Abstract
The interaction between α-lactalbumin (a-LA) with resveratrol (RES) and curcumin (CUR) was studied by multiple spectroscopic as well as the zeta potential and molecular modeling techniques. Fluorescence spectroscopy showed that RES could quench the a-LA fluorescence with a static mechanism and this quenching effect became more significant when both CUR and RES coexisted. The synchronized fluorescence spectroscopy indicated that the conformation of a-LA and the distance between RES and Trp residues was altered. On the other hand, in the binary system, the RES site was closer to Trp than to Tyr, which proved the significant contribution of the Trp residues to the fluorescence of a-LA. Using red edge excitation shift, we found that the mobility restriction around the Trp residues was unchanged, demonstrating that the rigidity of the environment of Trp residue remained the same. The binding distance between RES and the Trp residues of a-LA was obtained according to Förster’s theory of nonradioactive energy transfer and was found to be 2.67 nm and 2.78 nm for the binary and ternary systems, respectively. This confirmed the existence of static quenching for the binary and ternary systems alike. The quantitative analysis data from the circular dichroism spectra demonstrated that the binding of RES to a-LA induced conformational changes in a-LA. Moreover, the α-helix content in a-LA greatly decreased in the presence of RES and in the ternary system this decrease was significant. Protein–ligand docking suggested that the affinity of RES to the (a-LA–CUR) complex was lower than for RES to a-LA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Trans-resveratrol (3,5,4-trihydroxystilbene) (RES) is a natural compound with medicinal properties. It is a hydroxyl-substituted stilbene (Scheme 1) and a common phytoalexin (plant antibiotic) produced by plants in response to fungus infections. It is found in grapes, nuts, fruits and various berries, and is a potent antioxidant with strong anti-proliferative properties. This compound has beneficial effects on human health including anti-inflammatory and anti-cancer activities [1,2,3,4]. Inhibitory effects on cyclooxygenase, platelet aggregation inhibition, rat liver mitochondrial ATPase, human F1 ATPase and tyrosinase have been reported [5,6,7]. It has demonstrated to prevent or slow the progression of a variety of diseases, including cancer, cardiovascular disease, ischemic injuries, degenerative neurological disorders and Alzheimer’s disease. The antioxidant potential of RES has been studied extensively and the substance is reported to have a high efficiency as a scavenger of hydroxyl and superoxide radicals, and also protects against lipid peroxidation and DNA damage caused by ROS (reactive oxygen species) [8]. The combination of these properties with the lack of harmful effects makes TRES attractive as a biological drug.
Curcumin (1,7-bis(4-hydroxy-3-methoxyphenyl)-1,6-heptadiene-3,5-dione) (Scheme 2) is the active substance in tumeric rhizomes (Curcuma longa Linn.) and has been found to have an antioxidant activity with no side effects [9]. Curcumin exhibits a wide range of potential therapeutic and preventive actions on numerous diseases, including cataract, wounds, allergies, gastric ulcer, inflammatory bowel disease, myelodysplastic syndromes, acquired immune deficiency syndrome, psoriasis, colon cancer, Alzheimer’s disease, atherosclerosis, myocardial infarction, osteoporosis, lung diseases, diabetes mellitus and a wide spectrum of cancers [10,11,12,13]. Its anti-tumor activity is attributed to its ability to induce apoptosis via caspase-3 activation [14]. A recent study showed that diabetic rats treated with curcumin had lower blood glucose and glycated hemoglobin levels, in association with lower oxidative stress [15].
Unfortunately, the solubility and bioavailability of curcumin in water is low thus hindering the potential of curcumin to improve the effectiveness of cancer treatment. Moreover, it has low bioavailability due to its rapid metabolism in the liver and intestinal wall. Numerous approaches for increasing its bioavailability have been explored, including the use of adjuvants such as piperine [12].
a-Lactalbumin (a-LA) is a protein present in the milk of all mammals. Its molecular weight is 14.2 kDa and its isoelectric point is between 4.2 and 4.5 [16]. Under native conditions, the tertiary structure of a-LA is composed of a large domain (α) and a small domain (β) divided by a cleft. It comprises eight cysteines which form four disulfide bridges [17] and showed that the a-LA molecule contains one strong binding site for Ca2+ and a weaker binding site for Zn2+.
Although a-LA is a major calcium-binding protein in milk, t sequence comparison shows a strong similarity to lysozymes, specifically the Ca2+-binding c-lysozyme [18]. a-LA forms the regulatory subunit of the lactose synthase (LS) heterodimer and β-1,4-galactosyltransferase (beta4Gal-T1) forms the catalytic component. Together, these proteins enable LS to produce lactose by transferring galactose moieties to glucose. a-LA strongly binds calcium ions and may possess a bactericidal or antitumor activity which can make it eligible for important applications in clinical and veterinary medicine, as well as functional food products [19]. Proteolytic digestion of a-LA produces three peptides with bactericidal properties that inhibit the growth of Gram-positive bacteria.
Multimeric a-LA was shown to bind to the cell surface, enter the cytoplasm, and accumulate in cell nuclei, which is consistent with its ability to induce apoptosis. This multimeric form appears to exist in milk [20]. It has been reported that a-LA has many physiological functions, such as reduction of stress, opioid activity, antihypertensive action, regulation of cell growth and immunomodulation [21,22,23].
Whey proteins are used as food ingredients due to their important functional properties. In addition, RES and CUR have many clinical properties, so we can enrich milk with these ingredients to improve such characteristics. Since a-LA constitutes a large part of whey, this study on the interaction of RES with a-LA in the presence of CUR should prove helpful determining. The effects of ligands on a-LA were estimated and the binding constant and binding parameters, the number of binding sites and protein conformational changes of ligand-LA complexes were determined.
The techniques used included UV–Vis absorption, fluorescence quenching, and circular dichroism spectroscopies, as well as zeta potential analysis. Thanks to these methods, the interactions between small molecules and a-LA were studied.
Materials and methods
Materials and solutions
Curcumin, resveratrol, a-LA and potassium phosphate were all purchased from Sigma-Aldrich (St. Louis, MO, USA) and used without further purification. A stock solution (0.01%) of a-LA was prepared by dissolving an appropriate amount of solid a-LA into potassium phosphate buffer (pH 7.4). The protein solution (7.04 × 10−4 mM) was freshly prepared in potassium phosphate buffer (50 mM, pH 7.4).
The resveratrol solution (0.5 mM) was prepared by dissolution in phosphate buffer and the curcumin solution (0.1 mM) was prepared by dissolution in DMSO. All solutions were stored in a refrigerator at 4 °C in the dark.
Apparatus
Absorption measurements
Absorption measurements were carried out with a Jasco spectrophotometer (V-630 model) equipped with a personal computer and 1.0-cm quartz cells. The optical system was based on a split-beam with a grating bandwidth of 5 nm, and the light source was a Xenon lamp. The absorption measurements of all samples were carried out using quartz cells with a 1-cm optical path. The slit width was set to 5 nm, and the wavelength range was 200–500 nm. All measurements were performed at room temperature.
Fluorescence measurements
All fluorescence spectra were recorded on an F-2500 fluorescence spectrophotometer (Hitachi Japan) equipped with a xenon lamp light source, and 1.0-cm quartz cells were used for the measurements. The excitation wavelengths were set to 280 nm and 295 nm, and the emission wavelength was recorded between 300 and 600 nm. Fluorescence intensities were corrected for inner filter and dilution effects before analysis of the binding and quenching data. The emission spectra of the free a-LA and a-LA–ligand mixtures were measured using the buffer solutions of the drugs with the corresponding concentrations as reference. Measurements were performed for each studied ligand using the a-LA solution with a single ligand (RES or CUR) at various concentrations and with the two ligands, which were added simultaneously to the a-LA solution.
RLS and synchronous fluorescence spectroscopy
Synchronous fluorescence spectroscopy was carried out by simultaneously scanning the excitation and emission monochromators. The spectra only showed the Tyr and Trp residues of a-LA when the wavelength interval (Δλ) was 15 nm and 60 nm, respectively, and the RLS spectra were recorded by simultaneously scanning the excitation and emission spectra from 280 to 600 nm with Δλ = 0 nm and a slit width of 5.0 nm.
Circular dichroism spectroscopy
Far-UV CD experiments were performed on a Jasco-815 spectropolarimeter equipped with a Jasco 2-syringe titrator using a quartz cell with a path length of 1 mm. The results were expressed as the mean residue ellipticity (MRE) in deg cm2 dmol−1, which is defined as [θobs (mdeg)/10 nlCp]. The θobs represents the CD in milli-degree, n is the number of amino acid residues, and l and Cp are the path length of the cell and the mole fraction, respectively. The alpha-helical content of a-LA was calculated from the MRE value at 208 nm using the equation α% helix = [MRE 208–4000/33,000–4000] × 100. In this case, MRE 208 is the observed MRE value at 208 nm, 4000 is the MRE where the form and random coil conformations cross at 208 nm, and 33,000 is the MRE value of the α-helix at 208 nm. To explore changes in the secondary structure, far-UV CD spectra were obtained over a wavelength range of 190–240 nm in the absence and presence of different concentrations of CUR and RES at the same conditions as described above for the a-LA solution.
Zeta potential
The zeta potential was determined by laser Doppler electrophoresis. The zeta potential measurements were performed on a Zeta Sizer Nano series-ZS (Malvern Instrument, UK) at 298 K using drugs and protein. The initial volume of a-LA was 2000 µl into which different injection volumes of the stock drug solution were added. After each injection, the solution was stirred for 4 min with a magnetic stirrer prior to the measurements.
Three-dimensional fluorescence spectra
Three-dimensional fluorescence spectra were performed on an FP-2600 fluorescence spectrophotometer (Jasco, Japan) under emission and excitation wavelengths set between 220 and 500 nm with increments of 5 nm. The other scanning parameters were identical to those of the fluorescence quenching spectra.
Molecular modeling
The three-dimensional coordinates of a-LA were obtained from the Protein Data Bank (PDB entry Code 1F6S). The structure of RES and CUR was designed and energy minimized using MM method in MOE 2015.10®. a-LA, CUR and RES were prepared (minor correction of structure such as charge and angles) and protonated with MOE 2015.10®. To determine the interaction of the protein with the ligands, CUR and RES were separately loaded in MOE panel while a-LA has been already loaded in MOE. Systems were protonated in T: 300, pH: 7, salt: 0.1 while the electrostatics functional form was set as GB/VI. Other protonation parameters were left as default. Docking procedure was executed with a-LA–CUR and a-LA–RES using the following parameters. Replacement method: triangle matcher; force filed: London DG; and 30 output were requested after refinement. For the tertiary system, we minimized docked CUR–a-LA complex and saved the file as PDB. The resulted file was transferred to GROMACS 5.1, 2016 package for further molecular dynamic simulation (MD). The time of MD was set for 20 ns while the Amber force filed was selected and temperature was raised to 350 K. The last frame of GROMACS trajectory was saved and the PDB file was transferred to MOE software. Then RES was loaded as the tired molecule and docked against the CUR–a-LA complex using the same parameters as above.
General procedure
a-LA and RES were dissolved in a potassium phosphate buffer (50 mM, pH 7.4) and CUR was dissolved in a DMSO solution. The concentrations of CUR and RES were 0.1 mM and 0.5 mM, respectively.
These RES and CUR concentrations were below usage doses. The usage doses of RES and CUR differ from one another. To a 1.0-cm quartz cell, a-LA solution was added to make-up 2 ml, and the range of the drug (RES, CUR) solutions was gradually titrated manually into the cell using a micro-injector for the binary and ternary systems. The reaction time was investigated and the results showed that 3 min was enough for the stabilization. The fluorescence spectra were measured (excitation wavelengths at 280, 295 and emission wavelengths of 300–600 nm) at room temperature. The binding of a drug to a-LA can be weakened or enhanced by other drugs.
The zeta potential measurement and UV–Vis absorbance spectra of a-LA, RES and CUR were recorded at 291 nm at room temperature. The far-UV CD spectra were obtained over a wavelength range of 240–190 nm in the absence and presence of different concentrations of CUR and RES at the same conditions as above.
Results and discussion
Fluorescence quenching of a-LA by RES in the absence and presence of CUR
The measurement of fluorescence signals provides a sensitive method for monitoring the biochemical environment of a fluorophore and can give us important information about the molecular level of binding of small molecular substances to the protein, such as the binding mechanism, binding constants, and intermolecular distances. Fluorescence spectral changes are the result of solvent relaxation of fluorescent amino acids, such as Trp and Tyr, and give us important information on protein structure and folding. The intensity, the position of the emission wavelength and the lifetime are some observables that are used to characterize a fluorophore. Trp fluorescence is the most sensitive to the local environment of the fluorophore. When a protein contains two classes of Trp residues, the resultant fluorescence emission spectrum will be the result of the contribution of each class.
a-LA has only one domain with four Trp and four Tyr residues. In proteins, when we excite at 280 nm, for example, we excite both Trp and Tyr residues. If we want to observe the emission from the Trp residues alone, the excitation should occur at and/or above 295 nm. At these wavelengths, the energy transfer from Trp and Tyr does not take place and the observed emission emanates from the Trp residues only. To determine which residue is located in or near the binding position, the excitation wavelength was selected to 280 and 295 nm.
A comparison of the fluorescence quenching of a protein excited at 280 and 295 nm renders it possible to estimate the participation of Trp and Tyr groups in the complex (Mauric, & Camillo, 1981).
Figure 1a, b shows the fluorescence spectra of, respectively, (a-LA–RES) and (a-LA–CUR) RES at the excitation wavelength 280 nm, (pH 7.4). It can be seen that a-LA had a strong fluorescence emission at 340 nm after being excited with wavelengths of 280 nm and 295 nm. The fluorescence emission intensity of a-LA decreased regularly with increasing concentration of CUR and RES. The strong fluorescence quenching indicated that the protein conformation may have become changed and that an inter-molecular energy transfer occurred between CUR, RES and a-LA [24].
a Fluorescence emission spectra of the a-LA–RES system at 280 nm. The concentration of a-LA was 0.04 mM and the RES concentration was increased from 0 to 8.3 × 10−2 mM; pH 7.4, 298 K. Inset: fluorescence emission spectra of the a-LA–RES system at 295 nm under similar conditions; b fluorescence emission spectra of the (a-LA–CUR) RES system. The CUR concentration was 4.7 × 10−3 mM and the concentration of RES was increased from 0 to 8 × 10−2 mM; pH 7.4, 298 K. Inset: fluorescence emission spectra of the (a-LA–CUR) RES system at 295 nm under similar conditions
Quenching is the decrease of the quantum yield of fluorescence from a fluorophore induced by a variety of molecular interactions with a quencher molecule [24]. A variety of processes may be responsible for the decrease in fluorescence intensity such as molecular rearrangement, energy transfer, collisional quenching and ground-state complex formation [25]. Quenching occurs through either a static or dynamic process. Static quenching is ascribed to the formation of a ground-state complex between protein and quencher, whereas dynamic quenching results from collision between a quencher and a fluorophore. Dynamic quenching thus refers to a process where the fluorophore and the quencher come into contact during the transient existence of the excited state. In general, dynamic and static quenching can be recognized by their different dependencies on temperature and viscosity [26].
To confirm the quenching mechanism, the procedure of the fluorescence quenching was first assumed to be dynamic quenching. The dynamic fluorescence quenching behavior could be analyzed using the Stern–Volmer equation which is expressed as
Here, F0 and F are the fluorescence intensities in the absence and presence of a quencher, and kq is the quenching rate constant of the biomolecule. The value of kq allows us to estimate the fluorophore availability for RES and CUR, τ0 is the lifetime of the fluorophore in the absence of a quencher and [Q] is the quencher concentration. KSV is the Stern–Volmer dynamic quenching constant and allows for the assessment of the affinity of RES and CUR to the a-LA.
The non-linearity of the Stern–Volmer plot may be interpreted in two ways. One possibility is that the dynamic quenching and static fluorescence quenching occur simultaneously. The other is that a non-linear binding isotherm involving a significant occupation of binding sites is present at the free quencher concentration [27].
Non-linear Stern–Volmer plots can also occur when fluorophores can be quenched by both collision and complex formation with the same quencher.
In this situation, the Stern–Volmer plots exhibited an upward curve, i.e., they were concave towards the y-axis at higher [Q] by the following modified form of the Stern–Volmer equation:
Here, KD and KS are the dynamic and static quenching constants, respectively. This equation is second order in [Q] and thus leads to an upward-facing curve in plots of F0/F versus [Q] at higher [Q], which arises from a combined quenching (both dynamic and static) process.
In a static quenching process, when small molecules are bound independently to a set of equivalent sites on a macromolecule, the equilibrium between free and bound molecules is given by the modified Stern–Volmer equation. Analysis of the modified Stern–Volmer equation allowed us to determine the quenching constants Ka and fa for the a-LA–drug complexes [28]. Here, fa is the fraction of accessible fluorescence of the protein and Ka is the effective quenching constant that can be determined by the slope of the (F0–F)−1 vs. [Q]−1 for the accessible fluorophores:
Figure 2a shows the Stern–Volmer plots for the a-LA fluorescence quenching by RES in the absence and presence of CUR at 280 nm and the calculated KSV and kq values are summarized in Table 1.
It can be seen that the plot of F0/F versus [Q] in the absence or presence of CUR gave a positive (concave) deviation, towards the y-axis at higher [Q]. This result may suggest that either an additional binding site existed or that combined quenching (static and dynamic) occurred at higher concentrations of RES. a-LA had two binding constants for RES in the binary and ternary systems.
As you can see in Fig. 2b at 295 nm for the binary system, the curve was linear within the studied concentration. This indicates that a single type of quenching, either static or dynamic [29], occurred. But in the ternary system, when we added RES to (a-LA–CUR), the Stern–Volmer plots exhibited an upward curvature, which may be attributed to two complicating factors: (1) the existence of more than one type of quenching mechanism at higher RES concentrations, or (2) attenuation of the Trp fluorescence due to an inner filter effect arising from significant absorption of RES in the Trp emission region with these quencher concentrations.
a-LA had one binding constant for RES: KSV = 1.29 × 104 L mol−1 (in the binary system and at 295 nm). Moreover, it had two binding constants for RES in the ternary system (a-LA–CUR) RES: KSV1 = 9.85 × 103, KSV2 = 2.43 × 104 L mol−1. These results indicate that, in the ternary system, RES had two different binding sites on the protein with different affinity. The obtained value of kq was much higher than the maximum dynamic quenching constant for various quenchers with biopolymers (2 × 1010 L mol−1 s−1), thus confirming that a static quenching was dominant in this situation [30].
Table 1 lists the KSV,fa and kq values for the binary and ternary systems. As can be seen, the presence of the drugs affected the values. When fa was equal to 1, all the Trp residues were accessible to the quencher. Consequently, a change in the value of fa signified that the fraction of fluorescent components accessible to the quencher became altered.
Synchronous fluorescence studies
Synchronous fluorescence spectra provide information about changes in the molecular microenvironment in the vicinity of functional fluorophores and are used to investigate the conformational changes of proteins [31]. When ∆λ between the excitation and emission wavelengths is stabilized at 15 or 60 nm, the synchronous fluorescence gives characteristic information of the Tyr or Trp residues in a-LA, respectively [32]. It was proved that the Trp residues presented a maximum emission wavelength at 332 nm, 342 nm and 352 nm when the Trp residue was, respectively, totally surrounded by hydrophobic media, partially surrounded by hydrophobic media and entirely exposed in the water phase. If the maximum emission wavelength (λmax) was 330–332 nm, the Trp residues were located in the non-polar region or were buried in the hydrophobic cavity. We can thus predict the conformational changes of the protein based on the changes in the synchronous fluorescence spectra.
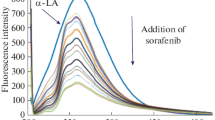
The effect of RES on the synchronous fluorescence spectra of a-LA in the absence and presence of CUR is shown in Fig. 3. The fluorescence intensity decreased regularly with the addition of RES in both figures, which further demonstrated the occurrence of fluorescence quenching in the binding process. The synchronous fluorescence changes in the polarities of the protein fluorophore environments did not occur when the concentration of the added RES was increased.
a Synchronous fluorescence spectra of a-LA–RES (open circle) and (a-LA–CUR) RES (filled circle) at ∆λ = 60. Inset: fluorescence spectra of a-LA–RES (open circle) and (a-LA–CUR) RES (filled circle) at ∆λ = 15; b a comparison of curves of F/F0 versus [Q] for a-LA–RES at ∆λ = 15 (filled circle) and ∆λ = 60 (open circle). Inset: a comparison of curves of F/F0 versus [Q] for the (a-LA–CUR) RES at ∆λ = 15 (filled circle) and ∆λ = 60 (open circle)
Figure 3a and the inset illustrate the slope of the plots of a-LA–RES and (a-LA–CUR) RES at Δλ = 60 and 15 nm, respectively. It can be seen that the first part of the a-LA–RES plot slope was higher than that of (a-LA–CUR) RES when Δλ = 60 nm. But when Δλ = 15 nm, the slope of the (a-LA–CUR) RES plot was not very different from the a-LA–RES plot which indicated that the conformation of a-LA and the distance between the RES and Trp residues had been altered.
In Fig. 3b, we can see that the slope of the a-LA–RES plot at Δλ = 60 was higher which demonstrated that the RES site was closer to Trp than to Tyr and that the Trp residues thus contributed significantly to the fluorescence of a-LA. In the ternary system, the slope of ∆λ = 15 and ∆λ = 60 was similar which means that the contributions of the Trp and Tyr residues in the ternary system did not differ markedly.
Three-dimensional fluorescence spectroscopy
Three-dimensional fluorescence spectroscopy is another powerful method for studying conformational changes of proteins [33]. The advantage of three-dimensional fluorescence spectroscopy is that the information regarding the fluorescence characteristics can be entirely acquired by simultaneously changing the excitation and emission wavelength [34]. The excitation wavelength, the emission wavelength and the fluorescence intensity can be used as the axes to investigate the synthetically generated information of the samples.
The three-dimensional fluorescence contour map of a-LA and a-LA–RES in the presence and absence of CUR are shown in Fig. 4a. Peak a (λex = λem) is the Rayleigh scattering peak, and peak b (λem = 2λex) is the second-order scattering peak. Peak 1 (λex = 275.0 nm, λem = 335 nm) is the fluorescence peak and mainly enunciates the spectral behavior of the Tyr and Trp residues, whereas the Phe residue fluorescence is negligible. Peak 2 (λex = 230 nm, λem = 345 nm) has the characteristic polypeptide backbone structure of a-LA (Ding et al. [32]), which was mainly caused by the π–π* transition of a-LA’s characteristic polypeptide backbone structure C=O.
The fluorescence intensity of peak a and peak b increased with the addition of a ligand. A possible reason is that an a-LA–RES complex came into being after the addition of RES (see Fig. 4b). Upon addition of CUR, the fluorescent intensity of peak 2 was omitted which indicated that the interaction between CUR and a-LA resulted in the polypeptides of a-LA becoming more extended and the conformation changing (see Fig. 4b).
Energy transfer from a-LA to the drugs
FRET is a nonradiative process where the energy is transferred between two fluorophores. FRET requires that the emission spectrum of one fluorophore (the donor) overlaps the absorption spectrum of a second fluorophore (the acceptor). This method can monitor the proximity and relative angular orientation of fluorophores and the distance between the two fluorophores can be quantified by spectrally resolving the relative fluorescence intensities of the donor and the acceptor.
The overlap of the UV absorption spectrum of the ligand molecule with the fluorescence emission spectrum of a-LA was used to evaluate the energy transfer. Its efficiency depends on (1) the extent of overlap between the donor emission and the acceptor absorption spectrum, (2) the orientation of the transition dipole of the donor and the acceptor, and (3) the distance between the donor and the acceptor [35]. Here, the donor and acceptor are a-LA and RES, respectively. The overlap of the absorption spectrum of RES with the fluorescence emission spectrum of a-LA and the a-LA–CUR complex is shown in Fig. 5.
The efficiency of energy transfer between the donor and acceptor, E, could be calculated by the following equation:
where r is the binding distance between donor and acceptor, and R0 is the critical distance at 50% transfer efficiency, and F0 and F are the fluorescence intensities of the donor in, respectively, the absence and presence of equal amount of acceptor. R0 is calculated by the following equation:
where K2 is the spatial orientation factor of the dipole that can range from 0 to 4. The extreme values require very rigid orientations; if both the donor and acceptor are tumbling rapidly and are free to assume any orientation, then K2 equals 2/3. Moreover, n is the refractive index of the medium in the wavelength range where spectral overlap is significant, ϕ is the fluorescence quantum yield of the donor and J is the overlap integral of the fluorescence emission spectrum of the donor and the absorption spectrum of the acceptor. J is given by
where F(λ) is the corrected fluorescence intensity of the donor in the wavelength range λ–λ + Δλ, and ε (λ) is the extinction coefficient of the acceptor when the wavelength is λ.
The energy transfer should most probably take place when the average distance between a donor fluorophore and an acceptor fluorophore is less than 10 nm [36]. The overlap of the absorption spectrum of RES with the fluorescence emission spectrum of a-LA is shown in Fig. 5a. The distance between RES and Trp residues of a-LA in the absence and presence of CUR was 2.67 and 2.78 nm, respectively (Fig. 5b). Obviously, the distances were lower than 7 nm when interaction between a-LA and RES occurred and also revealed the presence of a static quenching mechanism. We can see that the distance between the drugs and a-LA in the binary systems was shorter than in their ternary counterpart.
The overlap integral J was calculated to be 1.22 × 10−14 (cm3 l)/mol for a-LA–RES and 1.35 × 10−14 for (a-LA–CUR) RES.
Circular dichroism and conformational analysis
To determine the effect of RES on the secondary structure of a-LA, we used far-UV CD spectroscopy in aqueous solution. Far-UV CD spectra (below 250 nm) of proteins are extremely sensitive toward protein structure and have been used to show that the secondary structures of the molten globules resemble those of the corresponding native proteins [37].The CD spectrum of free a-LA exhibited two negative bands in the UV region, at 209 and 222 nm, both contributing to the n → p* transfer for the peptide bond of α-helixes, and revealed a high content of α-helix [38].
The helical content of free and bound a-LA was calculated from the mean residue ellipticity (MRE) values at 208 nm.
where CP is the molar concentration of the protein, n is the number of a-LA amino acid residues and l is the path length of the cell (in cm):
where MR 208 is the observed MRE value at 208 nm, 4000 is the MRE of a-form and random coil conformation cross at 208 nm and 33,000 is the MRE value of a pure a-helix at 208 nm.
Conformational transitions of a-LA induced by RES were analyzed by far-UV CD spectroscopy at varying drug concentrations.
As can been seen from Fig. 6a, the values of negatives bands at [θ]208 and [θ]222 decreased, which suggests the loss of α-helical content in a-LA, demonstrating that RES interacts with amino acid residues of the main polypeptide chain of the protein and destabilizes their hydrogen bonding network making the proteins adopt a less compact conformational state [39]. Table 2 shows the percentage of α-helix, β-sheet, turn and unordered coil for a-LA as induced by RES in binary and ternary systems.
A decrease in the tendency for the alpha-helix and beta-sheet contents, and an increase in the tendency for unordered structure contents were observed with an enhancement of RES to the a-LA–RES complex. In the ternary system, the induction of conformational changes was more significant than in the binary ones (Fig. 6b). These results suggested that the a-LA molecule adopted a looser conformation with the extended polypeptide structure.
Red edge excitation shift (REES)
By changing the excitation wavelength towards the red edge of the absorption band, a shift normally occurs in the emission wavelength. The REES occurs between the Trp indole rings and the neighboring dipoles as a result of electronic binding. This excitation shift (REES) of fluorescence can be used as a parameter for studying the photophysical and -chemical properties of proteins and enables us to compare the environment and mobility of the Trp residues in biomolecules [40].
Red shifts in the fluorescence emission spectrum of a fluorophore occur because long wavelength excitation results in photoselection of those fluorophores that interact strongly with the polar solvent molecules and give rise to slow rates of solvent relaxation around a fluorophore in an excited state. This depends on the motional restriction imposed on the solvent molecules in the immediate vicinity of the fluorophore [40].
In this study, we investigated the effect of REES upon excitation of Trp in a-LA, a-LA–RES and (a-LA–CUR) RES complexes at both 295 and 305 nm. The results are listed in Table 3.
The value of REES for a-LA was 3 while in the presence of RES, there was no difference in REES value which demonstrated a maintained rigidity of the environment of the Trp residue. When comparing the ternary and binary systems, it was found that the REES value for the binary system in the presence of CUR remained unchanged.
Resonance light scattering spectroscopy
During recent years, light scattering has been widely applied to study the aggregation, size, shape, and microscopic amounts of biomolecules, metal ions, and drugs as well as the distribution of particles in solution [41].
Figure 7a, b shows the RLS spectra of a-LA–RES and (a-LA–CUR) RES and demonstrates a sharp and maximum peak at about 306 nm and a wide band centered on 372 nm. The RLS intensity increased with increasing RES concentration, indicating that an interaction had occurred between RES and a-LA. It was thus concluded that larger particles were produced and resulted in the enhancement of RLS signals.
a RLS spectra of a-LA in the presence of increasing concentrations of RES from 0 to 4.3 × 10−2 mM in binary system. b RLS spectra of a-LA in the presence of increasing concentrations of RES in the ternary system. c The overlap of the ∆IRLS curves versus [RES] for a-LA–RES (open circle), (a-LA–CUR) RES (filled circle) systems and zeta potential for LA–RES (open circle), (a-LA–CUR) RES (filled circle) systems. Conditions: T = 298 K, pH 7.4, C (CUR) = 4.7 × 10−3 mM
Figure 7c displays the overlap of ∆IRLS versus the ligand concentration represented as ∆IRLS = IRLS − I0RLS (where IRLS and I0RLS are the RLS intensities of the systems with and without ligands, respectively) [42] and the zeta potential of the binary and ternary systems.
As can be seen in Fig. 7c, when the RES concentration reached about 6.1 × 10−3 mM, RES aggregated on the a-LA, and this value thus corresponded to the critical induced aggregation concentration (CCIAC) of the drug [43]. The CCIAC values for RES in the binary and ternary systems were found to be approximately 6.1 × 10−3 mM and 1.2 × 10−2 mM, respectively. Moreover, the comparison between binary and ternary systems indicated that the start of aggregate formation of RES on a-LA in the presence of CUR occurred at a higher concentration. This meant that CUR caused a change in the behavior of interaction between RES and a-LA in the ternary system.
The zeta potential, which is related to surface charge, is one of the parameters used in predicting the stability of protein-based preparations. The size of a protein is an important physical characteristic that provides useful information including the presence of monomers, dimers and trimers, changes in conformation, aggregation state, and denaturation. Much interest is also devoted to the surface charge or valence of proteins, and so the zeta potential is another measurement used by protein chemists. The most common particle size analysis technique for protein characterization is dynamic light scattering. The zeta potential can be measured using either electrophoretic light scattering or electro acoustic spectroscopy.
The zeta potential value of a-LA in the absence of RES was − 2.07 mV. As the RES concentration increased, the initial negative charge of the protein became reduced, which suggests the formation of a-LA–RES complexes. This reduction demonstrates that the initial interaction for a-LA and RES was mainly electrostatic. When the concentration of RES was increased to 1.3 × 10−2 mM, the zeta potential value became more negative. This suggests that the a-LA net charge was nearly neutralized by the formation of precipitation of RES on a-LA. This could be the result of a possible expansion of the protein structure induced by high drug concentrations and the presence of hydrophobic interactions in this process.
In Fig. 7c, we can see the critical induced aggregation concentration for RES in the binary and ternary systems overlapping with the most positive zeta potential value, which both point at the concentration of RES up to which RES aggregates on a-LA.
Molecular modeling
The experimental observations were followed by docking studies where RES was docked to a-LA and in both the binary and ternary systems to determine the changes in binding affinity, binding site of RES, etc., in the presence of CUR. a-LA secondary structure is made of nine helices and three anti-parallel beta strands while four disulfide bridges fix the protein in its tertiary structure (Fig. 8a). As a first step, CUR was docked to the protein after which RES was added. The best docking results of the interaction between RES and a-LA in the binary and ternary systems are shown in Fig. 8b.
a a-LA secondary structure. Nine helices and three anti-parallel beta strands make the protein. Four disulfide bridges fix the protein; b the best docking results of the interaction between RES and a-LA in the binary and ternary systems. Green molecule: RES in binary system. Red molecule: RES in ternary system. Brown molecule: CUR molecule in binary and ternary system. CUR is located near helix 3, RES is located between beta 3 and helix 7, and 8 in both binary and ternary systems. RES orientation is vise versa in binary and ternary systems; c three mail interaction sites of CUR and a-LA; d site view of a-LA and CUR surrounded by mostly polar residues. Pi–H interaction with Glu11 is seen by dashed line; e five sites of interaction of a-LA and RES in binary system. Site-1 has the most population of interaction but site-2 has the most favorable interaction with the lowest binding energy. None of them can be located in CUR (green) site; f two main interaction sites of a-LA and RES in the presence of CUR (ternary system). They are mainly located near RES (site-1) where RES (cyan) interacts in its binary system. None of RES interacts with CUR (green) site in ternary system; g the interaction of RES in binary system. Dashed arrow shows H-bound. Polar residues are in pink and non-polar residues are in green circles; h the distances of RES with Trp in ternary systems. Unit is as angstrom
Thirty results of docking of CUR and a-LA show mainly three sites (80% of them in site-1) of interaction between the protein and the ligand that is shown in Fig. 8c. We selected the most stable ligand of site-1 for further simulations. CUR is located near helix 3 and has a Pi–H interaction with Glu11 while Polar residues such as Lys122, Cys120, Glu121, Cys6, Arg10, Glu11 and Lys13 surround it (Fig. 8d).
Twenty-three results of docking of RES and a-LA in binary system show five sites (80% of them in site-1) of interaction between the protein and the ligand that is shown in Fig. 8e. The best binding energy for RES in binary system is − 5.06 Kcal/mol. It has one H-bound between a phenol ring and Thr33 while a mostly hydrophilic pocket surrounds the ligand (Fig. 8g).
The superimposition of normal a-LA and molecular dynamic simulated a-LA (after docking with CUR) shows RMSD 1.4 Å that is negligible. Almost all secondary structures remain conserved and some loops show variation in their situations.
Twenty-five results of docking of RES and a-LA in ternary system show two sites (80% of them in site-1) of interaction between the protein and the ligand that is shown in Fig. 8f. RES does not favor CUR site in binary and ternary systems. In fact, the presence of CUR does not affect the interaction site of RES even though the conformation of a-LA slightly changes in the presence of CUR. The interaction binding energy of RES in the binary and ternary system is − 5.06 and − 4.79 Kcal/mol, respectively; that means CUR decreases the affinity of RES to a-LA. The main difference of interaction of RES in binary and ternary system is the orientation of the ligand. Even though the site of interaction does not change, the ligand totally opposes in ternary system (Fig. 8b). The property of the binding site does not change but because of losing the H-bound of phenol ring with Thr33 its binding energy decreases. Although each drug was located in a different position and the interaction was non-competitive, they affected each other’s affinity. The distances between the Trp residues such as Trp 26, Trp 60, Trp 104 and Trp 118 of a-LA and RES in ternary complex are shown in Fig. 8h. Our theoretical data were supported by experimental results such as far-UV CD.
As the efficient energy transfer from Trp to the ligands was 2–8 nm, the distance of the Trp residues from the docked ligands was also measured. This was applicable with the high efficient fluorescence quenching of the a-LA emission in the presence of the second drug.
Conclusions
This manuscript describes the exploration of the binding of RES and CUR to a-LA by different spectroscopic and molecular dynamic techniques. The changes of the protein conformation upon binding were determined by fluorescence and CD spectroscopies. It was shown that the fluorescence of a-LA became quenched when reacting with RES in the absence and presence of CUR. The binding constant and the number of binding sites were calculated according to relevant fluorescence data. The fluorescence quenching mechanism for a-LA through RES binding in the absence and presence of CUR was mainly a static quenching process. Changes in the environment of the aromatic residues were also observed by synchronous fluorescence and far-UV CD. According to Förster’s non-radioactive resonance energy transfer theory, energy transfer from a-LA to RES in binary and ternary systems occurred with high probability. The results obtained from three-dimensional fluorescence spectra showed that the structure of the a-LA molecules changed remarkably in the presence of RES in binary and ternary systems. The CCIAC values of the drugs on a-LA in binary and ternary systems were determined by RLS. The results showed that the CCIAC values increased in the ternary complex. On the other hand, hydrophobic interactions played important roles in inducing aggregation concentrations of drugs as determined by the zeta potential technique. These results should be of help to determine optimal drug doses and prevent irreversible structural alterations in protein molecules which can lead to a diminution of their biological functions. Thanks to theoretical docking experiments, the binding site of RES on a-LA was determined in the binary and ternary systems.
References
B. Burstein, A. Maguy, R. Clément, H. Gosselin, F. Poulin, N. Ethier, J.-C. Tardif, T.E. Hébert, A. Calderone, S. Nattel, Effects of resveratrol (trans-3, 5, 4′-trihydroxystilbene) treatment on cardiac remodeling following myocardial infarction. J. Pharmacol. Exp. Ther. 323(3), 916–923 (2007)
M. Okawara, H. Katsuki, E. Kurimoto, H. Shibata, T. Kume, A. Akaike, Resveratrol protects dopaminergic neurons in midbrain slice culture from multiple insults. Biochem Pharmacol 73(4), 550–560 (2007)
S. Pervaiz, A.L. Holme, Resveratrol: its biologic targets and functional activity. Antioxid. Redox Signal. 11(11), 2851–2897 (2009)
F.Z. Marques, M.A. Markus, B.J. Morris, Resveratrol: cellular actions of a potent natural chemical that confers a diversity of health benefits. Int. J. Biochem. Cell Biol. 41(11), 2125–2128 (2009)
M. Mutoh, M. Takahashi, K. Fukuda, Y. Matsushima-Hibiya, H. Mutoh, T. Sugimura, K. Wakabayashi, Suppression of cyclooxygenase-2 promoter-dependent transcriptional activity in colon cancer cells by chemopreventive agents with a resorcin-type structure. Carcinogenesis 21(5), 959–963 (2000)
C.R. Pace-Asciak, O. Rounova, S.E. Hahn, E.P. Diamandis, D.M. Goldberg, Wines and grape juices as modulators of platelet aggregation in healthy human subjects. Clin. Chim. Acta 246(1–2), 163–182 (1996)
J.L. Kipp, V.D. Ramirez, Effect of estradiol, diethylstilbestrol, and resveratrol, on F0F1-ATPase activity from mitochondrial preparations of rat heart, liver, and brain. Endocr. Basingstoke 15(2), 165–176 (2001)
P.S. Ray, G. Maulik, G.A. Cordis, A.A. Bertelli, A. Bertelli, D.K. Das, The red wine antioxidant resveratrol protects isolated rat hearts from ischemia reperfusion injury. Free Radic. Biol. Med. 27(1), 160–169 (1999)
H.P. Ammon, M.A. Wahl, Pharmacology of Curcuma longa. Planta Medica 57(01), 1–7 (1991)
S. Barthelemy, L. Vergnes, M. Moynier, D. Guyot, S. Labidalle, E. Bahraoui, Curcumin and curcumin derivatives inhibit Tat-mediated transactivation of type 1 human immunodeficiency virus long terminal repeat. Res. Virol. 149(1), 43–52 (1998)
M. Ramırez-Tortosa, M. Mesa, M. Aguilera, J. Quiles, L. Baro, C. Ramirez-Tortosa, E. Martinez-Victoria, A. Gil, Oral administration of a turmeric extract inhibits LDL oxidation and has hypocholesterolemic effects in rabbits with experimental atherosclerosis. Atherosclerosis 147(2), 371–378 (1999)
P. Anand, C. Sundaram, S. Jhurani, A.B. Kunnumakkara, B.B. Aggarwal, Curcumin and cancer: an “old-age” disease with an “age-old” solution. Cancer Lett. 267(1), 133–164 (2008)
H. Hatcher, R. Planalp, J. Cho, F. Torti, S. Torti, Curcumin: from ancient medicine to current clinical trials. Cell. Mol. Life Sci. 65(11), 1631–1652 (2008)
A. Khar, A.M. Ali, B. Pardhasaradhi, Z. Begum, R. Anjum, Antitumor activity of curcumin is mediated through the induction of apoptosis in AK-5 tumor cells. FEBS Lett. 445(1), 165–168 (1999)
H. Maier, J. Ilich, J. Kim, M. Spicer, Nutrition supplementation for diabetic wound healing: a systematic review of current literature. Skinmed 11(4), 217–224 (2012) (quiz 224–215)
A. Belatik, C. Kanakis, S. Hotchandani, P. Tarantilis, M. Polissiou, H. Tajmir-Riahi, Locating the binding sites of retinol and retinoic acid with milk β-lactoglobulin. J. Biomol. Struct. Dyn. 30(4), 437–447 (2012)
R.J. FitzGerald, B.A. Murray, D.J. Walsh, Hypotensive peptides from milk proteins. J. Nutr. 134(4), 980S–988S (2004)
K. Acharya, D. Stuart, N. Walker, M. Lewis, D. Phillips, Refined structure of baboon α-lactalbumin at 1.7 Å resolution: comparison with C-type lysozyme. J. Mol. Biol. 208(1), 99–127 (1989)
B.S. Horton, Commercial utilization of minor milk components in the health and food industries. J. Dairy Sci. 78(11), 2584–2589 (1995)
M. Svensson, H. Sabharwal, A. Håkansson, A.-K. Mossberg, P. Lipniunas, H. Leffler, C. Svanborg, S. Linse, Molecular characterization of α-lactalbumin folding variants that induce apoptosis in tumor cells. J. Biol. Chem. 274(10), 6388–6396 (1999)
C.R. Markus, B. Olivier, G.E. Panhuysen, J. Van der Gugten, M.S. Alles, A. Tuiten, H.G. Westenberg, D. Fekkes, H.F. Koppeschaar, E.E. de Haan, The bovine protein α-lactalbumin increases the plasma ratio of tryptophan to the other large neutral amino acids, and in vulnerable subjects raises brain serotonin activity, reduces cortisol concentration, and improves mood under stress. Am. J. Clin. Nutr. 71(6), 1536–1544 (2000)
H. Teschemacher, G. Koch, V. Brantl, Milk protein-derived opioid receptor ligands. Pept. Sci. 43(2), 99–117 (1997)
L.G. Sternhagen, J.C. Allen, Growth rates of a human colon adenocarcinoma cell line are regulated by the milk protein alpha-lactalbumin, in Bioactive Components of Human Milk, ed. by D.S. Newburg (Springer, Boston, 2001), pp. 115–120
Z. Omidvar, K. Parivar, H. Sanee, Z. Amiri-Tehranizadeh, A. Baratian, M.R. Saberi, A. Asoodeh, J. Chamani, Investigations with spectroscopy, zeta potential and molecular modeling of the non-cooperative behaviour between cyclophosphamide hydrochloride and aspirin upon interaction with human serum albumin: binary and ternary systems from the view point of multi-drug therapy. J. Biomol. Struct. Dyn. 29(1), 181–206 (2011)
J.R. Lakowicz, Plasmonics in biology and plasmon-controlled fluorescence. Plasmonics 1(1), 5–33 (2006)
Z.-D. Qi, B. Zhou, X. Qi, S. Chuan, Y. Liu, J. Dai, Interaction of rofecoxib with human serum albumin: determination of binding constants and the binding site by spectroscopic methods. J. Photochem. Photobiol., A 193(2), 81–88 (2008)
M. Borisover, Y. Laor, N. Bukhanovsky, I. Saadi, Fluorescence-based evidence for adsorptive binding of pyrene to effluent dissolved organic matter. Chemosphere 65(11), 1925–1934 (2006)
J. Równicka-Zubik, A. Sułkowska, J. Pożycka, K. Gaździcka, B. Bojko, M. Maciążek-Jurczyk, W. Sułkowski, Fluorescence analysis of sulfasalazine bound to defatted serum albumin in the presence of denaturating factors. J. Mol. Struct. 924, 371–377 (2009)
M. Ranjan, P. Diffley, G. Stephen, D. Price, T.J. Walton, R.P. Newton, Comparative study of human steroid 5α-reductase isoforms in prostate and female breast skin tissues: sensitivity to inhibition by finasteride and epristeride. Life Sci. 71(2), 115–126 (2002)
T. Wang, B. Xiang, Y. Wang, C. Chen, Y. Dong, H. Fang, M. Wang, Spectroscopic investigation on the binding of bioactive pyridazinone derivative to human serum albumin and molecular modeling. Colloids Surf. B 65(1), 113–119 (2008)
J. Lloyd, Synchronized excitation of fluorescence emission spectra. Nature 231(20), 64–65 (1971)
F. Ding, J. Huang, J. Lin, Z. Li, F. Liu, Z. Jiang, Y. Sun, A study of the binding of CI Mordant Red 3 with bovine serum albumin using fluorescence spectroscopy. Dyes Pigm. 82(1), 65–70 (2009)
H.-M. Zhang, Y.-Q. Wang, M.-L. Jiang, A fluorimetric study of the interaction of CI solvent red 24 with haemoglobin. Dyes Pigm. 82(2), 156–163 (2009)
M. Rodrıguez-Cuesta, R. Boqué, F. Rius, D.P. Zamora, M.M. Galera, A.G. Frenich, Determination of carbendazim, fuberidazole and thiabendazole by three-dimensional excitation–emission matrix fluorescence and parallel factor analysis. Anal. Chim. Acta 491(1), 47–56 (2003)
L. Stryer, Fluorescence energy transfer as a spectroscopic ruler. Ann. Rev. Biochem. 47(1), 819–846 (1978)
F.-Y. Wu, L.-N. Zhang, Z.-J. Ji, X.-F. Wan, Spectroscopic investigation of the interaction between thiourea-zinc complex and serum albumin. J. Lumin. 130(7), 1280–1284 (2010)
G.D. Fasman, Circular dichroism and the conformational analysis of biomolecules (Springer, Berlin, 2013)
P. Yang, F. Gao, The principle of bioinorganic chemistry. Science, Beijing 349 (2002)
J. Kang, Y. Liu, M.-X. Xie, S. Li, M. Jiang, Y.-D. Wang, Interactions of human serum albumin with chlorogenic acid and ferulic acid. Biochim. Biophys. Acta (BBA) Gen. Subj. 1674(2), 205–214 (2004)
M.C. Tory, A.R. Merrill, Determination of membrane protein topology by red-edge excitation shift analysis: application to the membrane-bound colicin E1 channel peptide. Biochim. Biophys. Acta (BBA) Biomembr. 1564(2), 435–448 (2002)
D. Gao, Y. Tian, F. Liang, D. Jin, Y. Chen, H. Zhang, A. Yu, Investigation on the pH-dependent binding of Eosin Y and bovine serum albumin by spectral methods. J. Lumin. 127(2), 515–522 (2007)
Z. Chen, J. Liu, Y. Han, Rapid and sensitive determination of proteins by enhanced resonance light scattering spectroscopy of sodium lauroyl glutamate. Talanta 71(3), 1246–1251 (2007)
X. Long, C. Zhang, J. Cheng, S. Bi, A novel method for study of the aggregation of protein induced by metal ion aluminum(III) using resonance Rayleigh scattering technique. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 69(1), 71–77 (2008)
Acknowledgements
The financial support of the Research Council of the Mashhad Branch, Islamic Azad University, is gratefully acknowledged. The authors thank Dr. Ljungberg for the English editing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jahanshahtalab, M., Kamshad, M., Rezaei, S. et al. New insights into alpha-lactalbumin behavior upon interaction with resveratrol and curcumin by spectroscopic and molecular modeling techniques: binary and ternary system comparison. J IRAN CHEM SOC 16, 1311–1326 (2019). https://doi.org/10.1007/s13738-019-01608-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13738-019-01608-3