Abstract
Genetic and neuroimaging research has identified neurobiological correlates of obesity. However, evidence for an integrated model of genetic risk and brain structural alterations in the pathophysiology of obesity is still absent. Here we investigated the relationship between polygenic risk for obesity, gray matter structure and body mass index (BMI) by the use of univariate and multivariate analyses in two large, independent cohorts (n=330 and n=347). Higher BMI and higher polygenic risk for obesity were significantly associated with medial prefrontal gray matter decrease, and prefrontal gray matter was further shown to significantly mediate the effect of polygenic risk for obesity on BMI in both samples. Building on this, the successful individualized prediction of BMI by means of multivariate pattern classification algorithms trained on whole-brain imaging data and external validations in the second cohort points to potential clinical applications of this imaging trait marker.
Similar content being viewed by others
Introduction
Obesity is a highly heritable condition that increases the risk for multiple somatic as well as psychiatric disorders.1, 2, 3 Recent meta-analyses of genome-wide association study (GWAS) data have identified up to 97 loci that showed genome-wide significant associations with body mass index (BMI).1 Analysis of tissue enrichment in these GWAS studies revealed a high expression of many genetic variants in the central nervous system,1, 4 a finding well in line with results from neuroimaging studies reporting associations between BMI and alterations in brain structure and function, both indicating a pivotal role of altered neural processing in the pathophysiology of obesity.5, 6, 7, 8, 9 In particular, gray matter deficits and neural processing bias in the medial prefrontal cortex and the striatum, structures involved in reward-related cognition and impulse inhibition, are among the most frequently observed results of structural and functional imaging studies in obesity.6, 7, 10, 11, 12 However, albeit genetic and neuroimaging studies independently report neurobiological correlates of obesity, evidence for a mechanistic relationship between genetic risk variants and imaging findings in obesity is sparse, hitherto. Nonetheless, considering the high heritability of brain structure and function, it seems plausible that genetic predisposition and neuroimaging findings in obesity are related.13, 14 This would shed more light on the question whether neural alterations in obesity reflect a—genetically shaped—trait marker that precedes weight gain or must rather be considered as an adverse consequence in terms of accelerated neurodegenerative processes in obese subjects.
If the influence of genetic risk for obesity on brain structures associated with increased weight was evidenced, the notion of BMI-related brain structural aberrations as a predisposing trait marker of obesity would be corroborated. Under this assumption, brain morphometric information could be used as an imaging trait marker to individually predict the risk of developing obesity in future, which might lead to new preventive strategies.
In this context, the combination of structural imaging data and machine learning techniques has evoked great promises, for example, by demonstrating successful application in the differential diagnosis of psychiatric disorders and in the detection of subjects at risk for psychiatric disorders.15, 16, 17, 18 Especially successful identification of multivariate patterns of brain structure indicating high risk would highlight the potential relevance of neuroimaging in obesity.
Thus, the aims of the present study were (a) to replicate previous findings of associations between BMI and gray matter changes in two large independent studies, (b) to analyze possible relationships between genetic risk for obesity, structural alterations in obesity-related brain areas and BMI and (c) to predict individual BMI through a machine learning approach.
MATERIALS and methods
Subjects
Our study initially comprised a sample of 345 healthy subjects from the Münster Neuroimaging Cohort (MNC) and a sample of 367 healthy subjects from the BiDirect (BD) study. After the exclusion of subjects with poor image quality detected by visual inspection and by using the check homogeneity using covariance function implemented in VBM8, final samples of N=330 for the MNC and N=347 for the BD study remained (see Table 1). Further details on rational, design and inclusion criteria of both studies have been described elsewhere19, 20, 21, 22 (see also Supplementary Methods).
Image acquisition and VBM
T1-weighted high-resolution anatomical images were acquired and preprocessed using the VBM8-toolbox (http://dbm.neuro.uni-jena.de/vbm) in both cohorts following published protocols, as described in our previous work6, 18, 21, 23, 24 (see also Supplementary Methods).
Polygenic risk score
The MNC sample was genotyped using the Infinium PsychArray-24 that includes 265 000 proven tag single-nucleotide polymorphisms (SNPs) found on the Infinium HumanCore-24 BeadChip, 245 000 markers from the Infinium Exome-24 BeadChip and 50 000 additional markers associated with common psychiatric disorders. These additional SNPs include genetic variants associated with the research of common psychiatric conditions such as schizophrenia, bipolar disorder, autism spectrum disorders, attention deficit hyperactivity disorder, major depressive disorder, obsessive compulsive disorder, anorexia and Tourette’s syndrome. The BD participants were genotyped with the Illumina HumanCore Exome chip va. 1.1 (Psych-chip, Illumina, San Diego, CA, USA).
Imputation of the two study cohorts was carried out using the ENIGMA 1000G reference panel (v.3.20101123), following the ENIGMA imputation protocol using quality control filtered SNPs 257 573 (MNC) and 264 278 (BD). Following the imputation, only the autosomal SNPs that passed the quality control criteria (minor allele frequency >0.01 and imputation quality score >0.5) were used for the polygenic score analysis. Linkage disequilibrium pruning was carried out using the clumping option in plink with r2>0.25 and physical distance threshold of 250 kb.
A polygenic risk score (PRS) was generated using the sets of SNPs selected based on a P-value threshold at 1.0 from the base GWAS data.1 The R program 'PRSice',25 which uses PLINK-1.926 in the background for linkage disequilibrium pruning and the PRS, was used for this analysis. Associations between the PRS and BMI values for each study were assessed via analysis of covariances including age and sex as covariates of no interest.
Mediation analyses
To test for possible mediation effects, a bootstrapping approach implemented in the SPSS macro PROCESS was applied (http://www.processmacro.org),27 which has been demonstrated to provide reliable results in neuroimaging research.28, 29 PROCESS estimates direct and indirect effects between a defined set of variables by applying an ordinary least-squares path analytic framework. For testing inference of an indirect or mediation effect, bootstrap confidence intervals are calculated. Significance of an observed indirect effect is assumed if the 95% confidence interval (CI) does not include zero (see also Supplementary Methods).
Pattern classification
Individualized prediction of BMI values was assessed by the use of support vector regression as implemented in the Neurominer Software Package (N. Koutsouleris, PRONIA Working Group, Department of Psychiatry, University of Munich, Munich, Germany).16, 30 For all multivariate analyses, VBM8 whole-brain gray matter images were used as classifier input. To reduce dimensionality by preserving maximal localized morphometric differences, gray matter images were resliced to a voxel size of 2 mm × 2 mm × 2 mm and subsequently smoothed with a Gaussian kernel of 8 mm full-width at half-maximum for multivariate analyses. To strictly separate the training process from the evaluation of the classifier’s generalizability, Neurominer uses a nested cross-validation (CV) framework as described previously.16, 31 A 5 × 5-fold CV cycle at the outer (CV2) and 1 × 5-fold cycle at the inner (CV1) levels of repeated double CV were generated including the subsequent analyses steps: the training subjects’ gray matter maps were initially corrected for age and sex effects using partial correlations.16 Features were then scaled from 0 to 1. Soft feature weighting was applied, which upweighted features highly predictive for the target label (BMI) by means of a Pearson's correlation. Finally, to reduce the maps’ dimensionality and discard noisy information, principal component analysis projected correlated voxel sets to uncorrelated eigenvariates, thus retaining 80% of the variance in each CV1 training partition.32 Correction, scaling, upweighting and principal component analysis parameters were applied to the CV1 test data. Then, in each training partition, principal component analysis features entered a recursive feature elimination algorithm that used a linear support vector machine to remove those eigenvariates that impaired separability on the respective CV1 test data (support vector machine penalty).33, 34
To obtain CV2 test predictions, the respective gray matter images were first processed using the correction, scaling, feature weighting and principal component analysis parameters of each CV1 training partition, and then classified using the learned decision rules. Classification produced decision scores measuring the neuroanatomical likeness of the BMI value of each subject. Finally, a CV2 test case’s BMI value was predicted by an ensemble classifier that averaged the decision scores of those CV1 base learners in the repeated double CV, in which the subject had not been involved in the training process. Permutation tests for each classifier were conducted to estimate the likelihood of obtaining classification performance by chance (at an α<0.05 for each mean-squared error (MSE) value), by using 1000 random permutations of the labels and applying CV to each permutation.
Results
Univariate regression of BMI on gray matter
Univariate regression analyses were calculated using statistical parametric mapping software (SPM12, http://www.fil.ion.ucl.ac.uk/spm) in two large independent samples of healthy subjects (MNC: n=330; BD: n=347).19, 20, 24
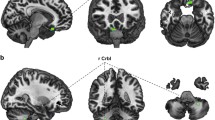
To investigate associations between BMI and whole-brain gray matter, multiple regression models including age and sex as nuisance covariates for the entire cerebrum with P<0.05 at a rigorous family-wise error (FWE) correction on the voxel level and an additional cluster threshold of k=100 voxels were carried out for both samples separately. The anatomical labeling was performed by means of the AAL-Toolbox (N. Tzourio-Mazoyer, Groupe d’Imagerie Neurofonctionnelle, UMR 6095 CNRS CEA, Université de Caen, Université de Paris, France).35 BMI was associated with significant gray matter decrease in medial prefrontal brain areas in both samples (see Figure 1).
Overlapping results of the univariate regression analysis of body mass index (BMI) values on whole-brain gray matter in blue color for the Münster Neuroimaging Cohort (MNC) and in red color for the BiDirect (BD) sample (Montreal Neurologic Institute (MNI) coordinates: x=−9, z=−14/x=−6, z=−12/x=−4, z=−10) at a voxel-threshold pFWE<0.05, minimum cluster volume threshold k⩾100. FWE, family-wise error.
In the MNC sample, the strongest BMI-related gray matter reductions emerged in the bilateral medial frontal gyrus and medial and orbital parts of the bilateral superior frontal gyrus (x=−8, y=69, z=−6, t(1,326)=6.26, k=488, Cohen's d=0.71; x=10, y=69, z=−8, t(1,326)=5.89, k=198, d=0.67) as well as in the temporal gyrus, the thalamus and the insula (for details see Table 2). The same pattern of results with only marginal differences could be observed when the analysis was repeated using BMI values corrected for possible self-report bias (see Supplementary Results and Supplementary Table 1).
BMI-associated gray matter volume decrease was even more pronounced in the BD sample, again involving large parts of the bilateral superior frontal gyrus (orbital and medial parts) and the bilateral medial frontal gyrus (x=−8, y=68, z=−3, t(1,343)=7.60, k=1963, d=0.85; x=8, y=69, z=−10, t(1,343)=7.26, k=1188, d=0.81)). Besides, volume decrease in medial prefrontal areas, additional BMI-related gray matter reductions emerged in the temporal cortex (x=69, y=−42, z=8, t(1,343)=7.62, k=4771, d=0.85) and BMI was furthermore significantly associated with volume reductions in the thalamus and the cingulate cortex (for details see Table 3).
Additional analyses stratified for different age groups in both the MNC and the BD study did not reveal a significant age-dependent effect of BMI on prefrontal gray matter (see Supplementary Results). No positive association between BMI and gray matter could be detected at the applied thresholds in both samples. Even at an exploratory threshold of P<0.001, uncorrected and k=250 voxels, no significant positive associations emerged.
Polygenic risk score and mediation analysis
To evaluate a potential effect of genetic risks for obesity on brain structural alterations associated with increased BMI, a PRS for obesity was calculated for all subjects of the MNC (n=328) and the BD (n=327) study for which GWAS data were available. The PRS was calculated using public available GWAS data as discovery sample, thus including a total of 169 167 SNPs for the MNC and 172 030 SNPs for the BD cohort (p-threshold of P=1.0) (https://www.broadinstitute.org).1 The polygenic risk scores calculated for the MNC and the BD study were significantly positively associated with BMI controlling for age and sex as nuisance regressors (MNC: F(1,327)=14.34, P<0.001, ηp2=0.042; BD: F(1,326)=6.52, P=0.011, ηp2 =0.020).
Multiple regression of PRS scores on gray matter volumes was performed, again including age and sex as covariates. For this step, following our aim to investigate the genetic influence on BMI-related gray matter reductions, for each study an region of interest based on a mask including the two peak clusters (MNC: k=488, x=−8, y=69, z=−6; k=198, x=10, y=69, z=−8/BD: k=1963, x=−8, y=68, z=−3; k=4771, x=69, y=−42, z=8) of the respective preceding regression analyses of BMI on whole-brain gray matter was used, applying a rigorous FWE correction within this ROI at P<0.05 on the voxel level.
In both the MNC and the BD study, regression analysis of the PRS on BMI-associated gray matter yielded a significant volume decrease in the orbitofrontal cortex. Again, the observed gray matter reductions were more pronounced in the BD study (BD: x=−14, y=60, z=−4, t(1,323)=4.40, k=325, pFWE=0.003, d=0.49; MNC: x=9, y=57, z=−6, t(1,324)=3.46, k=9, pFWE=0.016, d=0.39). Mean gray matter values of the entire resulting clusters were extracted for further mediation analyses for each study using a bootstrapping approach as implemented in the PROCESS macro for SPSS. Age and sex were included as nuisance regressors of no interest in each model (number of bootstrap samples: n=5000). Unstandardized regression coefficients are presented for each effect (coeff). In both studies, significant total effects of polygenic risk for obesity on BMI could be observed (MNC: coeff=98944.25, s.e.=26 125.14, t(3,324)=3.79, P<0.001, 95% CI=47 547.93–150 340.57; BD: coeff=57 488.70, s.e.=22 519.67, t(3,323)=2.55, P=0.011, 95% CI=13 184.94–10 1792.45; see Figure 2, path c), which decreased if mean prefrontal gray matter was included in the model (MNC: coeff=79 151.56, s.e.=25 900.06, t(3,324)=3.06, P=0.002, 95% CI=28 197.44–130 105.68; BD: coeff=26 992.92, s.e.=22080.49, t(3,323)=1.22, P=0.222, 95% CI=−16 447.33–70 433.17; path c′). Moreover, in both studies a significant positive mediation effect of polygenic risk for obesity on BMI through medial prefrontal gray matter could be observed (MNC: coeff=19 792.69, s.e.=7886.24, 95% CI=7223.55–38 293.78; BD: coeff=30 495.77, s.e.=7934.50, 95% CI=17 071.99–47 941.82) (see also Supplementary Results).
(a) Plots depicting correlations between the polygenic risk score, body mass index (BMI) values and orbitofrontal cortex (OFC) gray matter volume for the Münster Neuroimaging Cohort (MNC) study (orbital part of the medial frontal gyrus at x=9, y=57, z=−6). (b) Results of the regression analysis of the polygenic risk score on prefrontal gray matter volumes, in blue for the MNC and in red for the BiDirect (BD) study projected on a standard brain template (for display reasons unthresholded values at P<0.05 uncorrected are presented). (c) Models of the mediation analysis of polygenic risk score on BMI through OFC gray matter volume for the MNC and the BD study, unstandardized regression coefficients and s.e. are presented. *Significance at P<0.05.
Additional exploratory regression analyses of PRS values on whole-brain gray matter in both the MNC and the BD sample revealed no significant effect of polygenic risk scores outside the prefrontal cortex (see Supplementary Results).
Individualized prediction of BMI by the use of pattern classification
To investigate a possible utilization of pattern recognition in the individualized prediction of BMI values, classifiers were trained on whole-brain gray matter images in each sample separately and subsequently tested in the respective sample including age and sex as nuisance covariates. Robust individualized prediction of BMI levels by using whole-brain gray matter data was demonstrated in both samples separately. In both the MNC and the BD sample, significant individualized prediction of BMI was achieved with comparable classifier performance (MNC: r=0.38 (correlation between predicted and true BMI), mean absolute error (MAE)=2.8, rMSE (root mean-squared error)=3.7, MSE=13.5, P<0.05; BD: r=0.45, P<0.001, MAE=2.9, rMSE=3.7, MSE=14.0, P<0.05).
Second, to demonstrate external validation, we aimed to transfer the classifiers between both samples. We therefore tested the performance of the above-described classifier trained on whole-brain data of the MNC sample on structural data of the BD sample. Conversely, the performance of the classifier trained on the BD data was subsequently tested on data of the MNC sample. As could be expected, classifier performance decreased but was maintained at robust and comparable level in both cohorts if the trained classifiers were transferred and applied to the imaging data of the respective other sample. The classifier trained on the BD sample was demonstrated to significantly predict BMI in the MNC sample (r=0.42, MAE=3.8, rMSE=4.5, MSE=20.1,P<0.05), whereas the classifier trained on the MNC sample and subsequently tested on the BD sample achieved individualized prediction of BMI values at a comparable error rate, which however failed to maintain significance in the permutation test (r=0.40, MAE=3.6, rMSE=4.5, MSE=20.4) (see also Supplementary Figures 1 and 2).
Discussion
Based on the results of two independently conducted large studies, we believe we provide first evidence for a relationship between genetic and neuroimaging correlates of obesity. The significant impact of polygenic risk for obesity on medial prefrontal gray matter suggests that alterations in prefrontal brain structures are an intermediate phenotype in the development of obesity. This relationship was further delineated by a significant mediation of polygenic risk for obesity on BMI through medial prefrontal gray matter. The clinical potential of this neuroimaging-derived trait marker in obesity was demonstrated through an individualized prediction of BMI by the use of structural brain scans and its external validation in an independent study.
Obesity has been linked to altered neural structure and function in several previous studies. Most reports suggest a primordial role of brain structures closely connected to the reward system in obesity.36, 37, 38 Among these findings, the medial prefrontal cortex as well as tightly connected subcortical structures such as the striatum, the insula and midbrain regions have repeatedly been evidenced to be associated with obesity in both functional and structural magnetic resonance imaging (MRI) studies.7, 12, 38, 39 In accordance with previous results,6, 7, 12 we found that increasing BMI was related to a volume decrease in the medial prefrontal cortex. The involvement of partially overlapping brain structures in structural and resting state fMRI studies in obesity40, 41 might furthermore be considered as supportive for the present results, especially considering reports that link prefrontal brain structure to altered connectivity in resting state fMRI studies.42
Given the pivotal role of the medial prefrontal cortex in reward processing, executive control and impulse inhibition its association with obesity does not surprise. Yet, it should be noted that the importance of this brain region has also been highlighted in affective disorders. Thus, the present findings might also confirm a possible neurobiological connection between obesity and psychiatric disorders.43, 44 It furthermore appears important to consider the evidence for structural alterations in reward-related brain areas in eating disorders such as anorexia nervosa,45, 46 which might point to the primordial role of altered reward processing in different forms of adverse eating behavior.
Regarding the ongoing discussion on state and trait characteristics of obesity-related brain changes, it seems important to emphasize that all of our findings have been corrected for age and sex. Furthermore, analyses stratified for different age groups did not yield any age-dependent effect of BMI on gray matter. Taken together, strong age-related effects, such as pronounced neurodegeneration in older subjects, should not have influenced the present results, strengthening the idea of gray matter decrease as a trait marker for obesity. This notion is further supported by previous findings of brain abnormalities associated with BMI in adolescent and early adulthood subjects.8, 9, 12, 47 We are furthermore aware of two longitudinal studies that investigated BMI-related brain structural changes in healthy individuals, which did not find significant changes in prefrontal brain volume during follow-up, thus supporting the hypothesis of a trait characteristic of medial prefrontal gray matter atrophy in obese individuals.10, 48
Building on this, our finding of a significant association between polygenic risk for obesity and medial prefrontal cortex gray matter volume gives further support to the notion of brain structural alterations as a genetically influenced predisposing trait marker in obesity. Furthermore, the significant mediation effect of medial prefrontal brain structure provides a first insight into the interplay of genetic and brain structural alterations in the development of obesity, even though results of this mediation analyses should be treated with caution considering the non-experimental design of the present study. A genetic influence on BMI-related brain structure is supported by previous reports on the heritability of brain structural correlates.13, 49 In our studies, we focused on the genetic impact on prefrontal brain structure, which was the core finding in our univariate whole-brain regression analyses and which is also one of the most frequently reported brain areas associated with obesity in the literature.6, 7, 10, 11, 12 The specificity of medial prefrontal gray matter as a correlate of genetic risks for obesity is further highlighted by the fact that associations between polygenic risk scores and gray matter were restricted to prefrontal brain areas in exploratory whole-brain analyses.
However, we would like to emphasize that the observed effect size in the PRS analyses makes it highly probable that other factors are contributing to the genesis of gray matter volume decrease in obesity. Different environmental and lifestyle factors, such as fitness level and cardiac function, have also been shown to influence the individual brain structural development. Thus, they are likely to contribute to BMI-related neurostructural alterations.50, 51 Other important factors, such as medication intake and history of psychiatric disorders that have the potential to bias brain structural findings, were controlled in the present study and did not contribute to the observed obesity-related effects.6, 52, 53
Yet, the notion of an environmental and genetic interplay in obesity does not necessarily contradict or weaken our results. Following the food addiction model, increased vulnerability predisposes to adverse behavior (food seeking and taking), which in turn aggravates neural control function followed by development of tolerance.54 Hence, findings of the present study refer to this model by pointing to decreased medial prefrontal gray matter as a vulnerability factor in the development of obesity.
The possible use of this vulnerability marker was demonstrated by robust individualized predictions of BMI by means of whole-brain gray matter in two independently conducted studies. We thus demonstrate that structural MRI-based machine learning techniques are applicable to the prediction of BMI. The idea to apply imaging techniques in obesity prevention is supported by studies demonstrating successful prediction of BMI development as well as successful weight loss during longitudinal follow-up by means of group-level analyses.8, 55 Our results provide an important first step to bridge the gap between research and clinical routine by successfully transferring imaging-derived biomarkers from the group level to individual prediction. This notion heavily relies on previous findings from psychiatric research indicating different applications of structural imaging-based machine learning, such as the differential diagnosis of psychiatric disorders, classification of high-risk subjects and prediction of therapy response.16, 17, 18, 56 From a method point of view, the predictive power of a classifier requires external replication in different, independent test studies to better allow generalizability. Thus, the replication of results with the successful transfer of trained classifiers between our two cohorts strongly supports the feasibility of machine learning approaches in future obesity research and prevention. Nonetheless, considering the classification performance with individually predicted BMI values at an error ranging between 3.7 to 4.5 BMI points, the presented classifiers of course do not allow immediate clinical application, but might indicate that a relatively small gain in classification performance would be sufficient to justify its implementation as an add-on tool, for example, in clinical studies on obesity prevention and outcome prediction following intervention. We thus emphasize that the present findings only provide a perspective for imaging applications in obesity. Further research, especially longitudinal studies applying structural MRI-based pattern recognition are required to analyze outcome prediction in obesity.
Strengths of our study include the combination of univariate and multivariate neuroimaging techniques, two completely different, complementary approaches, the replication of all findings in an independently conducted, robustly powered cohort and the transfer of trained classifiers between both samples.
A possible limitation in the interpretation of our findings might arise from differences between both study cohorts. Mean age was higher in the BD study and weight and height was directly measured, while in the MNC study BMI scores were calculated from self-reported weight and height. Besides ruling out possible age-related effects, we attempted to control for differences in BMI assessment by carrying out additional analyses (a) for the MNC sample based on BMI values corrected for self-report bias and (b) for the BD sample based on self-reported weight and height. Yet, only marginal differences between different types of BMI assessment occurred in these analyses, suggesting that different BMI assessment might not have been a major confounder in the present study. However, it still appears important to state that we cannot fully suspend an error between assessed, corrected and real BMI in the MNC sample. Finally, the assessment of the medical history of each participant was more extensive in the BD study compared with the MNC study, which might have resulted in a higher prevalence of subclinical or undiscovered medical comorbidities in the MNC study. Especially the two latter mentioned issues (assessment of BMI and medical comorbidities) might have biased our brain structural findings and are thus susceptible to explain the observed pattern of results with stronger associations in the BD sample in all imaging analyses.
Moreover, considering the cross-sectional design of the present study, the time of onset of the observed BMI-related brain structural alterations cannot be settled. Interpretation of our findings regarding the chronology of neurostructural alterations in obesity should thus be treated with caution. It should furthermore be noted that application of mediation analysis in non-experimental settings in general is highly problematic because of its strict assumptions, including among other causalities, temporal precedence and the assumption that no further variables exist that could affect the relations in the mediation model. While we aimed to address the majority of these issues in the present study, we clearly state that given the non-experimental design of the present study, we cannot suspend that further undiscovered factors might have biased the presented relationships in the mediation analysis; interpretations of this analysis should thus be drawn with great caution; we especially underline that causality cannot be deduced from these results.
In summary, this study provides first evidence for a relationship of genetic and imaging correlates in obesity. The successful prediction of BMI by multivariate classifiers trained on whole-brain imaging data indicate the potential of imaging techniques in the future prevention of obesity. Longitudinal studies are required to further investigate possible utilizations of imaging applications and pattern recognition before the development and during the course of obesity.
References
Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015; 518: 197–206.
Luppino FS, de Wit LM, Bouvy PF, Stijnen T, Cuijpers P, Penninx BW et al. Overweight, obesity, and depression: a systematic review and meta-analysis of longitudinal studies. Arch Gen Psychiatry 2010; 67: 220–229.
Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010; 42: 937–948.
Willer CJ, Speliotes EK, RJF Loos, Li S, Lindgren CM, Heid IM et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 2009; 41: 25–34.
Opel N, Redlich R, Grotegerd D, Dohm K, Haupenthal C, Heindel W et al. Enhanced neural responsiveness to reward associated with obesity in the absence of food-related stimuli. Hum Brain Mapp 2015; 36: 2330–2337.
Opel N, Redlich R, Grotegerd D, Dohm K, Heindel W, Kugel H et al. Obesity and major depression: Body-mass index (BMI) is associated with a severe course of disease and specific neurostructural alterations. Psychoneuroendocrinology 2015; 51: 219–226.
Raji CA, Ho AJ, Parikshak NN, Becker JT, Lopez OL, Kuller LH et al. Brain structure and obesity. Hum Brain Mapp 2010; 31: 353–364.
Burger KS, Stice E . Greater striatopallidal adaptive coding during cue-reward learning and food reward habituation predict future weight gain. Neuroimage 2014; 99: 122–128.
Batterink L, Yokum S, Stice E . Body mass correlates inversely with inhibitory control in response to food among adolescent girls: an fMRI study. Neuroimage 2010; 52: 1696–1703.
Bobb JF, Schwartz BS, Davatzikos C, Caffo B . Cross-sectional and longitudinal association of body mass index and brain volume. Hum Brain Mapp 2014; 35: 75–88.
Pannacciulli N, Del Parigi A, Chen K, Le DSNT, Reiman EM, Tataranni PA . Brain abnormalities in human obesity: a voxel-based morphometric study. Neuroimage 2006; 31: 1419–1425.
Marqués-Iturria I, Pueyo R, Garolera M, Segura B, Junqué C, García-García I et al. Frontal cortical thinning and subcortical volume reductions in early adulthood obesity. Psychiatry Res Neuroimag 2013; 214: 109–115.
Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N et al. Common genetic variants influence human subcortical brain structures. Nature 2015; 520: 224–229.
Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 2012; 44: 552–561.
Kambeitz J, Kambeitz-Ilankovic L, Leucht S, Wood S, Davatzikos C, Malchow B et al. Detecting neuroimaging biomarkers for schizophrenia: a meta-analysis of multivariate pattern recognition studies. Neuropsychopharmacology 2015; 40: 1742–1751.
Koutsouleris N, Meisenzahl EM, Borgwardt S, Riecher-Rössler A, Frodl T, Kambeitz J et al. Individualized differential diagnosis of schizophrenia and mood disorders using neuroanatomical biomarkers. Brain 2015; 138: 2059–2073.
Opel N, Zwanzger P, Redlich R, Grotegerd D, Dohm K, Arolt V et al. Differing brain structural correlates of familial and environmental risk for major depressive disorder revealed by a combined VBM/pattern recognition approach. Psychol Med 2016; 46: 277–290.
Redlich R, Almeida JJR, Grotegerd D, Opel N, Kugel H, Heindel W et al. Brain morphometric biomarkers distinguishing unipolar and bipolar depression. JAMA Psychiatry 2014; 71: 1222.
Teismann H, Wersching H, Nagel M, Arolt V, Heindel W, Baune BT et al. Establishing the bidirectional relationship between depression and subclinical arteriosclerosis—rationale, design, and characteristics of the BiDirect Study. BMC Psychiatry 2014; 14: 174.
Dannlowski U, Grabe HJ, Wittfeld K, Klaus J, Konrad C, Grotegerd D et al. Multimodal imaging of a tescalcin (TESC)-regulating polymorphism (rs7294919)-specific effects on hippocampal gray matter structure. Mol Psychiatry 2015; 20: 398–404.
Opel N, Redlich R, Zwanzger P, Grotegerd D, Arolt V, Heindel W et al. Hippocampal atrophy in major depression: a function of childhood maltreatment rather than diagnosis? Neuropsychopharmacology 2014; 39: 2723–2731.
Redlich R, Grotegerd D, Opel N, Kaufmann C, Zwitserlood P, Kugel H et al. Are you gonna leave me? Separation anxiety is associated with increased amygdala responsiveness and volume. Soc Cogn Affect Neurosci 2015; 10: 278–284.
Dannlowski U, Stuhrmann A, Beutelmann V, Zwanzger P, Lenzen T, Grotegerd D et al. Limbic scars: long-term consequences of childhood maltreatment revealed by functional and structural magnetic resonance imaging. Biol Psychiatry 2012; 71: 286–293.
Teuber A, Sundermann B, Kugel H, Schwindt W, Heindel W, Minnerup J et al. MR imaging of the brain in large cohort studies: feasibility report of the population- and patient-based BiDirect study. Eur Radiol 2017; 27: 231–238.
Euesden J, Lewis CM, O’Reilly PF . PRSice: Polygenic Risk Score Software. Bioinformatics 2015; 31: 1466–1468.
Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ . Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7.
Hayes AF . Introduction to Mediation, Moderation, and Conditional Process Analysis: A Regression-Based Approach 2013; doi:10.1111/jedm.12050. Methodology in the Social Sciences. The Guilford Press: New York, NY; www.guilford.com.
Birdsill AC, Carlsson CM, Willette AA, Okonkwo OC, Johnson SC, Xu G et al. Low cerebral blood flow is associated with lower memory function in metabolic syndrome. Obesity (Silver Spring, MD) 2013; 21: 1313–1320.
Mackey S, Chaarani B, Kan K-J, Spechler PA, Orr C, Banaschewski T et al. Brain regions related to impulsivity mediate the effects of early adversity on antisocial behavior. Biol Psychiatry 2016; pii: S0006-3223(16)00043-3.
Chang C, Lin C . LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2011; 2: 1–27.
Filzmoser P, Liebmann B, Varmuza K . Repeated double cross validation. J Chemom 2009; 23: 160–171.
Hansen LK, Larsen J, Nielsen FA, Strother SC, Rostrup E, Savoy R et al. Generalizable patterns in neuroimaging: how many principal components? Neuroimage 1999; 9: 534–544.
Fan R-E, Chang K-W, Hsieh C-J, Wang X-R, Lin C-J . LIBLINEAR: a library for large linear classification. J Mach Learn Res 2008; 9: 1871–1874.
Guyon I, Weston J, Barnhill S, Vapnik V . Gene selection for cancer classification using support vector machines. Mach Learn 46: 389–422.
Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002; 15: 273–289.
Blum K, Thanos PK, Gold MS . Dopamine and glucose, obesity, and reward deficiency syndrome. Front Psychol 2014; 5; doi:10.3389/fpsyg.2014.00919.
García-García I, Horstmann A, Jurado MA, Garolera M, Chaudhry SJ, Margulies DS et al. Reward processing in obesity, substance addiction and non-substance addiction. Obes Rev 2014; 15: 853–869.
Burger KS, Berner LA . A functional neuroimaging review of obesity, appetitive hormones and ingestive behavior. Physiol Behav 2014; 136: 121–127.
Kishinevsky FI, Cox JE, Murdaugh DL, Stoeckel LE, Cook EW, Weller RE . fMRI reactivity on a delay discounting task predicts weight gain in obese women. Appetite 2012; 58: 582–592.
Zhang B, Tian D, Yu C, Li M, Zang Y, Liu Y et al. Altered baseline brain activity differentiates regional mechanisms subserving biological and psychological alterations in obese men. Sci Rep 2015; 5: 11563.
Geha P, Cecchi G, Todd Constable R, Abdallah C, Small DM . Reorganization of brain connectivity in obesity. Hum Brain Mapp 2017; 38: 1403–1420.
van Tol M-J, Li M, Metzger CD, Hailla N, Horn DI, Li W et al. Local cortical thinning links to resting-state disconnectivity in major depressive disorder. Psychol Med 2014; 44: 2053–2065.
Ansell EB, Rando K, Tuit K, Guarnaccia J, Sinha R . Cumulative adversity and smaller gray matter volume in medial prefrontal, anterior cingulate, and insula regions. Biol Psychiatry 2012; 72: 57–64.
Phillips ML, Drevets WC, Rauch SL, Lane R . Neurobiology of emotion perception I: the neural basis of normal emotion perception. Biol Psychiatry 2003; 54: 504–514.
Seitz J, Walter M, Mainz V, Herpertz-Dahlmann B, Konrad K, von Polier G . Brain volume reduction predicts weight development in adolescent patients with anorexia nervosa. J Psychiatr Res 2015; 68: 228–237.
Friederich H-C, Walther S, Bendszus M, Biller A, Thomann P, Zeigermann S et al. Grey matter abnormalities within cortico-limbic-striatal circuits in acute and weight-restored anorexia nervosa patients. Neuroimage 2012; 59: 1106–1113.
Stice E, Yokum S, Burger KS, Epstein LH, Small DM . Youth at risk for obesity show greater activation of striatal and somatosensory regions to food. J Neurosci 2011; 31: 4360–4366.
Driscoll I, Beydoun M a, An Y, Davatzikos C, Ferrucci L, Zonderman AB et al. Midlife obesity and trajectories of brain volume changes in older adults. Hum Brain Mapp 2012; 33: 2204–2210.
Fjell AM, Grydeland H, Krogsrud SK, Amlien I, Rohani DA, Ferschmann L et al. Development and aging of cortical thickness correspond to genetic organization patterns. Proc Natl Acad Sci USA 2015; 112: 15462–15467.
Jefferson AL . Cardiac output as a potential risk factor for abnormal brain aging. J Alzheimer’s Dis 2010; 20: 813–821.
Hayes SMM, Alosco MLL, Forman DEE . The effects of aerobic exercise on cognitive and neural decline in aging and cardiovascular disease. Curr Geriatr Rep 2014; 3: 282–290.
Arnone D, McKie S, Elliott R, Juhasz G, Thomas EJ, Downey D et al. State-dependent changes in hippocampal grey matter in depression. Mol Psychiatry 2013; 18: 1265–1272.
Pan A, Sun Q, Czernichow S, Kivimaki M, Okereke OI, Lucas M et al. Bidirectional association between depression and obesity in middle-aged and older women. Int J Obes 2012; 36: 595–602.
Smith DG, Robbins TW . The neurobiological underpinnings of obesity and binge eating: a rationale for adopting the food addiction model. Biol Psychiatry 2013; 73: 804–810.
Sweet LH, Hassenstab JJ, McCaffery JM, Raynor HA, Bond DS, Demos KE et al. Brain response to food stimulation in obese, normal weight, and successful weight loss maintainers. Obesity (Silver Spring, MD) 2012; 20: 2220–2225.
Redlich R, Opel N, Grotegerd D, Dohm K, Zaremba D, Bürger C et al. Prediction of individual response to electroconvulsive therapy via machine learning on structural magnetic resonance imaging data. JAMA Psychiatry 2016; 73: 557–564.
Acknowledgements
The study was supported by grants from the German Research Foundation (DFG; Grants FOR 2107, DA1151/5-1 to UD, and SFB-TRR58, Project C09 to UD), and the Interdisciplinary Centre for Clinical Studies (IZKF, Grant Dan3/012/17 to UD). The BiDirect Study is supported by the German Federal Ministry of Education and Research (BMBF, Grants FKZ-01ER0816, FKZ-01ER1506 and FKZ-01ER1205).
Author contributions
Study concept and design: NO, RR, DG, KD, WH, VA, BTB, NK, KB and UD. Acquisition, analysis or interpretation of data: NO, RR, KD, AT, CK, HW, NK, BTB, KB, VA, HK and UD. Drafting of the manuscript: NO and UD. Critical revision of the manuscript for important intellectual content: RR, DG, CK, KD, WH, VA, AT, NK, AT, HW, BTB, KB, HK and UD. Statistical analysis: NO, RR, CK, DG and UD. Obtained funding: UD and KB. Administrative, technical or material support: RR, NO, DG, CK, KD, AT, NK, AT, HW, BTB, KB, VA, HK and UD. Study supervision: NO, RR, DG, UD, WH, HK, KB, BTB and VA.
Ethical statement
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Declaration of Helsinki of 1975, as revised in 2008.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
V Arolt is a member of advisory boards and/or gave presentations for the following companies: Astra-Zeneca, Eli Lilly, Janssen-Organon, Lundbeck, Otsuka, Servier and Trommsdorff. These affiliations have no relevance to the work covered in the manuscript. The remaining authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
PowerPoint slides
Rights and permissions
About this article
Cite this article
Opel, N., Redlich, R., Kaehler, C. et al. Prefrontal gray matter volume mediates genetic risks for obesity. Mol Psychiatry 22, 703–710 (2017). https://doi.org/10.1038/mp.2017.51
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2017.51
- Springer Nature Limited
This article is cited by
-
Multivariate patterns of brain functional connectome associated with COVID-19-related negative affect symptoms
Translational Psychiatry (2024)
-
Neuroanatomical correlates of genetic risk for obesity in children
Translational Psychiatry (2023)
-
Brain functional and structural magnetic resonance imaging of obesity and weight loss interventions
Molecular Psychiatry (2023)
-
Higher body weight-dependent neural activation during reward processing
Brain Imaging and Behavior (2023)
-
Mechanisms linking obesity and its metabolic comorbidities with cerebral grey and white matter changes
Reviews in Endocrine and Metabolic Disorders (2022)