Abstract
Apple chlorotic leaf spot virus (ACLSV) is one of the most common viruses infecting apple and pear trees, and Iran is among the top ten apple-producing countries in the world. In the present study, incidence and genetic diversity of ACLSV were investigated in the main apple-growing regions of Iran. To achieve this purpose, a total of 1053 leaf samples were collected from orchards located in nine Iranian provinces. ACLSV infection was detected by DAS-ELISA in 48 samples (4.55%) from seven provinces and was confirmed by RT-PCR. Based on the geographical origin, 19 representative isolates were selected for phylogenetic analysis. Nineteen amplicons, with a size of 677 base pairs (bp), containing the 3′ end of the movement protein (MP), the coat protein (CP) gene, and 3′-UTR sequence, were sequenced. Sequence analysis, using data of 45 isolates from 12 different countries, including the 19 Iranian isolates, showed that CP gene among the Iranian isolates were 80–99% identical at both nucleotide and amino acid levels, and these isolates were placed in the B6 (AB326224) and P-205 (D14996) ACLSV groups. This is the first genetic analysis of ACLSV in Iran.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apple chlorotic leaf spot virus (ACLSV) is the type species of the genus Trichovirus in the family Betaflexiviridae, order Tymovirales (Adams et al. 2004). Although ACLSV infection is typically latent, in some cases, it causes severe symptoms. The virus is recognized as economically important due to its worldwide occurrence and to the effects in some infected fruit tree species. ACLSV naturally infects many Prunus species, apples (Malus domestica Borkh.) and pears (Pyrus communis L.), and other Rosaceae species (Paduch-Cichal et al. 2005; Salem et al. 2005; Keshavarz et al. 2009, Keshavarz and Shams-Bakhsh 2014; Mahfoudhi et al. 2013). The severity of symptoms caused by ACLSV is dependent on plant species and virus strains (Németh 1986). ACLSV could be responsible for symptomatic diseases in stone fruits, such as plum bark split and cherry fruit necrosis (Desvignes and Boyé 1989; Desvignes et al. 1999). It may also induce top-working disease in apple trees and graft incompatibility in apricot (Desvignes and Boyé 1989).
ACLSV virions are flexuous filaments, 640–760 × 12 nm in size. The genome of ACLSV contains three open reading frames (ORFs) encoding respectively, a 216.5-kDa protein (ORF1) involved in genome replication, a 50.4-kDa movement protein (MP) (ORF2), and a 21.4-kDa coat protein (CP) (ORF3). The genome of ACLSV consists of 7474–7561 nt with a 3′ poly adenine tail (Guo et al. 2016). The genomic organization of ACLSV shows gene overlapping of 82 nt between ORF1 and ORF2 and 316 nt between ORF2 and ORF3 (Sato et al. 1993). The complete genome sequences of some ACLSV isolates have been determined from, e.g., apple (P-205, A4, B6, and MO-5) (Sato et al. 1993; Yaegashi et al. 2007), plum (P863 and PBM1) (German et al. 1990; Jelkmann and Kunze 1995), peach (Ta Tao 5, Z1, and Z3) (Marini et al. 2008; Niu et al. 2012), and cherry (ball) (German et al. 1997), and their molecular characteristics, such as genetic diversity and phylogenetic relationship, have been investigated.
Sequence analysis of the genomes revealed that there is significant molecular diversity among ACLSV isolates, especially at the 5′ end of the CP gene which is partially overlapped by the 3′ end of the MP gene (Al Rwahnih et al. 2004; Rana et al. 2010; Song et al. 2011). In addition, based on amino acid sequences of CP, ACLSV isolates are clustered into P205, B6, and Ta Tao 5 groups (Yaegashi et al. 2007; Chen et al. 2014; Liu et al. 2014).
In Iran, a previous survey using DAS-ELISA indicated that 18.55% of pome fruit trees are infected with ACLSV (Keshavarz and Shams-Bakhsh 2014). No information is currently available regarding gene sequences and molecular variation among Iranian ACLSV isolates. The aim of the present study was therefore to determine the current status of ACLSV distribution in the most important apple-growing regions of Iran and also to investigate the genetic diversity of the virus, based on a fragment of 677 nt including the 3′ end of the MP, the CP gene, and 3′-UTR sequence.
Materials and methods
Survey for ACLSV
A survey was carried out during 2012 and 2013, in 110 commercial orchards located in nine geographical districts of Iran, from which a total of 1053 samples were randomly collected from apple trees (Table 1, Fig. 1). Many of the surveyed orchards were 5- to 10-year-old, and generally included more than one variety. The most frequent varieties were ‘Golden Delicious’, ‘Red Delicious’ and ‘Granny Smith’ grafted on Malling (m-7, m-9, and m26), Malling-Merton 111 (MM111), and MM106 rootstocks. Some local varieties, i.e., ‘Golab-e Isfahan’, ‘Shafi Abadi’ and ‘Golab-e Kohanz’, were also present which are propagated by grafting on to wild local apple. Leaf sampling was performed randomly from different parts of the canopy to minimize effects of uneven virus distribution. Symptoms of chlorosis along the veins of the leaves were observed in the orchards surveyed. Composite leaf samples (0.2 g) were subjected to double-antibody sandwich enzyme-linked immunosorbent assays (DAS-ELISA) with ACLSV-specific antibodies (Bioreba, Switzerland) according to the manufacturer’s instructions. Absorbance values at a wavelength of 405 nm were measured in an AD340 Beckman ELISA plate reader (USA).
Map of Iran showing the distribution of ACLSV in 11 provinces. Provinces where ACLSV was not detected are indicated by a caret symbol in this research and by a currency sign in (Keshavarz et al. 2009). Provinces on which ACLSV was identified in this research are shown with an asterisk while a black triangle is used to identify provinces in which Keshavarz and Shams-bakhsh (2014) reported ACLSV. Provinces are (1) Alborz, (2) West Azarbaijan, (3) Qazvin, (4) Hamedan, (5) Isfahan, (6) Lorestan, (7) Markazi, (8) Mazandaran, (9) Tehran, (10) East Azarbaijan, and (11) Khorasan
RNA isolation, RT-PCR, cloning, and sequencing
Total RNA was extracted from leaf tissue of the ELISA-positive samples, according to Chang et al. (1993) with some modifications. A 1 μl aliquot of total RNA was subjected to complementary DNA (cDNA) synthesis and the rest was kept at − 70 °C for future use. Specific primers amplifying a 677 nt fragment, between nucleotide 6860 (primer ACLSV-s 5′-TTCATGGAAAGACGGGGCAA-3′) and 7536 (primer ACLSV-as 5′-AAGTCTACAGGCTATTTATTATAAGTCTAA-3′) (P-250 isolate, GenBank accession No. D14996), were used for detection purposes (Menzel et al. 2002). Synthesis of cDNA was performed using 0.4 pmol reverse primer and the MMLV reverse transcriptase (Promega, USA) according to the manufacturer’s instructions. Total RNAs obtained from Italian ACLSV-infected and healthy apple leaves were used as positive and negative controls, respectively. The cDNA product was subjected to PCR using 0.4 pmol of ACLSV primers and the GoTaq®Green Master Mix (Promega, USA) according to the manufacturer’s instructions. Amplification cycles were as follows: denaturation step for 5 min at 94 °C, then 30 cycles for 1 min at 94 °C, 1 min at 60 °C and 1 min at 72 °C, and a final extension step at 72 °C for 10 min. A total of 19 ACLSV isolates were selected for sequencing and downstream analysis. The fragments amplified by RT-PCR were ligated into the pGEM-T Easy Vector (Promega, USA) following the manufacturer’s instructions and transformed into Escherichia coli strain M1022 electrocompetent cells using a 2510 Electroporator (Eppendorf, Germany) as recommended by the manufacturer. Eight white colonies resulting from transformation with each isolate were selected randomly and used as the templates in the colony PCR screening (Sambrook and Russell 2001) using the M13 universal primers. At least two independent clones for each amplicon were sequenced in both directions by a commercial company (Eurofins MWG Operon, Germany).
Phylogenetic and genetic analysis
Sequence data were edited using Vector NTI Advance 11.5 software (Invitrogen Corp., USA). The Blast-n algorithm, available through the NCBI web server (http://www.ncbi.nlm.nih.gov), was used to identify the closest matches (relatives) to putative ACLSV sequences from samples collected in the present study. Based on the Blast-n results, 45 full-length/partial ACLSV isolates/strains were identified that collectively had matches to isolates previously described from three continents: Asia (India, China, Japan, Taiwan, Turkey), Europe (Czech Republic, Italy, Lithuania, Albania), and North America (USA, Canada). Nucleotide and amino acid partial sequences were aligned using CLUSTAL W, implemented in MEGA6 software (Tamura et al. 2011), with the default parameters. The sequences were subjected to identity matrix estimation and recombination analysis, phylogenetic reconstruction, and genetic variability analysis. BioEdit version 7.1.3.0 (Hall 1999) was used to determine identities based on deduced amino acid. Pairwise distances were calculated using the PASC algorithm available at the GenBank database (http://www.ncbi.nlm.nih.gov/sutils/pasc/) (Bao et al. 2012). Using MEGA6, a neighbor-joining (NJ) tree was reconstructed, with the evolutionary model Kimura 2-parameter +G and 1000 bootstrap iterations (Kimura 1980) at the nucleotide level.
Results and discussion
ACLSV detection and distribution
All of the 1053 tested apple trees were individually inspected. Out of the 1053 collected leaf samples tested by DAS-ELISA, ACLSV was detected in 48 samples (4.55%) of Malus domestica cv. “Shafi Abadi,” cv. ‘Shafi Abadi’, cv. ‘Golab-e Kohanz’, and cv. ‘Gholab-e Isfahan’ from seven provinces, i.e., Alborz, Hamedan, Lorestan, Qazvin, Markazi, Mazandaran, and Tehran (Fig. 1, Table 1). The infection rate of ACLSV was particularly significant in the local varieties ‘Shafi Abadi’, ‘Golab-e Kohanz’, and ‘Gholab-e Isfahan’ (100%). Among the apple trees infected with ACLSV, cv. ‘Shafi Abadi’ showed symptoms of chlorosis along the veins of the leaves, while other apple cultivars were symptomless (Table 1). Our results showed that ACLSV is distributed in the northern and the central regions of Iran (Fig. 1). Rates of infected trees varied from no infection in West Azarbaijan and Isfahan provinces to 13.8% in Alborz province.
Genetic diversity and selection pressure analysis
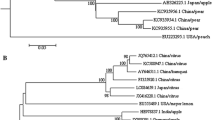
All sequences obtained in the present work, after deletion of primer regions, were deposited in the GenBank under the accession Nos. KM586358 to KM586376. Sequence analysis data revealed that the identities of the partial CP gene among all 19 Iranian ACLSV isolates were 79.8–99.2% and 79.8–98.8% at the nucleotide and amino acid levels, respectively. The 180T5Te (KM586369), T4Te (KM586368), T1Ma (KM586366), and R1T7Ga (KM586362) isolates were the most distantly related of the Iranian isolates (Fig. 2). When nucleotide sequences were considered, the ACLSV isolates were divided into three major groups (Fig. 2). A total of 45 ACLSV isolates, including 19 from Iran (determined in this study) and 26 from GenBank, were separated into B6, P205, and Ta Tao 5 groups (Fig. 2). Group I, the equivalent of B6 group, contained 20 isolates, including 15 of the 19 apple isolates under study, three apple isolates published in GenBank, one pear isolate, and one peach isolate. Twenty isolates clustered into group II equivalent to P-205 group, containing four of the 19 apple isolates under study, 13 apple isolates published in GenBank, one plum isolate, and one pear isolate. These isolates came from different continents of Asia (China, Japan, and Taiwan) and Europe (Albania, Italy, and the Czech Republic). The remaining five peach isolates which were classified into group III, equivalent to Ta Tao 5 group, from China and USA. Comparisons of the Iranian isolates with the available ACLSV sequences in GenBank based on amino acid sequence showed that the T4Te, T1Ma, and R1T7Ga isolates were most closely related to the Lch isolate from China (KF134397) with 92.2%, 89.4%, and 89.5% identity, respectively. The isolates T5Te and T4R4Ma were the closest to the Albanian isolate M62 (AJ596639) with 92.2% and 91.2% identity, respectively. The isolate T9Ja was the closest to the Turkish isolate (AM409323) (> 95.1% identity) while other isolates were closest to Lithuanian isolate (HQ398250) with 85.7–87.5% identity (Supplementary Fig. S1). In all cases, the identities among the isolates based on amino acid sequences were in accordance with those of nt sequences. Collectively, these results serve as indicators of high genetic variability among the Iranian ACLSV isolates when compared to the ACLSV isolates from the GenBank from different countries resulting from a high rate of synonymous mutations.
Phylogenetic tree based on partial CP gene nucleotide sequence of 19 ACLSV isolates under study and 26 isolates retrieved from the NCBI database. Isolates are indicated in the tree by accession number/host/country. The tree was constructed by MEGA 6.0 using the neighbor-joining method and Kimura two-parameter model with 1000 bootstrap replicates. Bootstrap values above 60% are shown. The pink, blue, and yellow bars denote the three groups of ACLSV, respectively. ACLSV isolates determined in this study are marked with pink circles
Results of the present study showed that the incidence of ACLSV in major apple production areas of Iran was 4.55%. This incidence could be considered as low when compared to those of other countries such as Greece, where the incidence of ACLSV in apple orchards has been found to be 65.7% (Mathioudakis et al. 2010), or China with 51.5% infection (Liu et al. 2014). In the present study, we detected ACLSV infection just in local cultivars such as ‘Gholab-e Isfahan’, ‘Shafi Abadi’, and ‘Golab-e Kohanz’, but not in imported apple cultivars (Table 1). This might be due to propagation method and/or the use of virus-infected scions or stock compared to certified imported stocks. In Iran, local cultivars are grafted on wild local apple rootstock without any certification schemes for the production of virus-free propagating material, so the orchards with local cultivars are not established with virus-indexed material propagated on virus-indexed rootstocks. On the other hand, international apple cultivars with virus-free certification have been imported. Moreover, imported apple trees are first kept in a special greenhouse for a period of time to check if they show disease symptoms, before introducing these cultivars to the nursery for apple trees. In the nursery for apple trees, apple scions are grafted on virus-indexed Malling rootstocks (m-7, m-9, and m26), Malling-Merton 111 (MM111), and MM106, so it is supposed that the international/imported cultivars in Iran are virus-free. Moreover, the extraordinary outbreak of ACLSV in Greece could be due to the low sanitary status of apple and pear orchards in that country (Mathioudakis et al. 2010). In this research, ACLSV was not detected in West Azarbaijan and Isfahan provinces; however, Keshavarz and Shams-Bakhsh (2014) have reported the infection from these provinces and they reported a higher incidence of ACLSV in Isfahan than other regions (30.9%). In a previous study (Keshavarz and Shams-Bakhsh 2014), only symptomatic plant leaves were tested, while in the present study, the symptomatic and asymptomatic plants were tested. Present evidence suggests that origin and spread are at least primarily due to vegetative propagation of infected germplasm.
High variability was evident in the N-terminal part of the CP, consistent with the C terminus. Sequence diversity has been reported in amino acids towards the central (37–100) and C-terminal domains of the CP in Indian isolates (Rana et al. 2010) suggesting that co-variation has occurred which may be due to host-correlated positive selection or genetic drift (Yaegashi et al. 2007). The high level of sequence homology observed among Iranian isolates could be explained by considering the source of trees used in propagation and the same host of isolates. All Iranian isolates came from local varieties grafted on wild local apple rootstock, and apple scions were produced in commercial orchards of the main pome fruits production area of such as Alborz and Tehran provinces and sent out to other provinces.
Based on analysis of the CP gene region, the studied Iranian isolates were categorized into B6 and P205 groups. The phylogenetic analysis did not discriminate ACLSV isolates according to geographic origin nor did it allow us to draw conclusions on their origin and dispersion, which is consistent with the report from Rana et al. (2010) who studied genetic diversity of 26 ACLSV isolates by sequencing of the CP gene.
Future studies are needed to evaluate the genetic diversity of Iranian ACLSV isolates based on the full-length sequences of the viral genome. For this reason, the first report of the level of incidence of infections emphasizes the need for certification schemes for the production of virus-free propagating material for local cultivars and warrants a comprehensive study on its sources, means of spread, and survival in Iran.
References
Adams MJ, Antoniw JF, Bar-Joseph M, Brunt AA, Candresse T, Foster GD, Martelli GP, Milne RG, Zavriev SK, Fauquet CM (2004) The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch Virol 149:1045–1060
Al Rwahnih M, Turturo C, Minafra A, Saldarelli P, Myrta A, Pallas V, Savino V (2004) Molecular variability of Apple chlorotic leaf spot virus in different hosts and geographical regions. J Plant Pathol 86:117–122
Bao Y, Chetvernin V, Tatusova T (2012) Pairwise sequence comparison (PASC) and its application in the classification of filoviruses. Viruses 4:1318–1327
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11:113–116
Chen S, Zhou Y, Ye T, Hao L, Guo L, Fan Z, Li S, Zhou T (2014) Genetic variation analysis of Apple chlorotic leaf spot virus coat protein reveals a new phylogenetic type and two recombinants in China. Arch Virol 159:1431–1438
Desvignes J, Boyé R (1989) Different diseases caused by the chlorotic leaf spot virus on the fruit trees. Acta Hortic 235:31–38
Desvignes JC, Boyé R, Cornaggia D, Grasseau N, Hurtt S, Waterworth H (1999) Virus diseases of fruit trees. Centre Technique Interprofessionnel des Fruits et Légumes (CTIFL), Paris, pp 239–252
German S, Candresse T, Lanneau M, Huet J, Pernollet J, Dunez J (1990) Nucleotide sequence and genomic organization of Apple chlorotic leaf spot closterovirus. Virology 179:104–112
German S, Delbos RP, Candresse T, Lannean L, Dunez J (1997) Complete nucleotide sequence of the genome of a severe cherry isolate of Apple chlorotic leaf spot trichovirus (ACLSV). Arch Virol 142:833–841
Guo W, Zheng W, Wang M, Xiaohong L, Ma Y, Dai H (2016) Genome sequences of three Apple chlorotic leaf spot virus isolates from Hawthorns in China. PLoS One 11:e0161099. https://doi.org/10.1371/journal.pone.0161099
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Oxford Journals 41:95–98
Jelkmann W, Kunze L (1995) Plum pseudopox in German plum after infection with an isolate of Apple chlorotic leaf spot virus causing plum line pattern. Acta Hortic 386:122–125
Keshavarz T, Shams-Bakhsh M (2014) Incidence and distribution of Apple chlorotic leaf spot virus in the main fruit growing areas of Iran. Arch Phytopathol Plant Protect 48:306–312
Keshavarz T, Shams-Bakhsh M, Norinejad SH (2009) First report of Apple chlorotic leaf spot virus infection of apple trees in Iran. J Plant Pathol 91:233
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Liu P, Li Z, Song S, Wu Y (2014) Molecular variability of Apple chlorotic leaf spot virus in Shaanxi, China. Phytoparasitica 42:445–454
Mahfoudhi N, Elair M, Moujahed R, Salleh W, Djelouah K (2013) Occurrence and distribution of pome fruit viruses in Tunisia. Phytopathol Mediterr 52:136–140
Marini D, Gibson P, Scott S (2008) The complete nucleotide sequence of an isolate of Apple chlorotic leaf spot virus from peach (Prunus persica (L.) Batch). Arch Virol 153:1003–1005
Mathioudakis MM, Maliogka VI, Katsiani AT, Katis NI (2010) Incidence and molecular variability of apple stem pitting and apple chlorotic leaf spot viruses in apple and pear orchards in Greece. J Plant Pathol 92:139–147
Menzel W, Jelkmann W, Maiss E (2002) Detection of four apple viruses by multiplex RT-PCR assays with co-amplification of plant mRNA as internal control. J Virol Methods 99:81–92
Németh M (1986) Virus, mycoplasma and rickettsia diseases of fruit trees. Akademiai Kiado, Budapest
Niu F, Pan S, Wu Z, Jiang D, Li S (2012) Complete nucleotide sequences of the genomes of two isolates of Apple chlorotic leaf spot virus from peach (Prunus persica) in China. Arch Virol 157:783–786
Paduch-Cichal E, Szyndel MS, Tomala K (2005) Preliminary results of the study on viruses occurring in ‘Mutsu’ apple cultivar trees. Phytopathol Pol 37:87–90
Rana T, Chandel V, Kumar Y, Ram R, Hallan V, Zaidi AA (2010) Molecular variability analyses of Apple chlorotic leaf spot virus capsid protein. J Biosci 35:605–615
Salem N, Mansour A, Al-Musa A (2005) Viruses of pome fruit trees in Jordan. J Plant Pathol 87:123–126
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Laboratory Press, New York
Sato K, Yoshikawa N, Takahashi T (1993) Complete nucleotide sequence of the genome of an apple isolate of Apple chlorotic leaf spot virus. J Gen Virol 74:1927–1931
Song YS, Hong N, Wang LP, Xu WX, Hu HJ, Tian R, Wang GP (2011) Molecular and serological diversity in Apple chlorotic leaf spot virus from sand pear (Pyrus pyrilofia) in China. Eur J Plant Pathol 130:183–196
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Yaegashi H, Isogai M, Tajima H, Sano T, Yoshikawa N (2007) Combinations of two amino acids (Ala40 and Phe75 or Ser40 and Tyr75) in the coat protein of Apple chlorotic leaf spot virus are crucial for infectivity. J Gen Virol 88:2611–2618
Acknowledgements
We would like to thank the Iranian Ministry of Science, Research and Technology, for supporting F. Abtahi’s internship in Italy.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 38 kb)
Rights and permissions
About this article
Cite this article
Abtahi, F., Shams-Bakhsh, M., Safaie, N. et al. Incidence and genetic diversity of apple chlorotic leaf spot virus in Iran. J Plant Pathol 101, 513–519 (2019). https://doi.org/10.1007/s42161-018-00224-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42161-018-00224-z