Abstract
A total of 20 fungal cultures were isolated from the rumen of cattle fed a high fibre-containing diet. All of the isolates showed polycentric growth patterns and were identified as different strains of Orpinomyces and Anaeromyces. Enzyme assays of most of the isolates showed the highest carboxymethylcellulase (CMCase) and xylanase activities after 96 h of growth and highest avicelase activity after 120 h. Among all enzymes tested, xylanase activity was the highest, followed by CMCase and avicelase. The results of the in vitro fibre digestibility and rumen fermentation analyses revealed that the addition of fungal cultures significantly increased acetate, in vitro dry matter digestibility, partition factor values and microbial biomass synthesis levels. Overall, Orpinomyces spp. were found to be the better enzyme producers and fibre degraders than Anaeromyces spp.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Ruminants in tropical countries such as India rely mainly on highly fibrous plant materials as feed stuff, which are utilized following their breakdown by a complex microbial population in the rumen. Only two groups of rumen microorganisms, i.e. bacteria and protozoa, were recognized up to the mid-1970s, when the existence of a third group (i.e. anaerobic rumen fungi) was reported by Orpin (1975). These fungi are classified into two different groups, i.e. polycentric (Orpinomyces, Anaeromyces and Cyllamyces) and monocentric (Neocallimastix, Piromyces and Caecomyces) fungi on the basis of sporangial development. Their respective occurrence is dependent upon host and diet (Liggenstoffer et al. 2010). These fungi show a preference for the thick-walled sclerenchyma and vascular tissues (Akin et al. 1983) and have been shown to contribute to the overall digestion of various forages (Trinci et al. 1994; Shelke et al. 2009). Moreover, the production of high levels of extracellular enzymes capable of degrading plant structural carbohydrates to simple sugars, with acetate, lactate, ethanol, formate, CO2 and H2 as end-products (Bauchop 1989; Paul et al. 2010b), is also reported. The possibility of increasing the digestibility of highly fibrous material ingested by ruminants to obtain high protein-containing products is of particular interest where these feed stuffs constitute the main dietary components for animals, with feeds of higher energy and protein value (cereals, legumes, etc.) being reserved for human needs. Therefore, the aim of our study was to isolate and study the effects of different fungal isolates on rumen fermentation parameters in vitro.
Materials and methods
Animal diet and feeding
Two rumen fistulated cross-bred (Karan fries) adult cattle aged 3 years with an average body weight (BW) of 450 ± 20 kg were used in this study, which was conducted at the cattle yard of the National Dairy Research Institute, Karnal, India. The animals were fed individually on a standard diet (concentrate/roughage ratio 40:60) in separate animal houses with free access to water. The daily allowances of the basal diets were offered in two equal portions (8:00 AM and 4:00 PM) at about 2.5 % of their BW on a dry matter basis. The experiments were conducted after approval from the institutional animal ethics committee.
Isolation and morphological characterization
Anaerobic fungi were isolated from the rumen of cattle as described by Dagar et al. (2011). Morphological features of the isolated fungi, such as thallus morphology, growth patterns and position of sporangia, were examined microscopically (Dagar et al. 2011).
Extraction of genomic DNA
The cultures were grown at 39 °C in cellobiose broth medium. Genomic DNA was extracted by the method of Brookman and Nicholson (2005) with minor modifications. The cultures were centrifuged at 5,000 rpm for 10 min and the pellet ground with a pestle and mortar. Approximately 100 mg of the ground fungi pellet was transferred to a 1.5-ml Eppendorf tube, followed by the addition of 0.8 ml CTAB DNA isolation buffer [100 mM Tris–HCl, pH 8.0, 1.4 M NaCl, 20 mM EDTA (sodium salt), 2 % CTAB]; the mixture was vortexed and incubated at 70 °C for 1 h, with continuous vortexing at each 15-min incubation period. After incubation, 0.5 ml chloroform was added and vortexed until a white emulsion was visible. The sample was centrifuged at 12,000 rpm for 20 min, and the aqueous layer was removed and precipitated by isopropanol (0.3 ml) for 10 min at room temperature. The precipitated pellet was collected by centrifugation and washed with ethanol (70 %). The resultant pellet was air dried, dissolved in 1× TE buffer (100 μl) and stored at −20 °C.
PCR amplification of the internal transcribed spacer region
The internal transcribed spacer (ITS) region was subjected to PCR amplification in a 50 μl reaction mixture containing 1 μl of template DNA (50 ng), 5 μl of 10× assay buffer (1.5 mM MgCl2, 50 mM KCl, 20 mM Tris-HCl, pH 8.0, 0.01 % gelatin), 1 μl (20 pmol) of forward primer (5′-TACACACCGCCCGTCGCTA-3′), 1 μl (20 pmol) of reverse primer (5′-TCCTCCGCTTATTGATATGC-3′), 100 μM each of dNTP mix, 0.25 μl of Taq DNA polymerase (0.75 U); sterile distilled water was added to reach 50 μl. The PCR was carried out using a thermal cycler (Bio-Rad, Hercules, CA) with an initial denaturation of 94 °C for 5 min followed by 35 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min, extension at 72 °C for 1 min and a final extension at 72 °C for 5 min.
Amplified DNA fragments were examined by horizontal electrophoresis in 1.5 % agarose gel containing ethidium bromide (0.5 μg/ml) at 100 V for 2 h in 1× TBE (Tris–Boric acid–EDTA) buffer with 5 μl aliquots of PCR products and digitized using gel documentation imaging system (Bio-Rad). All PCR products were purified by using the QIAquick PCR purification kit according to the manufacturer’s instructions (Qiagen, Hilden, Germany) and sequenced (Xceleris Genomics, Mumbai, India).
Sequence analysis and phylogenetic tree construction
Sequences obtained after sequencing were aligned by using the Clustal W (Thompson et al. 1994) program in BioEdit (Hall 1999). A Basic Local Alignment Search Tool (BLAST) search was performed with the obtained sequences in order to determine the homology with already available fungal sequences in the GenBank database. All sequences were submitted to the GenBank database. Six reference sequences of the ITS gene (FJ501287, AY429664, FJ501296, GQ857641, JF974109, FJ483845) representing all gut fungal genera were included in the phylogenetic analyses, and the evolutionary history was inferred using the Neighbor-Joining (NJ) method (Saitou and Nei 1987). Evolutionary analyses were conducted in MEGA5 (Tamura et al. 2011).
In vitro gas production and dry matter digestibility
Effects of the fungal isolates on in vitro gas production (Menke and Steingass 1988) and in vitro dry matter digestibility (IVDMD) were estimated using neutral detergent fibre (NDF) as substrate. The NDF (375 ± 5.0 mg) was weighed into 100 ml calibrated glass syringes (FORTUNA®; Optima glass syringes; Sigma-Aldrich, St. Louis, MO) along with 30 ml medium containing buffered rumen liquor and 2 ml of each fungal isolate. The syringes were shaken gently and the air bubbles removed. The level of the piston was recorded, and syringes were incubated at 39 °C for 48 h, after which the contents were drained through the tube in the sample collection tube. The contents were filtered through the pre-weighed sintered glass crucible and kept in a hot air oven for drying. The percentage DMD, total gas production, partition factor (PF) and microbial biomass (MBM) yield (Blummel et al. 1997) were calculated and compared with the control (autoclaved cultures).
The PF is calculated as the ratio of substrate truly degraded in vitro (mg) to the volume of gas (ml) produced by it. The substrate provides important information on the partitioning of fermentation products. The MBM yield was calculated using data on the degradability of the substrate and gas volume and a stoichiometrical factor as follows (Blummel et al. 1997): Microbial biomass (mg) = substrate truly degraded − (gas volume × stoichiometrical factor); the stoichiometrical factor in this case was 2.25.
Individual volatile fatty acid profiles
One millilitre of supernatant was treated with 25 % metaphosphoric acid (4 ml), kept at 4 °C for 3–4 h and centrifuged at 3,000 rpm for 10 min. The supernatant was used for estimating individual volatile fatty acid (IVFA) profiles using a gas chromatograph (model 5700; Nucon Industries, Hyderabad, India) equipped with flame ionization detector (FID) and stainless steel column (length 4 feet; width 1/4 inch; i.d. 3 mm) packed with chromosorb-101. The temperature of the injection port, column and detector was set at 200, 180 and 210 °C, respectively. The flow rate of the carrier gas (nitrogen) through the column was 40 ml/min, and the flow rate of hydrogen and air through the FID was 30 and 300 ml/min, respectively. Different VFAs were identified in the samples on the basis of their retention time, and their concentrations (mmol) were calculated by comparing the retention time and the peak area of standards after deducting the corresponding blank values.
Enzymatic profiles
The enzyme profiles of three enzymes, namely carboxymethylcellulase (CMCase), avicelase and xylanase were determined. Both CMCase (endo-cellulase) and avicelase (exo-cellulase) are important cellulolytic enzymes that function synergistically to remove the amorphous and crystalline regions of cellulose, respectively, whereas xylanase is one of the hemi-cellulolytic enzymes, degrading xylan. Inoculum (2 %) of each isolate (from a 3-day-old culture) was added to 60 ml anaerobic medium containing 300 mg wheat straw and 0.3 % cellobiose (w/v) as the carbon source. After 24 h of incubation at 39 °C, 10 ml medium was centrifuged at 5,000 rpm for 10 min and the supernatant used as a source of crude enzyme. All assays were carried out in triplicate at 39 °C in a waterbath using the dinitrosalicylic acid method of Miller (1959). The enzyme activities were calculated by comparison with the respective standard curve and presented as micromoles per millilitre per hour.
Results and discussion
Isolation and morphological characterization
A total of 20 fungal cultures (NFRI-1 to -20) were isolated and purified using the repeated roll tube culturing method. All of the isolates showed typical filamentous rhizoids and polycentric exogenous sporangial development. However, based on the presence of constricted hyphae and pointed sporangia (Fig. 1a), we identified 13 isolates as Anaeromyces spp. and seven as Orpinomyces spp. (oval sporangia; Fig. 1b). Their identities were also confirmed by zoospore flagellation patterns, i.e. uniflagellate (Fig. 2a) in Anaeromyces spp. and polyflagellate (Fig. 2b) in Orpinomyces spp.
PCR amplification, sequencing and phylogenetic analysis of ITS region
A product size of approximately 800 bp was obtained for each isolate. BLAST search results after sequencing showed similarities between the 13 Anaeromyces isolates and seven Orpinomyces isolates, as also indicated by microscopic analysis. The sequences reported here have been deposited in the GenBank database under accession no. JN560942–JN560955 and JN887638–JN887643. The phylogenetic analysis showed separate groups for both genera (Fig. 3). Moreover, all of the isolates also showed groupings with their respective reference cultures, whereas separate lineages were obtained for the remaining genera, further validating their identities (Table 1).
Phylogenetic tree of the fungal isolates obtained from the internal transcribed spacer (ITS) gene sequences. Symbols indicate reference cultures. The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987). The optimal tree with the sum of branch length = 0.69037436 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method (Tamura et al. 2004) and are in the units of the number of base substitutions per site. The analysis involved 26 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 412 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al. 2011)
IVDMD and fermentation parameters
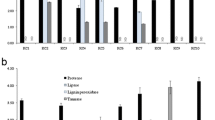
All the isolates were found to digest wheat straw fiber and to increase its digestibility (Table 1). The percentage IVDMD was found to be highest with isolate Orpinomyces NFRI-17 (60.98 ± 0.33 %) followed by Orpinomyces NFRI-3 (60.71 ± 0.60 %) and Orpinomyces NFRI-13 (60.44 ± 3.30) in comparison to control (45.73 ± 0.69 %). These results indicate higher fibre digestibility with Orpinomyces spp. than with Anaeromyces spp. isolates and are in accordance with those from previous studies (Theodorou et al. 1990; Rezaeian et al. 2005; Tripathi et al. 2007a; Paul et al. 2010a, 2011; Saxena et al. 2010) in which an increase in IVDMD after supplementation with anaerobic fungi, in vivo or in vitro, was reported. Our results are also in agreement with those of Nagpal et al. (2011) who reported an increase in IVDMD over the control after 48 and 72 h of incubation with Orpinomyces sp. RB2 and Orpinomyces sp. RC1. Kumar et al. (2004) also reported increased IVDMD of wheat straw after 48 h incubation with strained rumen liquor and Orpinomyces sp. Our results are also supported by Thareja et al. (2006) who found Neocallimastix sp. to be a better fibre degrader than Anaeromyces sp.
In addition to increased IVDMD, we also observed an increase in the PF and an up to twofold increase in MBM synthesis (Table 1). The increase in the PF and MBM yield clearly indicates that some of the energy diverted to microbial protein synthesis originated from accelerated fermentation of wheat straw fibre by fungal isolates. Fatty acid production patterns showed that acetate was the major end fermentation product (Table 1), and fatty acid production increased significantly in all fungal supplemented isolates incubated with pure wheat straw fibre as substrate. None of the isolates significantly increased propionate or butyrate production. These results are in agreement with those reported previously (Theodorou et al. 1996; Gordon and Phillips 1998).
Enzyme assays
All of the isolates showed CMCase, avicelase and xylanase activities (Table 2). Maximum xylanase activity was also obtained after 96 h of incubation by most of the isolates, with a maximum production of 11.00 ± 0.29 μmol/ml/h by isolate Orpinomyces NFRI-1. Similarly, CMCase enzyme activity also increased gradually and peaked at 96 h for most of the isolates with a maximum production of 9.03 ± 0.25 μmol/ml/h by isolate Orpinomyces NFRI-18. However, maximum avicelase activity was obtained after 120 h in almost all isolates, with isolates Anaeromyces NFRI-4 (2.53 ± 0.01 μmol/ml/h) and Orpinomyces NFRI-7 (2.41 ± 0.05 μmol/ml/h) being the maximum avicelase producers. Overall, Orpinomyces spp. were found to produce the maximum amounts of enzyme. In terms of enzyme activities, xylanase activity was found to be highest, followed by CMCase and avicelase, which is consistent with data reported by several researchers (Borneman et al. 1989; Novotna et al. 2010; Comlekcioglu et al. 2011; Nagpal et al. 2011). Novotna et al. (2010) reported xylanase as the most active Anaeromyces enzyme in comparison to all other enzymes, including cellulolytic enzymes. Maximum xylanase activity by a strain of Orpinomyces sp. (P4) was also observed by Comlekcioglu et al. (2011) on different lignocellulosic materials. Our results are in agreement with those of Borneman et al. (1989), who also reported slower and lower enzymatic activities by polycentric isolate PC-1 (classified as Anaeromyces sp. based on the presence of uniflagellated zoospores) for up to 96 h of incubation in comparison to other isolates. Our results are well supported by the study of Nagpal et al. (2011), where Orpinomyces spp. isolates were observed to be better enzyme producers than Anaeromyces isolates. Similar results were also observed by Tripathi et al. (2007b) who found higher CMCase, filter paper activity (FPase) and xylanase activities in a few isolates of Orpinomyces sp., with strains to strain variations. Hodrova et al. (1998) also reported higher cellulolytic enzyme production by Orpinomyces than Caecomyces. However, our results are not in agreement with those of Paul et al. (2010b), who reported much better CMCase and xylanase activities for Anaeromyces than Orpinomyces. The enzymatic activities of different anaerobic fungi have been studied in detail by numerous research groups (Tripathi et al. 2007b; Paul et al. 2010b; Comlekcioglu et al. 2011; Nagpal et al. 2011; Yang and Yue 2012). Borneman et al. (1989) even compared the various hydrolytic activities of monocentric and polycentric isolates and found that monocentric isolates produce enzymes faster and at higher levels than their polycentric counterparts. However, we could not isolate any monocentric fungi in our attempts.
Based on culture dependent method, polycentric filamentous fungi were found to be dominant in the collected rumen samples, which is well in agreement with the results reported by Sridhar et al. (2010) and Griffith et al. (2009). Sridhar et al. (2010) cultured and reported a large number of Orpinomyces spp. in the rumen and faecal samples of Indian cattle. Griffith et al. (2009) used the MPN method to study the diversity of anaerobic fungi and observed different fungal populations in digesta, fresh faeces and frozen- thawed faecal samples. They reported polycentric morphotypes as the most abundant fungal population in the rumen digesta and frozen–thawed faeces and the bulbous morphotypes in fresh faeces. However, Liggenstoffer et al. (2010) and Fliegerova et al. (2010) reported the prevalence of bulbous morphotypes (Caecomyces and Cyllamyces) in cattle faeces using culture-independent techniques. Since the isolation of these fungi has been shown to be influenced by factors such as animal diet, geographic locations, media composition, carbon source, among others (Griffith et al. 2009), comparison of culture-dependent and independent methods might not be justified. Moreover, culture-independent methods do not give any indication of the viability of cultures (Fliegerova et al. 2010). Furthermore, the abundance of polycentric isolates in Indian cattle may also be due to high lignocellulosic content in the animal diet, as the rhizoidal system of polycentric fungi have a better penetrating ability than that of monocentric/bulbous morphotypes (Borneman et al. 1989).
In conclusion, Orpinomyces spp. were found to be better fiber degraders than Anaeromyces spp., and the in vitro administration of these fungi was positively correlated with increased fibre digestibility and other rumen fermentation parameters.
References
Akin DE, Gordon GL, Hogan JP (1983) Rumen bacterial and fungal degradation of Digitaria pentzii grown with or without sulfur. Appl Environ Microbiol 46:738–748
Bauchop T (1989) Biology of gut anaerobic fungi. Biosystems 23:53–64
Blummel M, Makkar HPS, Becker K (1997) In vitro gas production: a technique revisited. J Anim Physiol Anim Nutr 77:24–34. doi:10.1111/j.1439-0396.1997.tb00734.x
Borneman WS, Akin DE, Ljungdahl LG (1989) Fermentation products and plant cell wall-degrading enzymes produced by monocentric and polycentric anaerobic ruminal fungi. Appl Environ Microbiol 55:1066–1073
Brookman J, Nicholson M (2005) Anaerobic fungal populations. In: Makkar H, McSweeney C (eds) Methods in gut microbial ecology for ruminants. Springer SBM, Dordrecht, pp 139–150
Comlekcioglu U, Aygan A, Yazdıc FC, Ozkose E (2011) Effects of various agro-wastes on xylanase and xylosidase production of anaerobic ruminal fungi. J Sci Ind Res 70:293–299
Dagar SS, Kumar S, Mudgil P, Singh R, Puniya AK (2011) D1/D2 domain of large-subunit ribosomal DNA for differentiation of Orpinomyces spp. Appl Environ Microbiol 77:6722–6725
Fliegerova K, Mrazek J, Hoffmann K, Zabranska J, Voigt K (2010) Diversity of anaerobic fungi within cow manure determined by ITS1 analysis. Folia Microbiol 55:319–325
Gordon GL, Phillips MW (1998) The role of anaerobic gut fungi in ruminants. Nutr Res Rev 11:133–168
Griffith GW, Ozkose E, Theodorou MK, Davies DR (2009) Diversity of anaerobic fungal populations in cattle revealed by selective enrichment culture using different carbon sources. Fungal Ecol 2:87–97
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hodrova B, Kopecny J, Kas J (1998) Cellulolytic enzymes of rumen anaerobic fungi Orpinomyces joyonii and Caecomyces communis. Res Microbiol 149:417–427
Kumar BM, Puniya AK, Singh K, Sehgal JP (2004) In vitro degradation of cell-wall and digestibility of cereal straws treated with anaerobic ruminal fungi. Indian J Exp Biol 42:636–638
Liggenstoffer AS, Youssef NH, Couger MB, Elshahed MS (2010) Phylogenetic diversity and community structure of anaerobic gut fungi (phylum Neocallimastigomycota) in ruminant and non-ruminant herbivores. ISME J 4:1225–1235
Menke KH, Steingass H (1988) Estimation of the energetic feed value obtained from chemical analysis and in vitro gas production using rumen fluid. Anim Res Dev 28:55
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Nagpal R, Puniya A, Sehgal J, Singh K (2011) In vitro fibrolytic potential of anaerobic rumen fungi from ruminants and non-ruminant herbivores. Mycoscience 52:31–38
Novotna Z, Prochazka J, Simunek J, Fliegerova K (2010) Xylanases of anaerobic fungus Anaeromyces mucronatus. Folia Microbiol 55:363–367
Orpin CG (1975) Studies on the rumen flagellate Neocallimastix frontalis. J Gen Microbiol 91:249–262
Paul SS, Deb SM, Punia BS, Singh D, Kumar R (2010a) Fibrolytic potential of anaerobic fungi (Piromyces sp.) isolated from wild cattle and blue bulls in pure culture and effect of their addition on in vitro fermentation of wheat straw and methane emission by rumen fluid of buffaloes. J Sci Food Agric 90:1218–1226
Paul SS, Kamra DN, Sastry VR (2010b) Fermentative characteristics and fibrolytic activities of anaerobic gut fungi isolated from wild and domestic ruminants. Arch Anim Nutr 64:279–292
Paul SS, Deb SM, Punia BS, Das KS, Singh G, Ashar MN, Kumar R (2011) Effect of feeding isolates of anaerobic fungus Neocallimastix sp. CF 17 on growth rate and fibre digestion in buffalo calves. Arch Anim Nutr 65:215–228
Rezaeian M, Beakes GW, Chaudhry AS (2005) Relative fibrolytic activities of anaerobic rumen fungi on untreated and sodium hydroxide treated barley straw in in vitro culture. Anaerobe 11:163–175
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Saxena S, Sehgal JP, Puniya AK, Singh K (2010) Effect of administration of rumen fungi on production performance of lactating buffaloes. Benefic Microbes 1:183–188
Shelke S, Chhabra A, Puniya AK, Sehgal J (2009) In vitro degradation of sugarcane bagasse based ruminant rations using anaerobic fungi. Ann Microbiol 59:415–418. doi:10.1007/bf03175124
Sridhar M, Kumar D, Anandan S, Prasad CS, Sampath KT (2010) Morphological and molecular characterization of polycentric rumen fungi belonging to the genus Orpinomyces isolated from Indian cattle and buffaloes. Res J Microbiol 5:581–594
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101:11030–11035
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. doi:10.1093/molbev/msr121
Thareja A, Puniya AK, Goel G, Nagpal R, Sehgal JP, Singh PK, Singh K (2006) In vitro degradation of wheat straw by anaerobic fungi from small ruminants. Arch Anim Nutr 60:412–417
Theodorou MK, Beever DE, Haines MJ, Brooks A (1990) The effect of a fungal probiotic on intake and performance of early weaned calves. Anim Prod 50:577 (Abstr)
Theodorou MK, Mennim G, Davies DR, Zhu WY, Trinci AP, Brookman JL (1996) Anaerobic fungi in the digestive tract of mammalian herbivores and their potential for exploitation. Proc Nutr Soc 55:913–926
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Trinci APJ, Davies DR, Gull K, Lawrence MI, Bonde Nielsen B, Rickers A, Theodorou MK (1994) Anaerobic fungi in herbivorous animals. Mycol Res 98:129–152
Tripathi VK, Sehgal JP, Puniya AK, Singh K (2007a) Effect of administration of anaerobic fungi isolated from cattle and wild blue bull (Boselaphus tragocamelus) on growth rate and fibre utilization in buffalo calves. Arch Anim Nutr 61:416–423
Tripathi VK, Sehgal JP, Puniya AK, Singh K (2007b) Hydrolytic activities of anaerobic fungi from wild blue bull (Boselaphus tragocamelus). Anaerobe 13:36–39
Yang HJ, Yue Q (2012) Effect of glucose addition and N sources in defined media on fibrolytic activity profiles of Neocallimastix sp. YQ1 grown on corn stover. J Anim Physiol Anim Nutr 96(4):554–562. doi:10.1111/j.1439-0396.2011.01177.x
Acknowledgment
The authors thank the National Agriculture Innovation Project, Indian Council of Agricultural Research, New Delhi-110012 for providing financial support for this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sirohi, S.K., Choudhury, P.K., Dagar, S.S. et al. Isolation, characterization and fibre degradation potential of anaerobic rumen fungi from cattle. Ann Microbiol 63, 1187–1194 (2013). https://doi.org/10.1007/s13213-012-0577-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-012-0577-6