Abstract
The type 2 interleukin-1 receptor (IL-1R2) is one of natural IL-1β singling inhibitors in mammals. We cloned and sequenced the IL-1R2 gene in V. variegatus (VvIL-1R2). The phylogenetic analysis showed that the molecular structure VvIL-1R2 is similar to that of its orthologues in other vertebrates. The expression levels of VvIL-1R2 are relatively high in the peripheral blood leukocytes (PBLs), gill, and spleen. In addition, peculiar expression patterns for his molecule were detected at various developmental stages, implying that in flatfishes the IL-1R2 may have be important for embryonic development and metamorphosis. In PBLs, the treatment with pathogen-associated molecular patterns (PAMPs) induced a significant and rapid up-regulation of VvIL-1R2, pointing at its involvement in the immune responses against bacterial and viral pathogens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Interleukin-1β (IL-1β), which is a member of the interleukin-1 cytokine family [1], plays a pivotal role in inflammatory responses and some related diseases. All of the known biological activities of IL-1β are mediated by its type 1 receptor (IL-1R1) [2]. In course of inflammation, IL-1β induces the expression of a large number of proinflammatory genes and proteins, therefore, enhancing inflammatory responses further. These genes and proteins include IL-1β itself, IL-6, IL-8, monocyte chemoattractant protein 1, and cyclooxygenase 2, which are usually not expressed in healthy body [3]. Due to associated damage to the tissue, excessive inflammatory cytokine production is harmful to the host [1, 2]. Accordingly, overexpression of IL-1β induces several natural IL-1β signaling inhibitors, in particular IL-1 receptor antagonist (IL-1Ra) and IL-1 receptor type 2 (IL-1R2) [4]. In mammals, IL-1Ra competes for IL-1R1 binding with IL-1β. Moreover, the recombinant human IL-1Ra analog, anakinra, is utilized to block the effect of IL-1β in patients [4]. IL-1R2, which lacks the Toll/interleukin-1 receptor (TIR) domain and, therefore, does not transduce signals, also competes with IL-1R1, thus serving as a decoy receptor for IL-1β [5].
IL-1R1 is expressed at low levels in nearly all cells [6, 7], whereas expression of IL-1R2 is predominant in neutrophils, B cells, and monocytes [8], pointing at the role of IL-1R2 as a naturally occurring inhibitor of IL-1β activity in mammals.
Among teleosts, IL-1R2 cDNA was first isolated from Oncorhynchus mykiss, where its inducible expression by LPS and TNF-α has been shown [9]. Subsequently, IL-1R2 sequences have been identified in Sparus aurata [10], Paralichthys olivaceus [11], Salmo salar [12] and Ctenopharyngodon idella [13]. In both S. aurata and P. olivaceus, a bacterial challenge markedly stimulates the IL-1R2 mRNA expression in immune-related tissues [10, 11], which also suggests the involvement of IL-1R2 in inflammatory responses. Importantly, the putative proteins of fish IL-1R2 also lack an intracellular TIR domain, implying the decoy function of fish IL-1R2.
Due to its high market value, Verasper variegatus has been widely recognized as a promising candidate for aquaculture and fishery enhancement in Asia. However, spontaneous maturation and ovulation of its broodfish have not yet been achieved in captivity [14]. Moreover, V. variegatus, which is an endangered flounder fish species, is susceptible to bacterial and virus infections. To improve the survival rate of V. variegatus in aquaculture stations, the function of immunity genes in this fish should be understood. For a better understanding of the function of VvIL-1R2, a full-length cDNA of this gene was reverse transcribed from the mRNA pool of the spleen and cloned, its expression levels were profiled in different adult tissues and at different developmental stages, including metamorphosis. In PBLs of V. variegatus, stimulated by pathogen-associated molecular patterns (PAMPs), the immune challenge responses were recorded.
EXPERIMENTAL
Collection of fish embryos and adult tissues. Fish samples of V. variegatus were obtained from a commercial hatchery in Weihai, Shandong Province, China. Adult tissues (heart, liver, spleen, kidney, brain, gill, muscle, and intestine) and PBLs were collected from five 1-year-old samples. The adult tissue and PBL samples were snap frozen in liquid nitrogen and then stored at –80°C until further use. Each of these samples was collected in triplicate. Embryos were collected from the same farm. Fertilized eggs were obtained by artificial fertilization and incubated at 11 ± 1°C in sterile seawater with an open recirculation water system and sufficient air supply. The different embryonic stages were observed under a stereomicroscope. Three pools of samples at different embryonic stages (i.e., unfertilized egg, 1-cell, 4-cell, morula, blastula, gastrula, neurula, somite, tail bud, and hatching stages) were separately collected from mixed families with a nylon net (100 mesh). The embryos were immersed in 1.5 mL of RNAwait liquid (Solarbio, Shanghai, China) overnight at 4°C and then stored at –80°C until further use.
RNA extraction and first-strand cDNA synthesis of VvIL-1R2. Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. The extracted total RNA was treated with RNase-free DNase I (TaKaRa, Dalian, China) to remove DNA contamination and then frozen at –80°C. Reverse transcription and cDNA synthesis were performed with 1 μg of total RNA and random hexamer primers using a reverse transcriptase M-MLV kit (TaKaRa) in accordance with the manufacturer’s protocol. The quality and quantity of the total RNA were evaluated by 1.5% agarose gel electrophoresis and spectrophotometry with the NanoPhotometer Pearl.
Molecular cloning and sequence analysis of VvIL-1R2. No genome information is available for V. variegatus. Thus, a pair of degenerate primers (Table 1) was designed based on the conserved sequences of IL-1R2 in other teleosts to find the conserved region of VvIL-1R2. PCR amplification was performed with an initial denaturation at 95°C for 5 min; 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and a final extension at 72°C for 7 min. The 5′- and 3′-rapid amplification of cDNA ends (RACE) were performed using the SMART RACE cDNA Amplification Kit (Clontech, CA, USA), in accordance with the manufacturer’s protocol, to isolate the full-length cDNA of IL-1R2 from the V. variegatus spleen samples. Gene-specific primers (GSPs) were designed based on the known cDNA sequence. The GSPs for the nested PCR assay were 5′RACE1 and 5′RACE2 for the 5′‑RACE (Table 1) and 3′RACE1 and 3′RACE2 for the 3′-RACE (Table 1). The PCR assay was conducted following the SMART RACE amplification method. The PCR products were separated by 1.5% agarose gel electrophoresis, purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research, CA, USA), cloned into the pMD-18T vector (TaKaRa), and then sequenced. The CDS region and open reading frame (ORF) were predicted by the ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
Quantitative real-time PCR of VvIL-1R2 in different tissues and at different developmental stages. A specific primer pair (IL-1R2-RT-FW/RV, Table 1) was designed based on the characteristics of IL-1R2. Pre-experiments were conducted to confirm the generation of single cDNA PCR products. The evaluation of the housekeeping genes of V. variegatus showed that β‑actin and 18S rRNA were the two most stable reference gene in different tissues and at different developmental stages. Thus, the relative expression of VvIL-1R2 was determined using both β-actin and 18S rRNA as the reference genes.
Total RNAs were extracted from different adult tissues and developmental stages, and cDNA synthesis was performed. Three biological replicates of each sample, running in triplicate, were analyzed. Quantitative real-time PCR (qRT-PCR) was performed in a 20 μL solution containing 10 ng of template cDNA and SYBR Premix Ex Taq II (TaKaRa) by using LightCycler 480 at 95°C for 5 min pre-incubation, followed by 45 cycles of 95°C for 15 s and 60°C for 45 s. Finally, the melting curve was analyzed to detect single amplification. Fluorescent signal accumulation was recorded at the 60°C 45 s phase during each cycle under the control of LightCycler 480 Software 1.5. The relative quantities of the target VvIL-1R2 expressed as fold variation over both β-actin and 18S rRNA were calculated using the 2−ΔΔCt comparative Ct method.
Sequence alignment and phylogenetic tree reconstruction. Homologous nucleotide and protein sequences were confirmed through a BLAST search against the NCBI and Ensembl databases. Multiple sequence alignments were conducted using ClustalX 2.1 and DNAMAN 7.0. A phylogenetic tree was constructed using MrBayes 3.2.3.
PAMPs-induced VvIL-1R2 expression in PBLs. Blood was collected from the caudal veins of the V. variegatus samples, and the PBLs were prepared with Percoll as previously reported [15]. V. variegatus PBLs were cultured at 24°C in a 24-well plate (Thermal Scientific, 3.6 × 104 cells/well) overnight and then stimulated with 50 μg/mL lipopolysaccharide (LPS), 50 μg/mL polyinosinic: polycytidylic acid (poly(I:C), Sigma), and PBS (control). The samples were obtained after 0, 0.5, 1, 2, 4, 6, 12, and 24 h of PAMPs administration. RNA isolation and cDNA synthesis were performed as described previously. The changes in VvIL-1R2 expression in response to LPS, poly(I:C), and PBS challenge were determined by qRT-PCR, with both β-actin and 18S rRNA as reference genes.
Protein structure analysis of VvIL-1R2. Pfam and CCD databases were used for searching the motifs of VvIL-1R2 and other vertebrate IL-1R2 sequences. The protein secondary and 3D structure of VvIL-1R2 were predicted using online prediction bioinformatics tools, such as Phyre2 (http://www.sbg.bio.ic.ac.uk/ phyre2/html/page.cgi?id=index) and PDBsum Generate (http://www.ebi.ac.uk/thornton-srv/databases/ pdbsum/Generate.html). Protein docking simulation was conducted with the ZDOCK server (http://zdock.umassmed.edu/).
Statistical analysis. qRT-PCR data were statistically analyzed using a one-way ANOVA followed by an LSD test using SPSS 20.0 (IBM, New York, USA). A difference of P < 0.05 between groups is considered statistically significant. The data are expressed as mean ± SD (n = 3).
RESULTS AND DISCUSSION
VvIL-1R2 Is Highly Conserved among Vertebrates
Given that no genome sequencing information for V. variegatus is available, we started with designing a pair of degenerate primers (Table 1) to the conserved region of VvIL-1R2 aftern aligning the sequences of IL-1R2 in other vertebrates. The 5′- and 3′-regions of the cDNA sequence of VvIL-1R2 were cloned using the RACE-PCR. Entire sequence of VvIL-1R2 cDNA was obtained using SeqMan by assembling all the cloned sequences (NCBI accession no. KY038172). The complete mRNA sequence of VvIL-1R2 consists of 1852 bp nucleotides with a 106 bp 5′-untranslated region (UTR), a 471 bp 3′-UTR including a poly(A) tail, and a 1275 bp ORF (Fig. 1). VvIL-1R2 has a complete polyadenylation (AATAAA) signal and TA-rich motifs (ATTTA) in the 3′-UTR (Fig. 1) [10, 11]. A previous research reported that the consensus sequence ATTTA is present in the 3′-UTR of both mouse and human IL-1 mRNAs, as well as the mRNAs encoding human and mouse TNF, human CSF, human lymphotoxin, human and rat fibronectin, and a majority of the sequenced human and mouse IFNs [16]. All these mRNAs lack homology to the IL-1R mRNAs in the coding region [16]. TA-rich motifs are particularly prevalent among mRNAs encoding proteins related to inflammatory responses [16]. The ATTTA instability motif is a characteristic feature of inflammatory mediator genes [17], and its presence suggests the transient expression of an inflammatory mediator for VvIL-1R2.
The mRNA structure of VvIL-1R2. Small letters stand for the UTR regions. Capital letters is used to indicate the CDS region. The predicted protein sequence is shown by single letter code of amino acids below the CDS region. Start and stop codons are in the frame and the stop codon is marked with the *. One instability motif (ATTTA) is underlined, two Ig-like domains are represented with wavy line, the transmembrane domain is noted with grey highlighting and polyadenylation signal (AATAAA) is represented with double line. The N-terminus signal peptide is marked with grey capital letters.
The ORF of VvIL-1R2 was predicted to encode a 424 amino acid residues long protein with a molecular weight of 47.38 kDa and a theoretical isoelectric point of 5.78. As in other vertebrates, a putative single transmembrane region with 23 amino acid residues (residue numbers 382–405, Fig. 1) is predicted at the C‑terminus by using Singer’s classification for membrane topology [18]. The transmembrane region separates the extracellular domain of 364 amino acids from the short intracellular domain of 19 amino acids. The predicted protein domains reveal a potential signal peptide, which comprises 18 amino acid residues at the N-terminus, a splicing site between amino acid residues 18 and 19 (VCG-KP), and a mature peptide of 406 amino acid residues (Fig. 1). The sequence identities of VvIL-1R2 with the IL-1R2 of P. olivaceus, Oreochromis niloticus, Cynoglossus semilaevis and Homo sapiens are 91, 80, 74, and 46%, respectively. These results imply that the IL-1R2 protein may be highly conserved among vertebrates.
Domain Structure of VvIL-1R2 Protein Is Consistent with that of the IL-1R2 Other Teleosts
The alignment of VvIL-1R2 with other vertebrate IL-1R2s shows that the translated VvIL-1R2 sequence possesses signature features of the IL-1R family of fish and higher vertebrate IL-1Rs. These features include two Ig-like domains in its extracellular region, one N-terminus signal peptide, one transmembrane domain, one short cytoplasmic tail of seventeen amino acids, four conserved proline residue sites, and six conserved cysteine residues [11]. The two Ig-like domains of VvIL-1R2 are observed in the extracellular region, but they lack an intracellular signaling Toll-interleukin-1 receptor (TIR) domain. The same set of features has been observed in S. salar, P. olivaceus, S. aurata and O. mykiss [9‒12]. Notably, human and mouse IL-1R2s also lack an intracellular signaling TIR domain, but they have three Ig-like domains in the extracellular region. Unlike the IL-1R2s of the compared species, the IL-1R1 of humans has a long intracellular signaling TIR domain and three Ig-like domains in the extracellular region. Thus, further studies should be conducted to confirm whether all teleost IL-1R2s possess two Ig-like domains. Moreover, in an extramembrane region, six cysteine residues are conserved at positions 112, 120, 156, 210, 290, and 358 in all of the species compared. However, among the six conserved cysteine residues, the last four residues are the backbone residues that form the interchain disulfide bonds of the Ig-like (C-x*-C) domains, and both domains form stable antiparallel β hairpin structures with disulfide bonds [19]. In addition, four proline residues at positions 101, 157, 267, and 272 are conserved in all the species examined.
Few studies have reported on the biological functions of IL-1R2. As IL-1R2 lacks the TIR domain, which is present in IL-1R1 and involved in signal transduction, it is believed to act as a decoy receptor for IL-1R1 [5]. However, the role of IL-1R2 in signal transduction cannot be discounted. A previous study showed that a monoclonal antibody against IL-1R2 could inhibit the thermogenic and pyrogenic responses to IL-1β released within the Rattus norvegicus brain [20]. The same antibody could weaken the IL-1β-induced prostaglandin E2 released from R. norvegicus hypothalamic explant [21]. These results suggested that IL-1R2 participated in the signal transduction through IL-1β in the brain and it was able to regulate the process. Moreover, the function of IL-1R2 in signal transduction is also confirmed in these studies [22, 23]. The mechanism for IL-1β signal transduction mediated by IL-1R2 may be similar to that proposed for IL-1R1. This mechanism may be accomplished by the binding of IL-1R2 to other transmembrane proteins [24]. The present study found that VvIL-1R2 possesses four conserved proline residues that are related to the signal transduction in IL-1R1. The signal-transducing capability of IL-1R1 significantly decreases when these conserved proline residues are mutated [25, 26]. We predict that the IL-1R2s in fishes probably have a signal-transducing function; however, further studies are required to confirm this hypothesis.
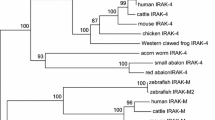
Analysis of MrBayes phylogenetic tree shows that IL-1R2 proteins of the vertebrates cluster into one group, which is separate from the H. sapiens IL-1R1 out-group (Fig. 2). This indicates that the ancestor gene of IL-1R has undergone gene duplication, leading to the formation of IL-1R1 and IL-1R2 before the species formation. The IL-1R2 encoding genes of the fishes cluster as a groip, and the IL-1R2 of V. variegatus, C. semilaevis, and P. olivaceus cluster into/group. This result implies that these flounder fishes have a close evolutionary distance from one another.
The protein sequence phylogenetic tree of IL-1Rs. Phylogenetic tree showing the relationship between V. variegatus IL-1R2 and other vertebrates proteins. A phylogram was constructed using MyBayes (mcmc = 200 000 generations, samplefreq = 10). The IL-1R1 sequence of H. sapiens served as the out-group. The species and GenBank accession numbers were as follows: H. sapiens IL-1R2 (NP_004624.1); M. musculus IL-1R2 (NP_034685.1); G. gallus IL-1R2 (XP_416914.3); X. tropicalis IL-1R2 (NP_001015713.1); P. olivaceus IL-1R2 (ABP99035.1); C. semilaevis IL-1R2 (XP_008326152.1); O. latipes IL-1R2 (XP_011487540.1); S. salar IL-1R2 (NP_001138892.1); X. maculatus IL-1R2 (XP_014329400.1); T. rubripes IL-1R2 (XP_003962211.1); O. niloticus IL-1R2 (XP_013132819.1); M. zebra IL-1R2 (XP_004569045.1); H. sapiens IL-1R1 (NP_000868.1). The Ig-like domains and other domains of IL-1Rs were showed by the CDD server [27‒30].
VvIL-1R2 Expression Exhibits a Significant Up-regulation in the PBLs, Gill, and Spleen and at the Hatching Stage
IL-1R2 expression can be regulated at the transcription level [31]; thus, we investigated the expression level of VvIL-1R2 in different tissues and at different embryonic developmental stages. The expression level was analyzed by qRT-PCR. The gene expression was normalized to the housekeeping genes, namely, 18S rRNA and β-actin. Consistent with the tissue distribution results for P. olivaceus, C. idella, and S. aurata [10, 11, 13], VvIL-1R2 is ubiquitously expressed in all the tested tissues (Fig. 3). The highest expression level is observed in the PBLs, followed by those in the gill and spleen. The other tissues, such as heart, liver, kidney, muscle, and intestine shows moderate expression levels of VvIL-1R2 transcripts. This result is similar to the phenomenon in C. idella and O. mykiss [9, 13]. The presence of the VvIL-1R2 transcript in the brain is of interest, given that a monoclonal antibody against IL-1R2 can inhibit the thermogenic and pyrogenic responses to IL-1β released within the R. norvegicus brain [20]. However, our result shows that, among the tested tissues, the brain exhibited the lowest expression level of VvIL-R2. Our data indicate that VvIL-1R2 may be one of the very important cytokines to maintain the immunological functions of the important fish immune organs, i.e., spleen, gill, and the PBLs.
Quantitative analysis of VvIL-1R2 expression in adult tissues. The relative expression variance is presented as a ratio (the amounts of V. variegatus IL-1R2 were normalized to the corresponding 18S rRNA and β-actin genes values). The data are shown as mean ± SD (n = 3). Columns with different letters show a significant difference (p < 0.05).
During embryonic development, fish embryos are usually protected by the egg envelope before they are hatched and exposed to the environment after hatching. Currently, our knowledge is rather limited regarding the defense mechanisms of fish against pathogenic attacks in their hostile environment at the earliest stages of their lives. Thus, we investigated how the expression pattern of VvIL-1R2 changes across the different embryonic stages. mRNA level was detected in the unfertilized eggs of V. variegatus, and it gradually decreased to an undetectable level until the blastula stage (Fig. 4). This phenomenon indicates that VvIL-1R2 is a maternally expressed gene. According to the literature, IL-1β may have several roles during development [32, 33]. As a receptor of IL-1β, IL-1R2 also plays an important role in cell proliferation and apoptosis [34‒36]. After the blastula stage, the embryonic cells are induced to differentiate. So we hypothesized that IL-1R2 might play a role in the differentiation of embryonic cells. The expression level maintains a relatively low level before the hatching stage. Interestingly, the highest expression level of VvIL-1R2 was observed at the hatching stage among the tested stages (Fig. 4). The embryos and hatchlings of most fishes are exposed to their aquatic environment, which is filled with thousands of various microorganisms, including potential pathogens, before their immune systems have fully developed [37]. Furthermore, at the hatching stage, embryos lose the protection of the egg envelope; consequently, they are more vulnerable to pathogens in their external environment. Therefore, the high expression of VvIL-1R2 in this period implies that this gene may have a role in combating the pathogenic factors in the environment. VvIL-1R2 has not been identified in the eggs of other fishes; thus, its role at the early developmental stages remains largely unclear. In the current study, we demonstrated that VvIL-1R2 is a maternal factor stored in the eggs of V. variegatus. Furthermore, its mRNA level gradually increases and peaks at the hatching stage. These results are similar to those for fish-egg lectin (FEL), which is an important immunity gene in fishes, in zebra fish and rock bream [37, 38].
Quantitative analysis of VvIL-1R2 expression at different embryonic developmental stages. The relative expression variance is presented as a ratio (the amounts of V. variegatus IL-1R2 were normalized to the corresponding 18S rRNA and β-actin genes values). The data are shown as mean ± SD (n = 3). Columns with different letters show a significant difference (p < 0.05).
The presented results show that the expression of VvIL-1R2, which may act as an inflammatory factor, is tissue and stage specific.
VvIL-1R2 Shows the Highest Expression at the Mid-Metamorphosis Stage
As the transition from larva to juvenile, metamorphosis is a crucial developmental phase in fish. Pleuronectiformes is an interesting group of teleosts for the study of metamorphosis, as members of this order undergo a dramatic morphological reorganization during this stage and change from a symmetrical larva to an asymmetrical juvenile. V. variegatus has evolutionary relatives with P. olivaceus [14, 39, 40], and they are all flatfish that undergo the metamorphic stage. We are interested in whether the expression patterns of VvIL-1R2 are associated with T3, T4, and TR at this developmental stage. The VvIL-1R2 expression gradually decreased during metamorphosis, then it increased sharply and transiently in mid-metamorphosis stage followed by another decline in late-metamorphosis stage (Fig. 5). VvIL-1R2 expression was predominantly detected in mid-metamorphosis, which is the most violent period of metamorphosis (Fig. 5).
Quantitative analysis of VvIL-1R2 expression during metamorphosis. The relative expression variance is presented as a ratio (the amounts of V. variegatus IL-1R2 were normalized to the corresponding 18S rRNA and β‑actin genes values). The data are shown as mean ± SD (n = 3). Columns with different letters show a significant difference (p < 0.05).
Metamorphosis arises from a series of regulated processes that involve cellular differentiation, apoptosis, biochemical, molecular, and physiological changes [41]. Thyroid hormones (T3 and T4) and thyroid hormone receptor (TR) have been reported to drive flatfish metamorphosis, and they are associated with apoptosis and cellular differentiation [41‒49]. Similarly, IL-1R2 also plays an important role in the cell proliferation, and apoptosis [34‒36]. More importantly, IL-1R2 has an interaction with T3, T4, and TR [50‒52].
Combined with the results of previous studies, the current results show that VvIL-1R2 has a basically consistent expression pattern compared with T3, T4, and TR in Scophthalmus maximus and P. olivaceus at the metamorphic stage [43, 44]. Furthermore, our results strongly imply that VvIL-1R2 may be involved in a series of regulated processes during flatfish metamorphosis. To the best of our knowledge, this current study is the first to show the relationship among the IL-1R2 gene, thyroid hormones, and metamorphosis in the development of pleuronectiform fishes. These results contribute to the further understanding of the molecular mechanism of flatfish metamorphosis.
Poly(I:C) and LPS Induce Significant and Rapid Up-Regulation of VvIL-1R2
IL-1R2 is an immunity gene that is involved in the immune defense against pathogens. Evidence has shown that mammalian IL-1β is a critical cytokine for the antibacterial immune response induced by various bacterial pathogens, including Listeria monocytogenes, Mycobacterium bovis, and Mycobacterium tuberculosis [53‒55]. This gene is known to be released from macrophages, monocytes, neutrophils, NK cells, and T-cells [56]. Thus, we investigated the expression level changes in IL-1R2 which is a receptor of IL-1β when the PBLs are exposed to PAMPs. PBLs of V. variegatus were co-incubated with LPS or poly(I:C). The expression level of VvIL-1R2 did not change significantly with PBS treatment except for 24 h. The incubation with PBS does not have a significant influence on VvIL-1R2 expression in the PBLs (Fig. 6). By contrast, LPS or poly(I:C) treatment has certain influences on the expression of this gene, as demonstrated by the differences between the experimental groups and the control groups. When the expression level was normalized to the control, the fold change levels of VvIL-1R2 were significantly and rapidly up-regulated within .5 h and 1 h with LPS and poly(I:C) treatments, respectively. This result indicates that IL-1R2 expression may be more sensitive to the PAMPs of LPS than to those of poly(I:C).
Quantitative analysis of VvIL-1R2 expression in peripheral blood leucocytes treated with 50 μg/mL (final concentration) LPS or poly(I:C). Total RNA was extracted at different time points (0, 0.5, 1, 2, 4, 6, 12, and 24 h post-stimulation). The relative expression variance is presented as a ratio (the amounts of V. variegatusIL-1R2 were normalized to the corresponding 18S rRNA and β-actin genes values). The data are shown as mean ± SD (n = 3). Columns with different letters show a significant difference (p < 0.05).
Studies in human models showed that PAMPs stimulated IL-1R2 expression in the human peripheral blood mononuclear cells, monocytes/macrophages, and lymphocytes [57]. Recent studies showed that LPS, poly(I:C), bacteria, virus, and parasites induced the high-level transcription of IL-1R2 in the different fishes [9‒11, 13]. Similarly, in our study, the VvIL-1R2 expression was detected in a wide range of tissues under normal physiological conditions, and VvIL-1R2 expression was significantly and rapidly up-regulated by LPS and poly(I:C) challenges. These results indicate that the VvIL-1R2 may be involved in host immune responses against bacterial and viral pathogens. In addition, the expression level began to decrease when it reached its peak within 24 h after co-incubation with the PAMPs. IL-1R2 can induce apoptosis [34]. Thus, we speculate that the decrease in the IL-1R2 expression may indicate the protection of PBLs from damage.
Protein 3D Modeling Analysis and Protein Docking Simulation of VvIL-1R2 and VvIL-1β Predict Its Potential Function
Given that IL-1β is the ligand of IL-1R2, we tested whether VvIL-1R2 and VvIL-1β interact with each other. We first predicted the secondary structures of VvIL-1R2 and VvIL-1β with Phyre2 tools. With the sequences of VvIL-1R2 and VvIL-1β, we predicted the 3D structures of VvIL-1R2 and VvIL-1β with SWISS-MODEL and illustrate the model with Phyre2 tools (Figs. 7a, 7b). The coverage of IL-1R2 and that of IL-1β could reach 72 and 60%, respectively. The PDBsum Generate software was used to illustrate the Ramachandran figure and predict the stability of the model. Only several amino acid residues were located in the forbidden area. The assessment indicates the high quality of the predicted 3D structure.
IL-1β can interact with IL-1R2 and IL-1R1, and this interaction is important for the immune response. IL-1R1 mediates all of the known biological responses to IL-1 [58], while IL-1R2 acts as a decoy receptor of IL-1 and appears to inhibit the activity of IL-1 [59, 60]. Although IL-1R2 is structurally similar to type 1 IL-1 receptor (IL-1R1), which is responsible for IL-1 signal transduction, its truncated cytoplasmic domain and lack of Toll-IL-1 receptor (TIR) region render IL-1R2 incapable of transmembrane signaling [61]. Thus, we tested the interaction of the predicted VvIL-1R2 and VvIL-1β by performing a protein–protein docking analysis. As shown in Fig. 7c, VvIL-1R2 could interact with VvIL-1β through the predicted receptor binding sites.
These results show that the 3D structure of VvIL-1R2 is similar to those of other vertebrate IL-1R2s and indicate that the interaction between VvIL-1R2 and VvIL-1β may mediate the reduction in immunological responses through the ligand–receptor interaction and dampen the signal transduction [62].
CONCLUSION
In this research, we cloned the mRNA of VvIL-1R2 from V. variegatus, and characterized its domain structure. The six conserved cysteine residues, four conserved proline residues, two Ig-like domains, N-terminus signal peptide sequence, and transmembrane domain of this gene, as well as the TA-rich motifs of its proteins related to inflammatory responses were similar to other reported fish IL-1R2 and mammalian IL-1R2. Interestingly, the spatial and temporal expression levels of VvIL-1R2 strongly imply that it may have some universal immunological functions and perform certain novel actions at the embryonic and metamorphic stages. The VvIL-1R2 expression level could significantly and rapidly respond to PAMPs, such as LPS or poly(I:C), thereby pointing that this gene may be involved in host immune responses against bacterial and viral pathogens. The 3D structure prediction and protein docking analysis provided additional information about how VvIL-1R2 mediates its function with VvIL-1β. This result is consistent with that obtained in a set of previous works, which asserts that the function of IL-1R2 relies on the interaction with IL-1β.
ACKNOWLEDGMENTS
This research was supported by the National Natural Sciences Foundation of China (Grant no. 31101891), and the Hitech Research and Development Program of China (Grant no. 2012AA10A408).
COMPLIANCE WITH ETHICAL STANDARDS
Conflict of interests. The authors declare that they have no conflict of interest.
Statement on the welfare of animals. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
REFERENCES
Netea M.G., Nold-Petry C.A., Nold M.F., Joosten L.A.B., Opitz B., van der Meer J.H.M., van de Veerdonk F.L., Ferwerda G., Heinhuis B., Devesa I., Funk C.J., Mason R.J., Kullberg B.J., Rubartelli A., van der Meer J.W.M., Dinarello C.A. 2009. Differential requirement for the activation of the inflammasome for processing and release of IL-1beta in monocytes and macrophages. Blood. 113, 2324‒2335.
Farasat S., Aksentijevich I., Toro J.R. 2008. Autoinflammatory diseases: Clinical and genetic advances. Arch. Dermatol. 144, 392‒402.
Buss H., Dorrie A., Schmitz M.L., Hoffmann E., Resch K., Kracht M. 2004. Constitutive and interleukin-1-inducible phosphorylation of p65 NF-κB at serine 536 is mediated by multiple protein kinases, including IκB kinase IKKα, IKKβ, IKKε, TRAF family member-associated (TANK)-binding kinase 1 (TBK1), and an unknown kinase, and couples p65 to TATA-binding protein-associated factor II31-mediated interleukin-8 transcription. J. Biol. Chem. 279, 55633‒55643.
Molto A., Olive A. 2010. Anti-IL-1 molecules: New comers and new indications. Joint Bone Spine. 77, 102‒107.
Li X., Qin J. 2005. Modulation of Toll-interleukin 1 receptor mediated signaling. J. Mol. Med. 83, 258‒266.
Dower S.K., Kronheim S.R., March C.J., Conlon P.J., Hopp T.P., Gillis S., Urdal D.L. 1985. Detection and characterization of high affinity plasma membrane receptors for human interleukin 1. J. Exp. Med. 162, 501‒515.
Chin J., Cameron P.M., Rupp E., Schmidt J.A. 1987. Identification of a high-affinity receptor for native human interleukin 1 beta and interleukin 1 alpha on normal human lung fibroblasts. J. Exp. Med. 165, 70‒86.
McMahan C.J., Slack J.L., Mosley B., Cosman D., Lupton S.D., Brunton L.L., Grubin C.E., Wignall J.M., Jenkins N.A., Brannan C.I. 1991. A novel IL-1 receptor, cloned from B cells by mammalian expression, is expressed in many cell types. EMBO J. 10, 2821‒2832.
Sangrador-Vegas A., Martin S.A., O’Dea P.G., Smith T.J. 2000. Cloning and characterization of the rainbow trout (Oncorhynchus mykiss) type II interleukin-1 receptor cDNA. Eur. J. Biochem. 267, 7031‒7037.
Lopez-Castejon G., Sepulcre M.P., Roca F.J., Castellana B., Planas J.V., Meseguer J., Mulero V. 2007. The type II interleukin-1 receptor (IL-1RII) of the bony fish gilthead seabream Sparus aurata is strongly induced after infection and tightly regulated at transcriptional and post-transcriptional levels. Mol. Immunol. 44, 2772‒2780.
Fan Y., Li S., Qi J., Zeng L., Zhong Q., Zhang Q. 2010. Cloning and characterization of type II interleukin-1 receptor cDNA from Japanese flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 157, 59‒65.
Morrison R.N., Young N.D., Nowak B.F. 2012. Description of an Atlantic salmon (Salmo salar L.) type II interleukin-1 receptor cDNA and analysis of interleukin-1 receptor expression in amoebic gill disease-affected fish. Fish Shellfish Immunol. 32, 1185‒1190.
Yang X., Wang S., Du L., Yang K., Wang X., Zhang A., Zhou H. 2013. Molecular and functional characterization of IL-1 receptor type 2 in grass carp: A potent inhibitor of IL-1β signaling in head kidney leukocytes. Dev. Comp. Immunol. 41, 738‒745.
Xu Y., Liu X., Liao M., Wang H., Wang Q. 2012. Molecular cloning and differential expression of three GnRH genes during ovarian maturation of spotted halibut, Verasper variegatus. J. Exp. Zool. A: Ecolog. Genet. Physiol. 317, 434‒446.
Zhou Z., Zhang B., Sun L. 2014. Poly(I:C) induces antiviral immune responses in Japanese flounder (Paralichthys olivaceus) that require TLR3 and MDA5 and is negatively regulated by Myd88. PLoS One. 9, e112918.
Caput D., Beutler B., Hartog K., Thayer R., Brown-Shimer S., Cerami A. 1986. Identification of a common nucleotide sequence in the 3'-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. U. S. A. 83, 1670‒1674.
Sachs A.B. 1993. Messenger RNA degradation in eukaryotes. Cell. 74, 413‒421.
Singer S.J. 1990. The structure and insertion of integral proteins in membranes. Annu. Rev. Cell Biol. 6, 247‒296.
Kishimoto T., Taga T., Akira S. 1994. Cytokine signal transduction. Cell. 76, 253‒262.
Luheshi G., Hopkins S.J., Lefeuvre R.A., Dascombe M.J., Ghiara P., Rothwell N.J. 1993. Importance of brain IL-1 type II receptors in fever and thermogenesis in the rat. Am. J. Physiol. 265, 585‒591.
Mirtella A., Tringali G., Guerriero G., Ghiara P., Parente L., Preziosi P., Navarra P. 1995. Evidence that the interleukin-1 beta-induced prostaglandin E2 release from rat hypothalamus is mediated by type I and type II interleukin-1 receptors. J. Neuroimmunol. 61, 171‒177.
Giri J.G., Kincade P.W., Mizel S.B. 1984. Interleukin 1-mediated induction of kappa-light chain synthesis and surface immunoglobulin expression on pre-B cells. J. Immunol. 132, 223‒228.
Horuk R., McCubrey J.A. 1989. The interleukin-1 receptor in Raji human B-lymphoma cells. Molecular characterization and evidence for receptor-mediated activation of gene expression. Biochem. J. 260, 657‒663.
Greenfeder S.A., Nunes P., Kwee L., Labow M., Chizzonite R.A., Ju G. 1995. Molecular cloning and characterization of a second subunit of the interleukin 1 receptor complex. J. Biol. Chem. 270, 13757‒13765.
Heguy A., Baldari C.T., Macchia G., Telford J.L., Melli M. 1992. Amino acids conserved in interleukin-1 receptors (IL-1Rs) and the Drosophila Toll protein are essential for IL-1R signal transduction. J. Biol. Chem. 267, 2605‒2609.
Kuno K., Okamoto S., Hirose K., Murakami S., Matsushima K. 1993. Structure and function of the intracellular portion of the mouse interleukin 1 receptor (type I). Determining the essential region for transducing signals to activate the interleukin 8 gene. J. Biol. Chem. 268, 13510‒13518.
Marchler-Bauer A., Derbyshire M.K., Gonzales N.R., Lu S., Chitsaz F., Geer L.Y., Geer R.C., He J., Gwadz M., Hurwitz D.I., Lanczycki C.J., Lu F., Marchler G.H., Song J.S., Thanki N., et al. 2015. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, 222‒226.
Marchler-Bauer A., Lu S., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., Hurwitz D.I., Jackson J.D., Ke Z., et al. 2011. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, 225‒229.
Marchler-Bauer A., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., He S., Hurwitz D.I., Jackson J.D., Ke Z., Lanczycki C.J., et al. 2009. CDD: Specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37, 205‒210.
Marchler-Bauer A., Bryant S.H. 2004. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 32, 327‒331.
Hawiger J. 2001. Innate immunity and inflammation: A transcriptional paradigm. Immunol. Res. 23, 99‒109.
Moro J.A., Carretero J., Alonso M.I., Martin C., Gato A., Mano A.D.L. 2008. Prenatal expression of interleukin 1beta and interleukin 6 in the rat pituitary gland. Cytokine. 44, 315‒322.
Bagu E.T., Gordon J.R., Rawlings N.C. 2010. Post-natal changes in testicular concentrations of interleukin-1 alpha and beta and interleukin-6 during sexual maturation in bulls. Reprod. Domest. Anim. 45, 336‒341.
West D.A., James N.H., Cosulich S.C., Holden P.R., Brindle R., Rolfe M., Roberts R.A. 1999. Role for tumor necrosis factor alpha receptor 1 and interleukin-1 receptor in the suppression of mouse hepatocyte apoptosis by the peroxisome proliferator nafenopin. Hepatology. 30, 1417‒1424.
Liu X., Min L., Duan H., Shi R., Zhang W., Hong S., Tu C. 2015. Short hairpin RNA (shRNA) of type 2 interleukin-1 receptor (IL1R2) inhibits the proliferation of human osteosarcoma U-2 OS cells. Med. Oncol. 32, 364.
Zieleniewski W., Zieleniewski J., Stepien H. 1995. Effect of interleukin-1a, IL-1b and IL-1 receptor antibody on the proliferation and steroidogenesis of regenerating rat adrenal cortex. Exp. Clin. Endocrinol. Diabetes. 103, 373‒377.
Wang Y., Bu L., Yang L., Li H., Zhang S. 2016. Identification and functional characterization of fish-egg lectin in zebrafish. Fish Shellfish Immunol. 52, 23‒30.
Kim B., Nam B., Kim J., Park H., Song J., Park C. 2011. Molecular characterisation and expression analysis of a fish-egg lectin in rock bream, and its response to bacterial or viral infection. Fish Shellfish Immunol. 31, 1201‒1207.
Li H., Fan J., Liu S., Yang Q., Mu G., He C. 2012. Characterization of a myostatin gene (MSTN1. from spotted halibut (Verasper variegatus) and association between its promoter polymorphism and individual growth performance. Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 161, 315‒322.
Li H., Jiang L., Han J., Su H., Yang Q., He C. 2011. Major histocompatibility complex class IIA and IIB genes of the spotted halibut Verasper variegatus: genomic structure, molecular polymorphism, and expression analysis. Fish Physiol. Biochem. 37, 767‒780.
Power D.M., Llewellyn L., Faustino M., Nowell M.A., Bjornsson B.T., Einarsdottir I.E., Canario A.V., Sweeney G.E. 2001. Thyroid hormones in growth and development of fish. Comp. Biochem. Physiol. C: Toxicol. Pharmacol. 130, 447‒459.
Campinho M.A., Silva N., Sweeney G.E., Power D.M. 2006. Molecular, cellular and histological changes in skin from a larval to an adult phenotype during bony fish metamorphosis. Cell Tissue Res. 327, 267‒284.
Marchand O., Duffraisse M., Triqueneaux G., Safi R., Laudet V. 2004. Molecular cloning and developmental expression patterns of thyroid hormone receptors and T3 target genes in the turbot (Scophtalmus maximus) during post-embryonic development. Gen. Comp. Endocrinol. 135, 345‒357.
Yamano K., Miwa S. 1998. Differential gene expression of thyroid hormone receptor alpha and beta in fish development. Gen. Comp. Endocrinol. 109, 75‒85.
Yamano K., Araki K., Sekikawa K., Inui Y. 1994. Cloning of thyroid hormone receptor genes expressed in metamorphosing flounder. Dev. Genet. 15, 378‒382.
Inui Y., Miwa S. 1985. Thyroid hormone induces metamorphosis of flounder larvae. Gen. Comp. Endocrinol. 60, 450‒454.
de Jesus E.G., Hirano T., Inui Y. 1991. Changes in cortisol and thyroid hormone concentrations during early development and metamorphosis in the Japanese flounder, Paralichthys olivaceus. Gen. Comp. Endocrinol. 82, 369‒376.
Liu Y., Chan W. 2002. Thyroid hormones are important for embryonic to larval transitory phase in zebrafish. Differentiation. 70, 36‒45.
Klaren P.H.M., Wunderink Y.S., Yufera M., Mancera J.M., Flik G. 2008. The thyroid gland and thyroid hormones in Senegalese sole (Solea senegalensis) during early development and metamorphosis. Gen. Comp. Endocrinol. 155, 686‒694.
Svenson M., Kayser L., Hansen M.B., Rasmussen Å.K., Bendtzen K. 1991. Interleukin-1 receptors on human thyroid cells and on the rat thyroid cell line FRTL-5. Cytokine. 3, 125‒130.
Van der P. T., Van Zee K.J., Endert E., Coyle S.M., Stiles D.M., Pribble J.P., Catalano M.A., Moldawer L.L., Lowry S.F. 1995. Interleukin-1 receptor blockade does not affect endotoxin-induced changes in plasma thyroid hormone and thyrotropin concentrations in man. J. Clin. Endocrinol. Metabolism. 80, 1341‒1346.
Zerek-Melen G., Zylinska K., Fryczak J., Mucha S., Stepien H. 1994. Influence of interleukin 1 and antihuman interleukin 1 receptor antibody on the growth and function of the thyroid gland in rats. Eur. J. Endocrinol. 131, 531‒534.
Meixenberger K., Pache F., Eitel J., Schmeck B., Hippenstiel S., Slevogt H., N’Guessan P., Witzenrath M., Netea M.G., Chakraborty T., Suttorp N., Opitz B. 2010. Listeria monocytogenes-infected human peripheral blood mononuclear cells produce IL-1beta, depending on listeriolysin O and NLRP3. J. Immunol. 184, 922‒930.
Bourigault M., Segueni N., Rose S., Court N., Vacher R., Vasseur V., Erard F., Le Bert M., Garcia I., Iwakura Y., Jacobs M., Ryffel B., Quesniaux V.F.J. 2013. Relative contribution of IL-1alpha, IL-1beta and TNF to the host response to Mycobacterium tuberculosis and attenuated M. bovis BCG. Immun. Infl. Disease. 1, 47‒62.
Zhou Y., Zhao D., Yue R., Khan S.H., Shah S.Z.A., Yin X., Yang L., Zhang Z., Zhou X. 2015. Inflammasomes-dependent regulation of IL-1beta secretion induced by the virulent Mycobacterium bovis Beijing strain in THP-1 macrophages. Antonie van Leeuwenhoek. 108, 163‒171.
Covello J.M., Bird S., Morrison R.N., Battaglene S.C., Secombes C.J., Nowak B.F. 2009. Cloning and expression analysis of three striped trumpeter (Latris lineata) pro-inflammatory cytokines, TNF-alpha, IL-1beta and IL-8, in response to infection by the ectoparasitic, Chondracanthus goldsmidi. Fish Shellfish Immunol. 26, 773‒786.
Jang C., Choi J., Byun M., Jue D. 2006. Chloroquine inhibits production of TNF-alpha, IL-1beta and IL-6 from lipopolysaccharide-stimulated human monocytes/macrophages by different modes. Rheumatology. 45, 703‒710.
Stylianou E., O’Neill L.A., Rawlinson L., Edbrooke M.R., Woo P., Saklatvala J. 1992. Interleukin 1 induces NF-kappa B through its type I but not its type II receptor in lymphocytes. J. Biol. Chem. 267, 15836‒15841.
Colotta F., Re F., Muzio M., Bertini R., Polentarutti N., Sironi M., Giri J.G., Dower S.K., Sims J.E., Mantovani A. 1993. Interleukin-1 type II receptor: A decoy target for IL-1 that is regulated by IL-4. Science. 261, 472‒475.
Sims J.E., Gayle M.A., Slack J.L., Alderson M.R., Bird T.A., Giri J.G., Colotta F., Re F., Mantovani A., Shanebeck K. 1993. Interleukin 1 signaling occurs exclusively via the type I receptor. Proc. Natl. Acad. Sci. U. S. A. 90, 6155‒6159.
Peters V.A., Joesting J.J., Freund G.G. 2013. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav. Immunity. 32, 1‒8.
Schreuder H., Tardif C., Trump-Kallmeyer S., Soffientini A., Sarubbi E., Akeson A., Bowlin T., Yanofsky S., Barrett R.W. 1997. A new cytokine-receptor binding mode revealed by the crystal structure of the IL-1 receptor with an antagonist. Nature. 386, 194‒200.
Author information
Authors and Affiliations
Corresponding authors
Additional information
The text was submitted by the author(s) in English.
Rights and permissions
About this article
Cite this article
Li, Z., Liu, X.M., Li, A.Y. et al. Teleost Type 2 Interleukin-1 Receptor (IL-1R2) from the Spotted Halibut (Verasper variegatus): 3D Structure and a Role in Immune Response. Mol Biol 53, 256–266 (2019). https://doi.org/10.1134/S0026893319020109
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893319020109