Abstract
The interaction of Nilotinib (NIL) with alpha lactalbumin (α-LA) were studied by spectrofluorimetry, UV-Vis spectroscopy, circular dichroism (CD) and molecular modeling methods. Static quenching procedure fluorescence spectroscopy revealed that the fluorescence quenching of α-LA by NIL was the reason of formation of complex) NIL–α-LA. Fluorescence, UV-Vis, and CD spectra of NIL–α-LA complex showed that the conformation of α-LA has been changed in the presence of NIL. Based on the fluorescence quenching analyses the binding constant is calculated which is (467.73 × 104 M–1 at 298 K) and number of ligand-binding site is equal to one then the thermodynamic parameters of this complex are (ΔG = –33.011 kJ mol−1, ΔS = –80.005 J mol−1 K−1; ΔH = −157.70 kJ mol−1) at 298 K which were computed by using van’t Hoff equation and revealed that the reaction between ligand and protein is spontaneous and hydrogen bonding and van der Waals forces played a key role in the binding of NIL to α-LA. The results of molecular docking investigations have good agreement with the results of fluorescence spectroscopy studies. The molecular dynamic simulation (MDS) showed that free α-LA and the NIL–α-LA complex reached equilibration after 20 ns according to monitoring their root mean square deviation (RMSD). Finally, analyzing the root mean square fluctuations (RMSF) show that interaction of NIL with α-LA did not cause a significant conformational change in α-LA during the simulation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Alpha lactalbumin (α-LA) is the second main whey protein in the milk of all mammals. α-LA is a small protein with molecular weight of about 14 kDa [1–3], with a single polypeptide chain of 123 amino acid residues which formed the large helical domain and the smaller β-sheet domains which are linked by a high-affinity Ca2+ binding site in the loop [4, 5]. The helical domain comprising of three main α-helices (residues 5–11, 23–24 and 86–98) and two short 310 helices (residues 18–20 and 115–118), and smaller βHdomain with three stranded antiparallel β-sheet (residues 41–44, 47–50 and 55–56), one short 310 helix (residues 77–80) and loops [6].
Drug delivery influenced by their binding ability of protein is one of the most important biological functions of the small whey proteins [7]. α-LA can act as natural carrier for some drugs and can show interactions with them. Therefore, by taking the drug during breastfeeding it is possible to pass the drug through α-LA to the baby [8]. If the baby is exposed to a drug in mother’s milk, the baby may be harmed by various factors (depend on timing of the dose, toxicity and oral bioavailability of drug) [9].
Prescribing medication during breastfeeding needs evaluating the benefits of taking medication for the mother against the risk of not breastfeeding the baby or the potential risk of exposing the infant to medications [10]. Nilotinib (NIL) is a new oral tyrosine-kinase inhibitor (TKI) with a molecular weight of 529.5 Da, with hydrogen chloride and hemihydrate salt is, amino-pyrimidine-derived tyrosine-kinase inhibitor [11]. NIL shows good solubility in an acidic milieu, moderate solubility in water (0.29 mg/mL), and poor solubility (<0.1 mg/mL) in a phosphate buffer at pH > 4.5 [12]. Nilotinib is stronger than Imatinib in inhibiting BCR/ABL and also designed to overcome Imatinib resistance in chronic myeloid leukemia (CML) [13]. Therefore, it is necessary to evaluate the Nilotinib binding to α-LA in order to evaluate its transmission to the breast milk and infant [14, 15]. Fluorescence spectroscopy is important methods for characterizing the interaction of small molecule with proteins such as, binding constants and binding numbers [13, 16]. Also the molecular modeling method as a quick and economical method has been performed to predict the drugs binding site on proteins [17–19]. Interactions of cationic surfactant with α-LA to find of binding sites by spectroscopic methods was done to elucidate unfolding of α-LA has been [20]. Moreover spectroscopic methods and molecular dynamics (MD) simulation have been used to carried out interaction behavior between drugs and nanoparticles in the viewpoint of pharmacokinetic and pharmacodynamics sciences [21–23].
In the present work, the binding affinity of NIL to α-LA, thermodynamic parameters of complexation, binding constants, binding numbers, structural changes of α-LA is investigated to provide useful information about interactions between NIL and α‑LA. These properties were studied in detail by using fluorescence quenching technique [22, 24], and UV-Vis [20, 25, 26], circular dichroism (CD) measurement [27, 28] molecular docking [29, 30] and molecular dynamics simulation [21, 31].
RESULTS AND DISCUSSION
Quenching Mechanism of \(\alpha \)-LA by NIL
Various dependence of the quenching process on temperature, viscosity and excited state lifetime are often used to recognize whether the quenching mechanism is static) or, dynamic [32]. Static quenching is usually associated with formation of ground state complex between the fluorophore (protein) and the quencher, while which in dynamic quenching there is a collision between the fluorophore (protein) and the quencher. The quenching rate depends on the diffusion rate of quenching to fluorophore and also the temperature and viscosity of the solution [33].
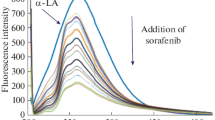
Quenching mechanism induced by a variety of molecular interactions such as excited state reactions, energy transfer, ground-state complex formation, and collisional quenching, which diminish the fluorescence intensity [34]. Intrinsic α-LA fluorescence spectra originate from emission of tryptophan (Trp), tyrosine (Tyr) and phenylalanine (Phe) residues. However, Trp residue alone plays the most important role due to the intrinsic fluorescence of the α-LA. As shown in Fig. 1, a gradual decrease in the fluorescence intensity of α-LA observed increasing the concentration of NIL at constant concentration of α-LA. To explore the type of fluorescence quenching in NIL–α-LA complexation, the data obtained from fluorescence quenching was analyzed by using the Stern–Volmer equation [35] at various temperatures.
In Eq. (1), F0 and F are the fluorescence intensities in the absence and presence of quencher (NIL), respectively; Kq is the bimolecular quenching rate constant; τ0 is the lifetime of the fluorophore in the absence of quencher (τ0 = 10−8 s) and [Q] is the concentration of quencher. The effect of temperature on lactalbumin is greatly influenced by the composition of the medium and the concentration of protein. Thus, these aspects should be considered in the design of heat treatments of lactalbumin in order to preserve its putative biological function when it is added to special food or pharmaceutical products. The heat of denaturation for lactalbumin has been shown to be dependent on protein concentration. Rate constants for denaturation of lactalbumin in phosphate buffer and in milk is increases by increasing the temperature therefore it is better to measure rate constant in the temperature range of 298 to 308 K [36].
The Stern–Volmer quenching constant is given by KSV which is equal to Kqτ0. If the quenching is identified to be dynamic, the Stern–Volmer constant is represented by KD. In static quenching the KSV and Kq values are decreased by increasing the temperature and vice versa in the case of dynamic quenching. In order to obtain the above parameters a plot of \(\frac{{{{F}_{0}}}}{F}~\) versus \(\left[ {\text{Q}} \right]\) at three different temperatures is plotted which is shown in Fig. 2. According to this plot KSV and values obtained from linear fittings of the experimental data and the values of KSV and Kq decreased when the temperature is raised from 25 to 35°C (Table 1). This quenching data prove that the quenching mechanism is not initiated by dynamic collision but resulting from the formation of a complex system and probably by static quenching [37]. By increasing the temperature, the dissociation of weakly NIL–α-LA complex bound is take place [32]. In dynamic quenching, bimolecular quenching constants are expected to increase as the temperature is raised. In contrast, by increasing in temperature a gradual decrease of complexes is occur, and thus lower values for the static quenching constant is obtained. In addition, KSV values decreased with increasing temperatures which indicating a static quenching mechanism.
Evaluation of the Binding Constant and Binding Site
Permission of drugs taking by a breastfeeding mother are basically dependent on the strength of interactions between drug and α-LA protein. The binding constant (Kb) and number of binding sites (n) can determine from the modified Stern–Volmer equation:
The values of Kb and n can be calculated from the intercept and slope of the plot of log [(F0 –F)/F] versus log[Q] (Fig. 3). According to this plot the value of Kb is 4.67 × 106 M–1 at 298 K, The moderate values of Kb show that NIL–α-LA complex formation is reversible and NIL can be saved and transferred by α-LA. Experimental investigation of Kb data indicating that the binding interaction of Nilotinib with α-LA is approximately strong, whereas the obtained KSV value indicating the high complexation rate of Nilotinib with α-LA. The obtained values of Stern–Volmer constant (KSV) and binding constant (Kb) show similar trend on.
Binding Forces and Thermodynamic Characteristics of NIL–\(\alpha \)-LA Complex
In a general sense, the acting forces between a drug (ligand) and biological macromolecule (receptor) in water mainly consist of four non-covalent interactions such as hydrophobic interactions, hydrogen bonding, van der Waals forces, and electrostatic interactions. The thermodynamic parameters of binding reaction such as, enthalpy change (ΔH) and entropy change (ΔS), free energy change (ΔG0) of the reaction, are the main evidence to proof nature of the binding forces.
From the thermodynamic standpoint, the values of ΔH > 0 and ΔS > 0 imply that hydrophobic interaction play important roles in the reaction. On the other hands the values of ΔH < 0 and ΔS < 0 reveals that the main force complexation would be van der Waals forces or hydrogen bond formation; and ΔH ≈ 0 and S > 0 suggest the existence of an electrostatic force [38]. Since studied temperature does not varies significantly therefore the enthalpy change (ΔH0) of a system did not change over the studied temperature range.
The values of thermodynamic parameters (enthalpy change (ΔH) and entropy change (ΔS)) were estimated from the van’t Hoff equation:
where Kb is the binding constant at temperature (T), and R (J mol–1 K–1) is the universal gas constant [36]. Slope and intercept of the linear van’t Hoff plot (Fig. 4) will give: ΔH and ΔS of complexation reaction, respectively. The value of standard free energy changes (ΔG) is calculated from the following relationship:
The obtained values of ΔH0, ΔS0, and ΔG0 for NIL–α-LA complex formation are calculated according to these data depicted in Table 2. As shown in this table, both negative sign of ΔH0 and ΔS0 lead to that van der Waals and hydrogen-bonding interactions are more important driving force of complexation. Furthermore, a negative value for ΔH0 advocates for the exothermic reactions and negative sign of ΔG0 reveal that the formation of NIL–α-LA complex is spontaneous reaction [39].
More over the negative value of ΔG0 demonstrate that the wild type form is more favorable than the mutant.
UV-Vis Spectroscopy
UV-Vis spectroscopy is one of the simplest and economical techniques for probing the structural changes of α-LA and provide information about formation of a molecular complex. UV-Vis absorption spectra of α-LA in the presence and absence of NIL were shown in Fig. 5. In dynamic quenching mechanism, peak positions in the UV-Vis spectrum of the α-LA should not change during complexation. In the case of static quenching, complex formation causes change in UV-Vis spectrum of α-LA, as shown in Fig. 5 [40]. Furthermore, blue shift was observed at maximum peak position of NIL–α-LA (280–250 nm). Maximum absorption at wavelength of 280 nm belonged to absorbance by the aromatic ring (π → π* transition) portion of aromatic substructure of some amino acid such as Trp, Tyr and Phe. In the case of macromolecules such as proteins, UV light absorption and scattering take place simultaneously. As absorption increases, 90° Rayleigh scattering intensity decreases but turbidity does not change [41]. Binding constant of complex can be calculated from to Benesi–Hildebrand equation [42, 43]:
where A0 and A are the absorbance of α-LA at 280 nm, in the absence and presence of complex, respectively. The parameters of εα-LA and εB are the molar extinction coefficient of α-LA and the bound complex, respectively, and b is the light path of the cuvette (1 cm). The value of A0/A – A0 is plot versus, 1/C complex in Fig. 6 the binding constant (K) values of this linear plot, can be calculated from the ratio of the intercept to the slope. The obtained K = 0.6 × 105 L/mol, demonstrating that strong binding exists between NIL to α‑LA and complex with lowest-energy state is formed.
Circular Dichroism and Conformational Analysis
CD spectroscopy is used in this work to investigate the secondary structure of α-LA after binding NIL to α-LA. The CD spectra of α-LA in the absence and presence of NIL under physiological condition (pH = 7.4) at room temperature are recorded and shown in Fig. 7. As can be seen in this figure the CD spectra of free α-LA exhibited two negative bands in the UV region, at 208 and 222 nm, which originated from the π → π* and n → p* transfer for the peptide bond of the α-helixes, respectively. The helical content of free and bounded form of α-LA was calculated from the mean residue ellipticity (MRE) values at 208 nm.
where Cp is the molar concentration of α-LA, n denotes the number of amino acid residues, l is the path length (1.0 cm), the value of 4000 is the MRE of the β-form and random coil conformation cross at 208 nm while 33 000 is the MRE value of a pure α-helix at 208 nm. It can be seen from Table 3 that the α-helical content of α-LA slightly decreases from 33.19 to 31.63%, while the concentration of NIL increased from 0 to 60 μM, suggesting that the conformational change of α-LA induced by NIL is slight.
Docking Studies
The molecular docking approach can predict the favored preferred posing of a small molecule (NIL) to target (α-LA) and estimate the energy of binding in interaction of NIL with α-LA. To form the NIL–α-LA complex, according to docking studies The value of the calculated binding energy for NIL–α-LA complex is 36.78 kJ mol−1 which is in agreement with experimental results that serve as a confirm for the binding mechanism and docking studies (Table 2).
The estimated values of binding free energy from molecular docking and fluorescence quenching studies were –36.78 mol−1 and –33.011 kJ/mol, respectively. In addition, the molecular modeling studies have been approved by all experimental results. These methods had previously been shown to produce binding models similar to experimentally observed counterparts [44, 45]. The reason for slight differences between these values is probably raised due to the differences between the X-ray structure of the protein and actual ones [45].
One of the main reasons to slight difference between the obtained values of ΔG0 experimental and docking methods is due to the effect of solvent interactions with NIL to α-LA. Also molecular docking study is used to explore the amino acid residues involved in binding site (Fig. 8). As can see in Fig. 8 NIL is surrounded by the hydrophobic residues amino acid such as: Ser34, Thr33, His32, Trp104, Val27, Thr30, Thr29, Ala106, Cys28, Tyr103, Trp26. Moreover the docking results also reveals that there is one hydrogen bonding interactions site between NIL and the carbonyl group of Gln54 residue to build a hydrogen bond with NH group in NIL with bond length of 3.24 Å. Therefore it can calculated that the NH group in the structure of the NIL have an important role in the NIL–α-LA complex formation. Therefore, as results of molecular docking studies show that hydrogen bonding and hydrophobic forces are the major forces in NIL interaction with α-LA which can stabilize the complex. Furthermore, the calculated interaction parameters and binding energy are in good agreement with those obtained from experiment.
Molecular Dynamics Simulation
To evaluate the stably of complex the molecular dynamics simulation (MDS) along 20 ns was performed. The value of root mean square deviation (RMSD) and root mean square of fluctuation (RMSF) are determined from MDS result. The analysis of RMSD was performed to investigate the stability of the system with time period of 20 ns. The RMSD values of backbone for non-liganded α-LA and NIL–α-LA complex with respect to starting structures were calculated and plotted from 0 to 20 ns, as shown in Fig. 9a as can be seen in this figure the RMSD of each system rapidly achieves stability and low fluctuations around their average values after nearly 4 ns. To investigate the flexibility and changes in the mobility of α‑LA residues in the absence and presence of NIL the analysis of root mean square fluctuations (RMSFs) of the backbone atoms are done. Results are shown in Fig. 9b, small fluctuation RMSF of the complex system reveals that NIL–α-LA complex were stable during the simulation time.
To further analyze the binding mode during the simulation, average conformation of the binding pocket of α-LA complexed with Nilotinib was obtained from MD simulation. As can be seen in Fig. 10 the most key residues have been appeared around the Nilotinib in the binding pocket on the basis of average MD-simulated complex structure are approximately similar to the results obtained from docking however. Analysis of MD results shows that two hydrogen bonds which are formed between NH group of Nilotinib and lle10 and amide group of Nilotinib and His32 remained stable throughout the simulation, which indicates that these hydrogen bond interactions were strong. In addition, hydrophobic interactions (Ala 109, lle41, Val21, Gly19, Gly20, Thr33) played an important role in stabilization of Nilotinib in the binding pocket.
EXPERIMENTAL
Materials and Methods
Alpha lactalbumin from bovine milk-type III-calcium depleted (L 6010) was obtained from Sigma-Aldrich. Nilotinib was procured from Baran Co. (Tehran, Iran).
Preparation of Stock Solution
A stock solution of α-LA (10 μM) is prepared in 0.1 M phosphate buffer at pH 7.4.
Stock solution of Nilotinib (4.5 mM) is prepared by dissolution in dimethyl sulphoxide (DMSO). All solutions were preserved in the refrigerator at 4°C in the dark. Experimental fresh solutions of α-LA and Nilotinib were made by diluting of their stock solutions.
Apparatus
UV-Vis measurements. UV spectra of all α-LA solutions in the absence and presence of Nilotinib were recorded from 200 to 400 nm at room temperature on a SQ4802 UV-Vis diode-array spectrophotometer equipped with 1.0 cm quartz cells.
Fluorescence measurements. Fluorescence measurements were performed on a personal computer based spectrofluorometer (JASCO Japan FP-8300) equipped with a Xenon lamp light and quartz cuvette (1.0 cm path length) were used for the measurements. Fluorescence spectra of α-LA in the absence and the presence of increasing NIL concentrations were recorded in the λem = 300–500 nm and λex = 280 nm at three temperatures (298, 303, and 308 K). In this experiment α-LA concentration was kept fix at (10 × 10–6 mol L–1), while the concentration of a NIL was varied in the range, 0–5 μM. The inner filter effects of any compound in fluorescence determination system were removed by using the following equation [46, 47]:
where \({{F}_{{{\text{cor}}}}}\) and \({{F}_{{{\text{obs}}}}}\) are corrected and observed fluorescence intensity, respectively, \({{A}_{{{\text{ex}}}}}\) and \({{A}_{{{\text{em}}}}}\) are the absorption of the system at excitation and emission wavelength, respectively.
Circular dichroism (CD). Far-UV CD measurements (190–240 nm) were carried out on a Jasco-815 spectro polarimeter with using a quartz cell with a path length of 1 mm (Tokyo, Japan) at room temperature. To explore changes in the secondary structure, the various concentrations of NIL (10, 20, 40, 60 μM) were added to the fixed concentration of α-LA (10 μM).
Docking Analysis and Structural Modeling of NIL–α-LA Complex
Protein structure selection and preparation. In order to estimate the conceivable orientation of the drugs that binds to the proper binding site of protein and also computes the binding free energy the Auto dock (version 4.2) and Auto dock tools (ADT) which is constructed on the Python Molecule Viewer (PMV) utilizing the Lamarckian genetic algorithm [48] were used.
The structure of NIL was sketched using HyperChem7 [49] and was optimized using the AM1 semi-empirical method. In the Protein Data Bank (PDB) the three-dimensional coordinates of a-LA was fetched (PDB ID 1HFZ) and ligand-free structure (chain A) was chosen, as primary protein structure. During the preparation of the protein input file water molecules were removed, then polar hydrogen atoms and partial Kollman charges were added using ADT to the protein structure also describing the rotatable bonds. The protein was held rigid receptor and the solvent molecules effect is not considered during the docking. Studies docking was carried out in two steps: first, high atomic affinity grid maps and analyze the surface characteristics of receptor by auto grid. The grid size was set to be 35, 33, and 47 Å along X, Y and Z axes with 0.375 Å between grid points. Then ligands are docked with Auto Dock using this information. Docking results through cluster analysis based on the root-mean-squared deviation (RMSD) tolerance of 2.0 Å was performed. Eventually according to the Autodock scoring function, best docked conformations were elected as initial structure for the molecular dynamics simulation.
Molecular dynamics simulation (MDS). MD simulation in the presence of an explicit solvent was carry out to evaluate the in-vivo stability of the docked complex. In this investigation GROMACS version 5.1.2 program [50] in Ubuntu Linux operating system was utilized to perform molecular dynamics simulations (MDS) and Potential energy minimization (PME). The AMBER99sb force field was used for simulation [51]. Similar conditions were considered for both protein and complex. Both of α-LA and α-LA-NIL were solvated with water model TIP3P in cubic box by utilizing periodic boundary condition to minimize edge effects in the system. In each system the negative charge was neutralized by adding sodium counter-ions [52]. Then energy minimization of system with a cut off 1 nm for van der Waals and Coulomb Forces can be done by using 50 000 steps of the steepest descent algorithm to determine the appropriate molecular arrangement in terms of energy. Then in order to prove the stability of conformation system we used equilibration using canonical ensembles (NVT) followed by isothermal isobaric ensembles (NPT) were carry out. In NVT thermostat technique is used to control the temperature by using the Nosé-Hoover thermostat and Langevin dynamics and in NPT in addition to a thermostat condition, a barostat is required. For this purpose, in the NPT phase the number of moles of molecules (N), pressure (P), and temperature (T), and in the NVT phase: number of moles of molecules (N), volume (V), and temperature (T) would constant. Both phase of equilibration was done by 50 000 steps and 100 ps in 310 K. Finally, full MDS of 20 ns were performed at constant temperature (310 K) and pressure (1 bar).
CONCLUSIONS
The present study conformational changes in α-LA upon binding to a NIL, interaction between them, binding sites, binding energies, binding forces and amino acid residues involved in bindings were investigated by combination of UV-Vis absorption, circular dichroism (CD), fluorescence quenching, and molecular docking method. The values of Kb indicate that strong binding form between NIL to α-LA, also it done at situation in vivo [51, 52]. The thermodynamic parameters and molecular docking analysis results reveals that involvement of hydrophobic interactions and hydrogen bonds as well as van der Waals forces in stabilizing complex formation. In addition, the value of obtained ΔG from fluorescence spectroscopy (‒33.01 kJ/mol–1) and those estimated from molecular docking (–36.78 kJ /mol–1) were compared which confirm each other. Changes of the UV absorption spectrum showed that the secondary structure of α-LA slightly changes after the formation of the NIL–α-LA complex. RMSD and RMSF of MD studies showed that the NIL–α-LA complex achieved a steady state after 4 ns.
REFERENCES
Baronea, G., Moloney, C., O’Reganb, J., Alan, L.K., and O’James A., J. Food Compos. Anal., 2020, vol. 92, article ID 103546. https://doi.org/10.1016/j.jfca.2020.103546
Belatik, A., Kanakis, C., Hotchandani, S., Tarantilis, P., Polissiou, M., and Tajmir-Riahi, H., J. Biomol. Struct. Dyn., 2012, vol. 30, pp. 437–447. https://doi.org/10.1080/07391102.2012.682209
Markus, C.R., Olivier, B., Panhuysen, G.E., Van der Gugten, J., Alles, M.S., Tuiten, A., Westenberg, H.G., Fekkes, D., Koppeschaar, H.F., and de Haan, E., Am. J. Clin. Nutr., 2000, vol. 71, pp. 1536–1544. https://doi.org/10.1093/ajcn/71.6.1536
Al-Hanish, A., Stanic-Vucinic, D., Mihailovic, J., Prodic, I., Minic, S., Stojadinovic, M., Radibratovic, M., Milcic, M., and Velickovic, T.C., Food Hydrocoll., 2016, vol. 61, pp. 241–250. https://doi.org/10.1016/j.foodhyd.2016.05.012
Fitz Gerald, R.J., Murray, B.A., and Walsh, D.J., J. Nutr., 2004, vol. 134 pp. 980–988. https://doi.org/10.1093/jn/134.4.980S
Jahanshahtalab, M., Kamshad, M., Rezaei, S., Beigoli, S., Sharifi Rad, A., Ehrzad, J., Khashkhashi Moghadam, S., Mokaberi, P., Gharebaghi, S., Saberi, M.R., Chamani, J., J. Iran. Chem. Soc., 2019, vol. 16, pp.1311–1326. https://doi.org/10.1007/s13738-019-01608-3
Mohammadi, F. and Moeeni, M., J. Funct. Foods, 2015, vol. 12, pp. 458–467. https://doi.org/10.1016/j.jff.2014.12.012
Bagheri, F. and Fatemi, M.H., Russ. J. Bioorg. Chem., 2021, vol. 47, pp. 864–873. https://doi.org/10.1134/s1068162021040038
Frank, J. and Luo, A.C., J. Am. Pharm. Assoc., 2012, vol. 52, pp. 86–94. https://doi.org/10.1331/JAPhA.2012.10139
Spencer, J.P., Gonzalez III, L.S., and Barnhart, D., Am. Fam. Physician., 2001, vol. 64, pp. 119–127.
Tanaka, C., Yin, O.Q.P., Sethuraman, V., Smith, T., Wang, X., Grouss, K., Kantarjian, H., Giles, F., Ottmann, O.G., Galitz, L., and Schran, H., Clin. Pharmacol. Ther., 2012, vol. 52, pp. 197–203 https://doi.org/10.1038/clpt.2009.208
Meyer, B. and Peters, T.A., Chem. Int., 2003, vol. 42, pp. 864–890. https://doi.org/10.1002/anie.200390233
Saad Tayyab, S., Izzudin, M.M., Zahirul Kabir, Md., Feroz, S.R., Tee, W.V., Saharuddin, B.M., and Alias, Z., J. Photochem. Photobiol., 2016, vol. 162, pp. 386–394. https://doi.org/10.1016/j.jphotobiol.2016.06.049
Quintas-Cardama, A. and Cortes, J., Drugs Today (Barc.), 2007, vol. 43, p. 691. https://doi.org/10.1358/dot.2007.43.10.1122218
Saglio, G., Kim, D.-W., Issaragrisil, S., Le Coutre, P., Etienne, G., Lobo, C., Pasquini, R., Clark, R.E., Hochhaus, A., Hughes, T.P., Gallagher, N., Hoenekopp, A., Dong, M., Haque, A., Larson, R.A., and Kantarjian, H.M., Engl. J. Med., 2010, vol. 362, pp. 2251–2259. https://doi.org/10.1056/NEJMoa0912614
Yanjun, Sh., Hongyan, L., Min X., Zhenpeng, L., Guoqiang, X., Liang H., and Zhengzhi, Z., Spectrochim. Acta A, 2012, vol. 87, pp. 251–257. https://doi.org/10.1016/j.saa.2011.11.048
Zahirul Kabir, M.D., Mukarramb, A.K., Mohamad, S.B., Alias, Z., and Tayyab, S., J. Photochem. Photobiol. B: Biol., 2016, vol. 160, pp. 229–239. https://doi.org/10.1016/j.jphotobiol.2016.04.005
Mousavi, S.F. and Fatemi, M.H., Struct. Chem., 2019, vol. 30 pp. 1–12. https://doi.org/10.1007/s11224-018-1178-1
Danesh, N., Navaee Sedighi, Z., Beigoli, S., Atena Sharifi-Rad, A., Saberi M.R., and Chamani, J., J. Biomol. Struct. Dyn., 2018, vol. 36, pp. 1747–1763. https://doi.org/10.1080/07391102.2017.1333460
Housaindokht, M.R., Chamani, J., Saboury, A.A., Moosavi-Movahedi A.A., and Bahrololoom, M., Bull. Korean Chem. Soc., 2001, vol. 22, pp. 145–148.
Atena Sharifi-Rad, A., Mehrzad, J., Darroudi, M., Saberi, M.R., and Chamani, J., J. Biomol. Struct. Dyn., 2021, vol. 39, pp. 1029–1043. https://doi.org/10.1080/07391102.2020.1724568
Mokaberi, P., Babayan-Mashhadi, F., Amiri Tehrani Zadeh, Z., Saberi, M.R., and Chamani, J., J. Biomol. Struct. Dyn., 2021, vol. 39, pp. 3358–3377. https://doi.org/10.1080/07391102.2020.1766570
Sadeghzadeh, F., Entezari, A.A., Behzadian, K., Habibi, K., Amiri-Tehranizadeh, Z., Asoodeh, A., Saberi, M.R., and Chamani, J., Protein Pept. Lett., 2020, vol. 27, pp. 1007–1021.
Kamshad, M., Jahanshah Talab, M., Beigoli, S., Sharifirad, A., Chamani, J., J. Biomol. Struct. Dyn., 2018, vol. 37, pp. 2030–2040. https://doi.org/10.1080/07391102.2018.1475258
Mokaberi, P., Reyhani, V., Amiri-Tehranizadeh, Z., Saberi M.R., Beigoli, S., Samandar, F., and Chamani, J., J. Chem., 2019, vol. 43, p. 8132. https://doi.org/10.1039/C9NJ01048C
Chamani, J. and Moosavi-Movahedi, A.A., J. Colloid Interface Sci., 2006, vol. 297, pp. 561–569. https://doi.org/10.1016/j.jcis.2005.11.035
Maryam Dareini, M., Amiri Tehranizadeh, Z., Marjani, N., Reza Taheri, R., Aslani-Firoozabadi, S., Talebi, A., Nayeb Zadeh Eidgahi, N., Saberi, M.R., and Chamani, J., Spectrochim. Acta A, 2020, vol. 225, article ID 117528. https://doi.org/10.1016/j.saa.2019.117528
Chamani, J., Moosavi-Movahedi, A.A., Saboury, A.A, Gharanfoli, M., and Hakimelahi, G.H., J. Chem. Thermodyn., 2003, vol. 35, pp. 199–207. https://doi.org/10.1016/S0021-9614(02)00312-9
Shakibapour, N., Dehghani Sani, F., Beigoli, S., Sadeghian., H., and Chamani, J., J. Biomol. Struct. Dyn., 2019, vol. 37, pp. 359–371. https://doi.org/10.1080/07391102.2018.1427629
Zhang, A.M., Wei, N., Liu, X.F., Wu, M.G., and Xuan, G.S., Russ. J. Bioorg. Chem., 2021, vol. 47, pp. 261–269. https://doi.org/10.1134/S1068162021010283
Sohrabi, T., Hosseinzadeh, M., Beigoli, S., Saberi, M.R., and Chamani, J., J. Mol. Liq., 2018, vol. 256, pp. 127–138. https://doi.org/10.1016/j.molliq.2018.02.031
Fraiji, L.K., Hayes, M.D., and Werner, T.C., J. Chem. Educ., 1992, vol. 69, p. 424. https://doi.org/10.1021/ed069p424
Lakowicz, J.R., Principles of Fluorescence Spectroscopy, Lakowicz, J.R., Ed., Boston, 1999.
Eftink, M.R., Fluorescence Quenching Reaction: Probing Biological Macromolecular Structures, Biophysical and Biochemical Aspects of Fluorescence Spectroscopy, Plenum Press, New York, 1991.
Żamojć, K., Bylińska, I., and Wiczk, W., Int. J. Mol. Sci., 2021, vol. 22, p. 885. https://doi.org/10.3390/ijms22020885
Wehbi, Z., Pérez, M.D., Sánchez, L., and Pocoví, C., J. Agric. Food Chem., 2005, vol. 53, pp. 9730−9736. https://doi.org/10.1021/jf050825y
Suryawanshi, V.D., Walekar, L.S., Gore, A.H., Anbhule, P.V., and Kolekar, G.B., J. Pharm. Anal., 2016, vol. 6, pp. 56–63. https://doi.org/10.1016/j.jpha.2015.07.001
Ross, P.D. and Subramanian, S., Biochemistry, 1981 vol. 20, pp. 3096–3102. https://doi.org/10.1021/bi00514a017
Paul, B.K. Guchhait, N., Photochem. Photobiol. Sci., 2011, vol. 10, pp. 980–991. https://doi.org/10.1016/j.molliq.2018.02.03110.1039/c0pp00309c
Madrakian, T., Bagheri, H., Afkhami, A., and Soleimani, M., J. Lumin., 2014, vol. 155, pp. 218–225. https://doi.org/10.1016/j.jlumin.2014.06.047
Kaur, J., Application of UV light scattering to detect reversible self-association and aggregation of proteins in solution, Doctoral Dissertation, 2017.
Bratty, M.A., Saudi Pharm. J., 2020, vol. 28, pp. 729–736. https://doi.org/10.1016/j.jsps.2020.04.015
Ariga, G.G., Naik, P.N., Chimatadar, S.A., and Nandibewoor, S.T., J. Mol. Struct., 2017, vol. 1137, pp. 485–494. https://doi.org/10.1016/j.molstruc.2016.12.066
Chamani, J., J. Mol. Struct., 2010, vol. 979, pp. 227–234. https://doi.org/10.1016/j.molstruc.2010.06.035
Dehghani Sani, F., Shakibapour, N., Beigoli, S., Sadeghian, H., Hosainzadeh, M., and Chamani, J., J. Lumin., 2018, vol. 203, pp. 599–608. https://doi.org/10.1016/j.jlumin.2018.06.083
Rashidipour, S., Naeeminejad, S., and Chamani, J., J. Biomol. Struct. Dyn., 2016, vol. 34, pp. 57–77. https://doi.org/10.1080/07391102.2015.1009946
Shi, J.H., Chen, J., Wang, J., and Zhu, Y.Y., Spectrochim. Acta A, 2015, vol. 149, pp. 630–637. https://doi.org/10.1016/j.saa.2015.04.034
Morris, G.M., Goodsell, D.S., Halliday, R.S., Huey, R., Hart, W.E., Belew, R.K., and Olson, A.J., J. Comput. Chem., 2009, vol. 30, pp. 2785–2791. https://doi.org/10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B
Taghipour, P., Zakariazadeh, M., Sharifi, M., Jafar Ezzati Nazhad Dolatabadi, J., Barzegar, A., J. Photochem. Photobiol. B., 2017, vol. 183 pp. 11–15. https://doi.org/10.1016/j.jphotobiol.2018.04.008
van der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A.E., and Berendsen, H.J.C., J. Comput. Chem., 2005, vol. 26 pp. 1701–1718. https://doi.org/10.1002/jcc.20291
Chen, A.A. and Pappu, R., J. Phys. Chem. B, 2007, vol. 111, pp. 11884–11887. https://doi.org/10.1021/jp0765392
Bussi, G., Donadio, D., and Parrinello, M., J. Chem. Phys., 2007, vol. 126, p. 14101. https://doi.org/10.1063/1.2408420
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
No animals were involved in this work. No human subjects were involved in this work.
Conflict of Interests
The authors declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Bagheri, F., Fatemi, M.H. Investigation of the Interaction between Nilotinib and Alpha-Lactalbumin by Spectroscopic Methods and Docking Studies. Russ J Bioorg Chem 48, 783–792 (2022). https://doi.org/10.1134/S1068162022040057
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1068162022040057