulose degradation and gas production are generally positive correlation.
The N. frontalis + M. gottschalkii co-culture YakQH5 degraded different roughages as substrates, the order of the lowest to highest IVDMD was: oat straw, wheat straw, sorghum straw, corn stalk, and rice straw; the order of the lowest to highest NDFD was: wheat straw, oat straw, sorghum straw, rice straw and corn stalk; and the order of the lowest to highest gas production was: corn stalk, sorghum straw, oat straw, rice straw and wheat straw. When the 5 kinds of roughages with different lignocellulose contents were used as substrates, there was not always a linear correlation between IVDMD, NDFD and gas production. This phenomenon may have been related to the different compositions of the five roughages. The lignocellulose degradation mechanism of the anaerobic fungi needs to be further studied to reveal the reason behind this phenomenon.
In 2015 and 2017, we first reported the natural fungus–methanogen co-culture N. frontalis + M. ruminantium Yaktz1 and Piromyces + M. ruminantium Yak-G18 that degraded straws with remarkable efficiency were isolated from the rumen of yaks grazing in Tianzhu Tibetan Autonomous County in Gansu Province of China (Wei et al. 2015, 2016, 2017). During the 7-day incubation, the N. frontalis + M. ruminantium co-culture Yaktz1 degraded 61.7% of wheat straw, 68.8% of corn stalk, and 71.9% of rice straw, with NDFD values of 56.0% on wheat straw, 61.7% on corn stalk, and 55.6% on rice straw, while exhibiting the highest enzyme activity values as follows: xylanase 12,500 mU/mL on wheat straw; FPase 430.3 mU/mL, FAE 11.4 mU/mL, AE 199.3 mU/mL and CAE 5.0 mU/mL on corn stalk, and FA release 24.1 μg/mL and PCA release 50.3 μg/mL on corn stalk as the peak values. Across the 7-day incubation, the Piromyces + M. ruminantium co-culture Yak-G18 degraded 60.5% of wheat straw, 65.0% of corn stalk, 65.9% of rice straw, 66.0% of Chinese wildrye, and 75.0% of alfalfa, with NDFD values of 40.8%–47.5% on the five substrates, showing peak values of xylanase activity ranging from 2750 to 5023 mU/mL (from alfalfa to Chinese wildrye), FPase ranging from 71.9 to 123.5 mU/mL(from rice straw to Chinese wildrye), and AE 66.3–118.1 mU/mL (from alfalfa to Chinese wildrye), releasing little FA and PCA. To date, 3 types of extremely effective fungus–methanogen co-cultures for straw degradation have been obtained from the rumen of yaks: the N. frontalis + M. ruminantium co-culture Yaktz1, the Piromyces + M. ruminantium co-culture Yak-G18, and the N. frontalis + M. gottschalkii co-culture YakQH5. According to degradation capability, the N. frontalis + M. ruminantium co-culture Yaktz1 and N. frontalis + M. gottschalkii co-culture YakQH5 showed the most prominent ability to degrade straws. The N. frontalis + M. gottschalkii co-culture YakQH5 from Qinghai yaks decomposed wheat straw, corn stalk, rice straw, oat straw, and sorghum straw to produce higher xylanase, FPase, and CAE activities than the N. frontalis + M. ruminantium co-culture Yaktz1 from Tianzhu yaks, other natural fungus-methanogen co-cultures (from rumen or faeces of ruminants and non-ruminants), and artificially mixed anaerobic fungus–methanogen co-cultures previously reported, with all kinds of roughages or fiter paper as substrates (Jin et al. 2011; Teunissen et al. 1992a, b; Wei et al. 2015, 2016, 2017). Specifically, the N. frontalis + M. gottschalkii co-culture YakQH5 showed FPase activity on corn stalk that was approximately 2.7 times higher than that of the N. frontalis + M. ruminantium co-culture Yaktz1 on corn stalk. The xylanase produced by N. frontalis + M. gottschalkii YakQH5 has good prospects for industrial application. Concurrently, the N. frontalis + M. gottschalkii co-culture YakQH5 could effectively degrade wheat straw, corn stalk and rice straw with IVDMD and NDFD values analogous to those of the N. frontalis + M. ruminantium co-culture Yaktz1 (Wei et al. 2016) but obviously higher IVDMD values than for other fungus–methanogen co-cultures from the rumen or faeces of ruminants and non-ruminants previously reported, with roughages as substrates (Jin et al. 2011). Values of 33.6%–53.1% IVDMD for wheat straw, corn stalk, bagasse, distiller’s dried grains with solubles (DDGS), wheat bran and rice straw by the Piromyces + M. thaueri CW co-culture from the rumen of goats; 26.8%–57.0% IVDMD for wheat straw, corn stalk, bagasse, DDGS, wheat bran, and rice straw by the Piromyces + Methanobrevibacter sp. Z8 co-culture from the rumen of goats; and 33.5%–48.3% IVDMD for rice straw by the Anaeromyces + M. gottschalkii strain PG co-culture from faeces of mules, the Piromyces + M. gottschalkii strain PG co-culture from faeces of mules, the Piromyces + Methanobrevibacter sp. Z8 co-culture from faeces of camel, the Neocallimastix + Methanobrevibacter sp. Z8 co-culture from feces of camel, and the Piromyces + Methanobrevibacter sp. 1Y co-culture from feces of buffalo, all after a 5-day incubation. The N. frontalis + M. ruminantium co-culture Yaktz1 degraded wheat straw, corn stalk, and rice straw to release the maximum values of FA 24.1 μg/mL and PCA 50.3 μg/mL on corn stalk, slightly lower than those of the N. frontalis + M. gottschalkii co-culture YakQH5 with corn stalk as substrate (Wei et al. 2016).
Our study showed that the N. frontalis + M. gottschalkii co-culture YakQH5 and the N. frontalis + M. ruminantium co-culture Yaktz1 from the rumen of grazing yaks degraded roughages more effectively than previously reported fungus-methanogen co-cultures from the digestive tract of herbivores, including ruminants and non-ruminants, and even some current industrial strains. This study also highlighted that a new-type fungus–methanogen combination, the N. frontalis + M. gottschalkii YakQH5, has been obtained from the rumen of Qinghai yaks, and it can superiorly degrade lignocellulosic materials. The N. frontalis + M. ruminantium co-culture Yaktz1 was isolated from yaks grazing in a Wushaoling pasture located in Tianzhu Tibetan Autonomous Prefecture in Gansu Province of China, an alpine meadow pasture with Kobresia myosuroides (Villars) Foiri as the main species, while the N. frontalis + M. gottschalkii co-culture YakQH5 was isolated from yaks grazing in Xinghai County located in Hainan Tibetan Autonomous Prefecture in Qinghai Province of China, where the pasture was alpine meadow with Festuca Ovina L. as the main species. It can be concluded that the combinations of fungus–methanogen co-cultures from the grazing yaks in different regions may vary, and these natural fungus–methanogen co-cultures had different characteristics and abilities to degrade lignocelluloses. Further studies on the host specificity or substrate specificity of anaerobic fungi are needed.
Meanwhile, the N. frontalis + M. gottschalkii co-culture YakQH5 degraded wheat straw straw, corn stalk, rice straw, oat straw, and sorghum straw to produce the highest yields of CH4 4.6 mmol/g DM on wheat straw and acetate 8.6 mmol/g DM (55.7 mM) on rice straw. These are slightly higher yields than the CH4 and acetate yields produced by the N. frontalis + M. ruminantium co-culture Yaktz1 with wheat straw, corn stalk, and rice straw as substrates during the 7-day incubation; markedly higher than those produced by the Piromyces + M. ruminantium co-culture Yak-G18 on wheat straw, corn stalk, rice straw, Chinese wildrye, and alfalfa during the 7-day incubation; and higher than those produced by most of natural fungus–methanogen co-cultures from the rumen or faeces of ruminants and non-ruminants, and artificially mixed anaerobic fungus–methanogen co-cultures, with roughages, fiter paper, cellulose or glucose as substrates (Jin et al. 2011; Teunissen et al. 1992a, b; Li et al. 2016; Nakashimada et al. 2000; Wei et al. 2015, 2016, 2017).
After methanogen inhibition, the pure fungus N. frontalis YakQH5 degraded wheat straw, corn stalk, rice straw, oat straw and sorghum straw to produce the highest yields of H2 3.9 mmol/g DM and ethanol 45.8 mmol/g DM (260.1 mM) on wheat straw, formate 2.5 mmol/g DM (15.0 mM) on sorghum straw, and lactate 2.5 mmol/g DM (15.0 mM) on sorghum straw. The yields of these end-products were generally higher than those produced by anaerobic fungi from the digestive tract of common ruminants and non-ruminants. The most interesting finding was that its ethanol yield was more greater than that produced by all reported anaerobic fungi with many kinds of roughages as substrates, even exceeding those of some industrial strains that produced ethanol (Jin et al. 2011; Sirohi et al 2013; Nagpal et al. 2011; Paul et al. 2010; Teunissen et al. 1992a, b; Sijtsma and Tan 1993; Thareja et al. 2006; Wei et al. 2016, 2017; Saye et al. 2021). Thus, the fungus N. frontalis YakQH5 is promising for use in the development of ethanol production.
Prospects for the fungus–methanogen co-cultures from the rumen of grazing yaks
Anaerobic fungi have been studied for more than 40 years and are considered to play a crucial role in degrading lignocelluloses. Some methanogens can further enhance the anaerobic fungi's ability to degrade lignocelluloses. Anaerobic fungus–methanogen co-cultures degrade lignocellulosic materials to produce CH4, acetate and lignocellulose degradation enzymes with high activities, even exceeding those of some industrial strains (Teunissen et al. 1993; Cheng et al. 2018; Mountfort and Asher 1989; Solomon et al. 2016; Yang and Xie 2010; Chang and Park 2020). However, until now, anaerobic fungi have not been used for widespread industrial applications due to their strict anaerobic growth requirements, limited preservation methods, difficulty in scale-up, and genetic intractability. Recently, with the improvement of culture media, more species of anaerobic fungi have been discovered and a large number of their nucleotides and proteins have been sequenced (Chang et al. 2020; Hess et al. 2020).
In the present study, the 31 fungus–methanogen co-cultures were first obtained from the rumen of yaks grazing in Qinghai Province of China. These co-cultures included 5 combination types. Among them, during the 5-day incubation, the new-type combination N. frontalis + M. gottschalkii co-culture YakQH5 degraded 59.0%–68.1% of the DM and 49.5%–59.7% of the NDF of wheat straw, corn stalk, rice straw, oat straw and sorghum straw to produce CH4 (3.0–4.6 mmol/g DM) and acetate (7.3–8.6 mmol/g DM) as end-products and released the most FA (4.8 mg/g DM) on corn stalk, PCA (11.7 mg/g DM) on sorghum straw. The peak values of enzyme activitie were as follows: xylanase 12,910 mU/mL on wheat straw, FAE 10.5 mU/mL on corn stalk and CAE 20.5 mU/mL on sorghum straw. The N. frontalis + M. gottschalkii co-culture YakQH5 degraded roughages to produce higher xylanase, CMCase, FAE, AE activities, IVDMD, NDFD, and more CH4 and acetate yields without any pretreatment, than reported for natural fungus–methanogen co-cultures isolated from the digestive tract of ruminants and non-ruminants. This study convincingly proved our original hypothesis that Yak-derived ruminal fungus–methanogen co-cultures have evolved to possess high efficiency to degrade plant lignocellulose.
In follow-up studies, it will be useful strategy to further construct a large-scale continuous culture facility to produce natural complex lignocellulose-degrading enzymes, CH4 and acetate by the N. frontalis + M. gottschalkii co-culture YakQH5, or express its xylanase, CMCase, FEA, AE and CAE genes under aerobic conditions using molecular biology techniques to realize large-scale industrial production and application. The study on combined multiple omics analysis including genomics, transcriptomics, proteomics and metabolomics of the N. frontalis + M. gottschalkii co-culture YakQH5 will help to reveal its lignocellulose degradation mechanism. The fermentation process of the fungus–methanogen co-cultures is mainly carried out in the cytoplasm and hydrogenosomes, and the detailed fermentation mechanism also needs to be further studied and optimized. The fungus–methanogen co-cultures can efficiently degrade and change lignocelluloses into CH4 and acetate, representing potential for biological energy production that cannot be ignored. Therefore, screening the dominant combination of fungus–methanogen co-cultures from grazing yaks for the production of high-quality cellulase, hemicellulase, esterase and CH4 will have broad application prospects. Improving the preservation methods of anaerobic fungi with high activity will further promote their practical application in industrial and agricultural production. In the future, the study of the fungus–methanogen co-cultures from the rumen of Qinghai yaks will potentially be highly important to address feed shortages, agricultural wastes utilization, environmental pollution and energy crises.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and the additional file.
Abbreviations
- ADF:
-
Acid detergent fiber
- ADFD:
-
Acid detergent fiber digestibility
- AE:
-
Acetyl esterase
- ATP:
-
Adenosine triphosphate
- ANOVA:
-
Analysis of variance
- BGI:
-
Beijing genomics institute
- BLAST:
-
Basic local alignment search tool
- CS:
-
Corn stalk
- CAE:
-
Coumaric acid esterase
- CMCase:
-
Carboxymethyl cellulase
- CGMCC:
-
China General Microbiological Culture Collection Center
- DGGE:
-
Denaturing gradient gel electrophoresis
- DDGS:
-
Distillers dried grains with solubles
- DM:
-
Dry matter
- FA:
-
Ferulic acid
- FAE:
-
Ferulic acid esterase
- FPase:
-
Filter paper ase
- GC:
-
Gas chromatography
- HPLC:
-
High-performance liquid chromatography
- ITS:
-
Internal transcribed spacer
- IVDMD:
-
In vitro dry matter disappearance
- NDF:
-
Neutral detergent fiber
- NDFD:
-
Neutral detergent fiber digestibility
- NCBI:
-
National center for biotechnology information
- OS:
-
Oat straw
- PBS:
-
Phosphate-buffered saline
- PCA:
-
p-Coumaric acid
- PCR:
-
Polymerase chain reaction
- PA:
-
Protocatechuic acid
- RS:
-
Rice straw
- SD:
-
Standard deviation
- SS:
-
Sorghum straw
- S.E.M:
-
Standard error of mean
- SEM:
-
Scanning electron microscopy
- VA:
-
Vanillic acid
- WS:
-
Wheat straw
References
AOAC (2005) Official Methods of Analysis, 18thed. Association of Analytical Chemists, Washington DC
Bauchop T, Mountfort DO (1981) Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl Environ Microb 42:1103–1110
Boonchuay P, Techapun C, Leksawasdi N, Seesuriyachan P, Hanmoungjai P, Watanabe M, Srisupa S, Chaiyaso T (2021) Bioethanol production from cellulose-Rich corncob residue by the thermotolerant Saccharomyces cerevisiae TC-5. J Fungi (basel) 7:7
Cao YC, Yang HJ (2011) Effect of roughage fibre content on fibrolytic activities and volatile fatty acid profiles of Neocallimastix sp. YAK11 isolated from rumen fluids of yak (Bos grunniens). Anim Feed Sci Tech 170:284–290
Cao YC, Yang HJ, Zhang DF (2013) Enzymatic characteristics of crude feruloyl and acetyl esterases of rumen fungus Neocallimastix sp. YAK11 isolated from yak (Bos grunniens). J Anim Physiol Anim Nutr 97(2):363–373
Chang J, Park H (2020) Nucleotide and protein researches on anaerobic fungi during four decades. J Anim Sci Technol 62:121–140
Cheng YF, Mao SY, Liu JX, Zhu WY (2009) Molecular diversity analysis of rumen methanogenic Archaea from goat in eastern China by DGGE methods using different primer pairs. Lett Appl Microbiol 48:585–592
Cheng YF, Shi QC, Sun RL, Liang D, Li YF, Li YQ, Jin W, Zhu WY (2018) The biotechnological potential of anaerobic fungi on fiber degradation and methane production. World J Microb Biot 34:155
Dijkerman R, Bhansing DCP, Op den Camp HJM, van der Drift C, Vogels GD (1997) Degradation of structural polysaccharides by the plant cell-wall degrading enzyme system from anaerobic fungi: An application study. Enzyme Microb Technol 21(2):130–136
Fan Q, Wanapat M, Hou F (2021) Rumen bacteria influence milk protein yield of yak grazing on the Qinghai-Tibet plateau. Anim Biosci 34(9):1466–1478
Ferraro A, Dottorini G, Massini G, Miriyana VM, Signorini A, Lembo G, Fabbricino M (2018) Combined bioaugmentation with anaerobic ruminal fungi and fermentative bacteria to enhance biogas production from wheat straw and mushroom spent straw. Bioresource Technol 260:364–373
Gasmi-Boubaker A, Kayouli C, Buldgen A (2005) In vitro gas production and its relationship to in situ disappearance and chemical composition of some Mediterranean browse species. Anim Feed Sci Tech 123–124:303–311
Getachew G, Robinson PH, DePeters EJ, Taylor SJ (2004) Relationships between chemical composition, dry matter degradation and in vitro gas production of several ruminant feeds. Anim Feed Sci Tech 111:57–71
Gilmore SP, Lillington SP, Haitjema CH, de Groot R (2020) Designing chimeric enzymes inspired by fungal cellulosomes. Synth Syst Biotechnol 5:23–32
Hanafy RA, Johnson B, Elshahed MS, Youssef NH (2018) Anaeromyces contortus, sp. nov., a new anaerobic gut fungal species (Neocallimastigomycota) isolated from the feces of cow and goat. Mycologia 110(3):502–512
Henske JK, StE W, Solomon KV, Smallwood CR, Shutthanandan V, Evans JE, Theodorou MK (2017) Metabolic characterization of anaerobic fungi provides a path forward for bioprocessing of crude lignocellulose. Biotechnol Bioeng 115(4):874–884
Hess M, Paul SS, Puniya AK, van der Giezen M, Claire S, Edwards JE, Kateřina F (2020) Anaerobic Fungi: Past, Present, and Future. Front Microbiol 11:584893–584893
Huang XD, Mi JD, Denman SE, Basangwangdui P, Zhang Q, Long RJ, McSweeney CS (2021a) Changes in rumen microbial community composition in yak in response to seasonal variations. J Appl Microbiol 132(3):1652–1665
Huang XD, Denman SE, Mi JD, Padmanabha J, Hao LZ, Long RJ, McSweeney CS (2021b) Differences in bacterial diversity across indigenous and introduced ruminants in the Qinghai Tibetan plateau. Anim Prod Sci 62(14):AN20204
Jin W, Cheng YF, Mao SY, Zhu WY (2011) Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. BioresourCe Technol 102:7925–7931
Joshi A, Lanjekar VB, Dhakephalkar PK, Callaghan TM, Griffith GW, Dagar SS (2018) Liebetanzomyces polymorphus gen. et sp. nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. MycoKeys 40:89–110
Leis S, Dresch P, Peintner U, Fliegerová K, Sandbichler AM, Insam H, Podmirseg SM (2014) Finding a robust strain for biomethanation: anaerobic fungi (Neocallimastigomycota) from the Alpine ibex (Capra ibex) and their associated methanogens. Anaerobe 29:34–43
Li YF, Jin W, Cheng YF, Zhu WY, Mao SY et al (2016) Effect of the associated methanogen Methanobrevibacter thaueri on the dynamic profile of end and intermediate metabolites of anaerobic fungus Piromyces sp. F1. Curr Microbiol 73:434–441
Li YQ, Sun MZ, Li YF, Cheng YF, Zhu WY (2020a) Co-cultured methanogen improved the metabolism in the hydrogenosome of anaerobic fungus as revealed by GC/MS analysis. Asian-Aust J Anim Sci 33(12):1948–1956
Li YQ, Hou ZS, Shi QC, Cheng YF, Zhu WY (2020b) Methane production from different parts of corn stover via a simple co-culture of an anaerobic fungus and methanogen. Front Bioeng and Biotech 8:314
Li YQ, Meng ZX, Xu Y, Shi QC, Ma YP, Aung M, Cheng YF, Zhu WY (2021) Interactions between anaerobic fungi and methanogens in the rumen and their biotechnological potential in biogas production from lignocellulosic materials. Microorganisms 9(1):190–190
Long RJ, Ding LM, Shang Z, Guo XS (2008) The yak grazing system on the Qinghai-Tibetan plateau and its status. Rangeland J 30:241–246
Lu WQ, Cao YH, Liu XH, Zhan TY, Liu YL, Cheng QH (2009) Determination of xylanase activity in feed additives-Spectrophotometric method. The State Standard of the People's Republic of China (GB/T 23874–2009)
Mi K, Song J, Ha JK, Park HS, Chang J (2009) Analysis of functional genes in carbohydrate metabolic pathway of anaerobic rumen fungus Neocallimastix frontalis PMA02. Asian-Aust J Anim Sci 22(11):1555–1565
Mi K, Song J, Park HS, Park H, Chang J (2016) Characterization of heterologously expressed acetyl xylan esterase1 isolated from the anaerobic rumen fungus Neocallimastix frontalis PMA02. Asian-Aust J Anim Sci 29:1576–1584
Mountfort DO, Asher RA (1989) Production of xylanase by the ruminal anaerobic fungus Neocallimastix frontalis. Appl Environ Microbiol 55(4):1016–1022
Nagpal R, Puniya AK, Sehgal JP, Singh K (2011) In vitro fibrolytic potential of anaerobic rumen fungi from ruminants and non-ruminant herbivores. Mycoscience 52:31–38
Nakashimada Y, Srinivasan K, Murakami Murakami M, Nishio N (2000) Direct conversion of cellulose to methane by anaerobic fungus Neocallimastix frontalis and defined methanohens. Biotechnol Lett 22(3):223–227
Paul SS, Kamra DN, Sastry VR (2010) Fermentative characteristics and fibrolytic activities of anaerobic gut fungi isolated from wild and domestic ruminants. Arch Anim Nutr 64:279–292
Peng XF, Swift CL, Theodorou MK, O’Malley MA (2018) Methods for genomic characterization and maintenance of anaerobic fungi. Methods Mol Biol 1775:53–67
Saye LMG, Navaratna TA, Chong JPJ, O’Malley MA, Theodorou MK, Reilly M (2021) The anaerobic fungi: challenges and opportunities for industrial lignocellulosic biofuel production. Microorganisms 9(4):694
Sebata A, Ndlovu LR, Dube JS (2011) Chemical composition, in vitro dry matter digestibility and in vitro gas production of five woody species browsed by Matebele goats (Capra hircus L.) in a semi-arid savanna. Zimbabwe Anim Feed Sci Tech 170:122–125
Sijtsma L, Tan B (1993) Degradation and utilization of grass cell walls by anaerobic fungi isolated from yak, llama and sheep. Anim Feed Sci Tech 44:221–236
Sirohi SK, Choudhury PK, Dagar SS, Puniya AK, Singh D (2013) Isolation, characterization and fibre degradation potential of anaerobic rumen fungi from cattle. Ann Microbiol 63:1187–1194
Solomon KV, Haitjema CH, Henske JK, Gilmore SP, Borges-Rivera D, Lipzen A, Brewer HM, Purvine SO, Wright AT, Theodorou MK, Grigoriev IV, Regev A, Thompson DA, O’Malley MA (2016) Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 351:1192–1195
Solomona KV, Henske JK, Theodorou MK, O’ Malley MA, (2016) Robust and effective methodologies for cryopreservation and DNA extraction from anaeroboc gut fungi. Anaerobe 38:39–46
Song JK, Wang D, Ren M, Yang F, Wang PX, Zou M, Zhao GH, Lin Q (2021) Seasonal prevalence and novel multilocus genotypes of giardia duodenalis in Yaks (Bos grunniens) in Qinghai Province. Western China Iran J Parasitol 16(4):548–554
Su Y, Yao W, Perez-Gutierrez ON, Smidt H, Zhu WY (2008) 16S ribosomal RNA-based methods to monitor changes in the hindgut bacterial community of piglets after oral administration of Lactobacillus sobrius S1. Anaerobe 14:78–86
Sun WC, Luo YH, Ma HQ (2011) Preliminary study of metal in yak (Bos grunniens) milk from Qilian of the Qinghai Plateau. Bull Environ Contam Toxicol 86(6):653–656
Sun MZ, Jin W, Li YF, Mao SY, Cheng YF, Zhu WY (2014) Isolation and identification of cellulolytic anaerobic fungi and their associated methanogens from Holstein cow. Acta Microbiol Sin 54(5):563–571
Teunissen MJ, Op den Camp HJM (1993) Anaerobic fungi and their cellulolytic and xylanolytic enzymes. Anton Leeuw Int J G 63:63–76
Teunissen MJ, de Kort GV, Op den Camp HJM, Huis in ’t Veld JHJ, (1992a) Production of cellulolytic and xylanolytic enzymes during growth of the anaerobic fungus Piromyces sp. on different substrates. J Gen Microbiol 138:1657–1664
Teunissen MJ, Kets EPW, Op den Camp HJM, Huis in ’t Veld JHJ and Vogels GD, (1992b) Effect of coculture anaerobic fungi isolated from ruminants and non-ruminants with methanogenic bacteria on cellulolytic and xylanolytic enzyme activities. Arch Microbiol 157:176–182
Thareja A, Puniya AK, Goel G, Nagpal R, Sehgal JP, Singh PK, Singh K (2006) In vitro degradation of wheat straw by anaerobic fungi from small ruminants. Arch Anim Nutr 60:412–417
Van Soest PJ, Robertson JB, Lewis BA (1991) Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 74:3583–3597
Wang R, Yang HJ, Yang X, Cao BH (2013) Four phenolic acids determined by an improved HPLC method with a programmed ultraviolet wavelength detection and their relationships with lignin content in 13 agricultural residue feeds. J Sci Food Agric 93:53–60
Wei YQ, Yang HJ, Luan Y, Long RJ, Wu YJ, Wang ZY (2015) Isolation, identification and fibrolytic characteristics of rumen fungi grown with indigenous methanogen from yaks (Bos grunniens) grazing on the Qinghai-Tibetan Plateau. J Appl Micribiol 120:571–587
Wei YQ, Long RJ, Yang H, Yang HJ, Shen XH, Shi RF, Wang ZY, Du JG, Qi XJ, Ye QH (2016) Fiber degradation potential of natural co-cultures of Neocallimastix frontalis and Methanobrevibacter ruminantium isolated from yaks (Bos grunniens) grazing on the Qinghai Tibetan Plateau. Anaerobe 39:158–164
Wei YQ, Yang HJ, Long RJ, Wang ZY, Cao BB, Ren QC, Wu TT (2017) Characterization of natural co-cultures of Piromyces with Methanobrevibacter ruminantium from yaks (Bos grunniens) grazing on the Qinghai-Tibetan Plateau: a microbial consortium with high potential in plant biomass degradation. AMB Express 120:1–12
Yang HJ, Xie CY (2010) Assessment of fibrolytic activities of 18 commercial enzyme products and their abilities to degrade the cell wall fraction of corn stalks in in vitro enzymatic and ruminal batch cultures. Anim Feed Sci Tech 159:110–121
Yang HJ, Yue Q, Cao YC, Zhang DF, Wang JQ (2009) Effects of crude feruloyl and acetyl esterase solutions of Neocallimastix sp. YQ1 and Anaeromyces sp. YQ3 isolated from Holstein steers on hydrolysis of Chinese wildrye grass hay, wheat bran, maize bran, wheat straw and corn stalks. Anim Feed Sci Tech 154:218–227
Young D, Dollhofer V, Callaghan TM, Reitberger S, Lebuhn M, Benz JP (2018) Isolation, identification and characterization of lignocellulolytic aerobic and anaerobic fungi in one- and two-phase biogas plants. Bioresource Technol 268:470–479
Acknowledgements
Not applicable.
Funding
This work was supported by the Grants from the National Natural Science Foundation of China (31860659), the Key Research and Development Project Plan of Gansu Provincial Science and Technology Department (20YF3NA020;18YF1NA077), the Application Research and Development Project of Gansu Academy of Sciences (2018JK-07; 2020JK-03; 2019HZ-03), the Science and Technology Industrialization Project of Gansu Academy of Sciences (2020–2022).
Author information
Authors and Affiliations
Contributions
YW conceived the project, performed analyzed, interpreted data, and drafted the manuscript. HY and ZW provided some intellectual content in the research design and interpretation. JD and HQ and MH contributed to research design and data collection. CW, JZ and TY performed statistical analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wei, Y., Yang, H., Wang, Z. et al. Roughage biodegradation by natural co-cultures of rumen fungi and methanogens from Qinghai yaks. AMB Expr 12, 123 (2022). https://doi.org/10.1186/s13568-022-01462-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-022-01462-2