Abstract
The Vibrio species causing major diseases in Litopenaeus vannamei are Vibrio harveyi, Vibrio alginolyticus, and Vibrio parahaemolyticus. For multiplex PCR primers, YeaD was used to detect the three Vibrio species. Bioinformatic analysis such as MultiPLX and primer-BLAST was used to design stable and species-specific multiplex PCR primers. Multiplex PCR results showed clear band patterns with bands at 185 bp for V. alginolyticus, 396 bp for V. harveyi, 805 bp for V. arahaemolyticus, and 596 bp for common Vibrio species. The minimum concentration of DNA was measured by PCR; the value for V. alginolyticus was 0.1 ng, that of V. harveyi was 0.03 ng, and that of V. parahaemolyticus was 0.003 ng. Taken together, YeaD showed stability and specificity in identifying Vibrio species. Our multiplex PCR amplification method is an effective and inexpensive tool for identifying Vibrio species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Shrimp accounts for 15.3% of internationally traded seafood, comprising the largest share of trades after tuna. There are various species of shrimp including Black Tiger Shrimp (Penaeus monodon), Pacific white shrimp (Litopenaeus vannamei), mantis shrimp (Oratosquilla oratoria), Alvinocaridid shrimps (Zhao et al. 2018; Xue et al. 2018; Wang et al. 2017a; Zhang et al. 2018). Although shrimp are extensively marketed worldwide, they have serious disease problem because antibiotics are not available for all crustaceans that don’t have specific immune system (Lu et al. 2017; Pourmozaffar et al. 2017). For example, Thailand was the world’s largest shrimp producer in 1992, but China, Norway, and Vietnam currently dominate the market because of continuous disease outbreaks since 1997 (FAO 2016). One of the most important diseases caused by gram-negative bacteria is vibriosis, a septicaemic disease transmitted by Vibrio species (Kinne 1993; Gomez-Gil et al. 1998; Soto-Rodriguez et al. 2010). Vibrio species have been studied, for example Pinctada fucata, before and after infection with Vibrio alginolyticus (Wang et al. 2017b). In Korea, Vibrio species were detected in 30% of sick shrimp Litopenaeus vannamei in 2008 (Jung et al. 2012). The Vibrio species causing major vibriosis in L. vannamei are Vibrio harveyi, Vibrio alginolyticus, and Vibrio parahaemolyticus (Vandenberghe et al. 1999; Shanmugasundaram et al. 2015; Min et al. 2015; Heenatigala and Fernando 2016).
These three species cause symptoms such as loose shell syndrome and red disease (Jayasree et al. 2006; Aftabuddin et al. 2017; Biju and Gunalan 2016). However, predominant symptoms caused by the three species differ. Vibrio harveyi predominantly causes loose shell syndrome and white gut syndrome, while V. alginolyticus causes shell disease with black spots. V. parahaemolyticus is responsible for red disease as well as tail necrosis (Jayasree et al. 2006). Vibrio harveyi typically infects adult and post-larval stage shrimp, while V. alginolyticus infects hatchery and larval stage shrimp. V. parahaemolyticus infects juvenile and adult stage shrimp (Vandenberghe et al. 1999). All three Vibrio species fall into the Harveyi clade, whose members are phylogenetically similar, but genetically little different (Sawabe et al. 2013; Sun et al. 2009). Different species can be identified by real-time PCR, microarray, and multiplex PCR methods (Lee et al. 2016; Lan et al. 2016). Multiplex PCR has advantages because it can distinguish species easily and quickly; limitations of the other methods include their costs and technical difficulties (Hossain et al. 2012).
An important aspect of multiplex PCR is selecting a target gene, which should be species-specific, widely distributed, and stable in the genome (Halder et al. 2009). The 16S ribosomal RNA (16S rRNA) gene is commonly used as a target gene for identification. However, sequencing of the 16S rRNA gene cannot distinguish between closely related Vibrio species (Gomez-Gil et al. 2004; Haldar et al. 2007). Therefore, another suitable gene in Vibrio species must be identified for multiplex PCR (Castroverde et al. 2006; Goarant et al. 2007; Kim et al. 2015; Pinto et al. 2005). Among Vibrio species chromosomes, putative d-hexose-6-phosphate mutarotase (YeaD) gene is not registered in the nucleotide database and is only registered in the protein database of NCBI.
YeaD encodes the important enzyme d-hexose-6-phosphate epimerase-like protein, which is involved in galactose metabolism (You et al. 2010). This enzyme converts α-galactose to β-galactose. The YeaD protein shows some differences in the substrate binding pocket, which may be important for enzyme specificity, but the catalytic residues and a few substrate-binding residues in the protein are conserved (Chittori et al. 2007; You et al. 2010).
In this study, YeaD was analysed by bioinformatics methods to evaluate both species-specificity and stability in Vibrio species. Next, YeaD was selected to design multiplex PCR primers for distinguishing V. harveyi, V. alginolyticus, and V. parahaemolyticus, while 16S rRNA gene-based primers were used to detect various bacteria including all Vibrio species. 16S rRNA-based primers reacted to various bacteria, including non-Vibrio species.
Materials and methods
Bacterial strains
Vibrio harveyi (PK-PVH), V. alginolyticus (PK-PVAL), and V. parahaemolyticus (PK-PVP) strains were used in this study. Vibrio vulnificus (PK-PW) and V. mimicus (PK-PVM) were used as controls. DNA of the strains were sourced and extracted from the Department of Fisheries Science, Pukyong National University. DNA were classified by direct sequencing for a specific region; cholera toxin transcriptional activator (toxR) gene for V. harveyi, thermolabile hemolysin (tlh) gene for V. alginolyticus and V. parahaemolyticus, cytolysin (vvh) gene for V. vulnificus. Vibrio parahaemolyticus, V. vulnificus, and V. mimicus were classified again using an API 20E kit (BioMérieux, Marcy-l’Étoile, France). Six unknown Vibrio species were additionally used and classified through direct sequencing (Supplementary Fig. 1).
Primer design
Four primer sets were designed using YeaD and the 16S rRNA gene. YeaD-based primers were used to identify the three major Vibrio species: V. harveyi, V. alginolyticus, and V. parahaemolyticus. The 16S rRNA gene-based primers were designed to detect various bacteria including all Vibrio species. Primers harv-F (5′-GTT CTG CAA GTA TCG ACA ACG-3′) and harv-R (5′-TAG AGC GAG TTC AAC GAT CAC-3′) generated a 396-bp (bp) product. Primers algi-F (5′-CGC CTG AAG GTC AAG AT-3′) and algi-R (5′-CAC ACC GTT ATG GTT CTC AC-3′) generated a 185-bp product. Primers para-F (5′-AAA GAG GCG GCT TGA TAG TC-3′) and para-R (5′-CAA CGT GCG GTT AAG AAC AG-3′) generated an 805-bp product. 16S rRNA gene-based primers universal-F (5′-CCA CAC TGG AAC TGA GAC A-3′) and universal-R (5′-TAA TCT TGC GAC CGT ACT CC-3′) generated a 596-bp product.
PCR assay and sensitivity test
PCR conditions were optimized as follows: 2.5 µL 10× Ex Taq buffer (Takara, Shiga, Japan), 10 pM of each primer (in case of Multiplex PCR, 5 pM of each primer), 2.5 mM each dNTP (Takara), 1.0 U Taq High-fidelity DNA polymerase (Takara), and 10 ng template DNA. PCR amplifications were performed in a thermal cycler (Eppendorf, Hamburg, Germany). Samples were heated to 94 °C for 3 min, followed by 30 cycles as follows: 94 °C for 40 s, 57 °C for 40 s, and 72 °C for 40 s. Cycling was followed by a final extension step at 72 °C for 3 min. The PCR products were subjected to 1.5% agarose gel electrophoresis. Additionally, PCR for checking the minimum concentration of templates was performed using 3, 1, 0.3, 0.1, 0.03, 0.01, and 0.003 ng template. PCR products were analysed quantitatively using the Image-J program (Fig. 1).
Targeting sequence gapA, YeaD, and 16 s rRNA gene
Sequences of chromosome 1 of V. harveyi (strain QT520), V. parahaemolyticus (strain K01M1), and V. alginolyticus (strain CHN25) from NCBI were used to design primers. The location of glyceraldehyde-3-phosphate dehydrogenase A (gapA) gene in chromosome 1 was obtained using another gapA from the three Vibrio species (strains LMG4044, ANC5-1, and LMG2850 in NCBI). Open Reading Frame Finder (ORF Finder) in NCBI was used to identify ORF regions in the gapA sequence (https://www.ncbi.nlm.nih.gov/orffinder/). An ORF of 850–900 bp was identified at the 3′ end of the gapA sequence. SmartBLAST was used to determine which protein is expressed by the ORF. The ORF showed a 100% match to YeaD protein. A region from gapA to YeaD in each Vibrio species was cut out and aligned using BioEdit for primer design. This was also conducted for V. anguillarum (strain NB10), V. campbellii (strain LMB29), V. cholera (strain E1162), V. fischeri (strain MJ11), V. fluvialis (strain ATCC 33809), V. mimicus (strain MB-451_contig43), V. splendidus (strain 5S-101_contig_13), and V. vulnificus (strain FORC_036) for comparison. For 16 s rRNA primer design, V. harveyi (strain LDP 1-1-10), V. alginolyticus (CECT 43), and V. parahaemolyticus (CECT 5305) were aligned by BioEdit.
Bioinformatics analysis
Targeted genes were compared using ClustalW in BioEdit software (http://www.mbio.ncsu.edu/BioEdit/page1.html) (Thompson et al. 1994). Primers were designed by comparing the three Vibrio species (Vibrio harveyi, V. alginolyticus, and V. parahaemolyticus) and eight control Vibrio species (V. anguillarum, V. campbellii, V. cholera, V. fischeri, V. fluvialis, V. mimicus, V. splendidus, and V. vulnificus). All primers for YeaD showed species-specificity. When comparing the strains of three Vibrio species in the YeaD gene sequences, the YeaD gene mutation rate in the homologous strains is low enough to design the primer (Supplementary Fig. 2). Targeted sequences for primer-design were the same in homologous strains. 16S rRNA gene-based primers were used for all strains.
Primer-BLAST program (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) is used to identify the species detected by primers (Ye et al. 2012). Each YeaD primer detected only target species in the program. Each primer corresponds to the sequence of Vibrio harveyi, V. alginolyticus, V. parahaemolyticus registered in NCBI (Supplementary Fig. 3). 16S rRNA-based primers detected various bacteria including 27 Vibrio species, Photobacterium species, and Escherichia coli. Primer-BLAST parameters were changed; Database to ‘nr’ and Organism to ‘Vibrio (taxid: 662)’ or ‘Bacteria (taxid: 2)’.
AutoDimer and MultiPLX 2.1 software (http://bioinfo.ebc.ee/multiplx/) were used to evaluate primer stability such as hairpin and dimer formation (Vallone and Butler 2004; Kaplinski et al. 2004). All designed primers were stable according to the results and could form multiplex primers.
The YeaD and 16S rRNA genes were analyzed by the neighbour-joining method using the MEGA7 program (Saitou and Nei 1987; Kumar et al. 2016). Thirty strains were detected by Primer-BLAST analysis from NCBI (Fig. 2). The nucleotides examined included approximately 1000 bp of 16S rRNA and YeaD. The phylogenetic tree of 30 strains revealed different stability and specificity characteristics between the two genes (Fig. 3).
Vibrio genes for multiplex PCR primers. a Chromosomes photograph of bacteria (Mašková 2011). b and c Two chromosomes present in Vibrio species. One is composed of approximately 3 million bp, while the other is composed of approximately 2 million bp. Other studies examining Vibrio detected gyrB, 16 s rRNA, collagenase, rpoB, toxR, tlh, and hutA. Their approximate locations on the chromosomes are indicated. The 16S rRNA gene and YeaD used in the study are shown on the chromosomes. 16S rRNA has several homologous genes. d The sizes of 16S rRNA and positions of the universal F and universal R primers. e The harv-F, harv-R, algi-F, algi-R, para-F, and para-R primers indicate the position detected by each. Para-F is located slightly outside of YeaD
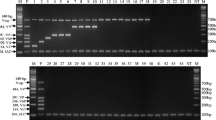
Multiplex PCR with multiple samples and primer sets. a Lane M—100-bp marker; lane 1—V. harveyi, V. alginolyticus, and V. parahaemolyticus with all primers (harv-F, harv-R, algi-F, algi-R, para-F, para-R, universal-F, and universal-R). b Lane M—100-bp; lane 1—Vibrio vulnificus with all primers; lane 2—V. mimicus with all primers; lane 3—V. harveyi with harv-F, and R; lane 4—V. alginolyticus with algi-F, and R; lane 5—V. parahaemolyticus with para-F, and R; lane 6—V. harveyi and V. alginolyticus with harv-F, R, algi-F, and R; lane 7—V. alginolyticus and V. parahaemolyticus with algi-F,R, para-F, and R; lane 8—V. harveyi and V. parahaemolyticus with harv-F, R, para-F, and R; lane 9 - V. harveyi, V. alginolyticus, and V. parahaemolyticus with harv-F, R, algi-F, R, para-F, and R
Results
PCR amplification and multiplex PCR
PCR was performed with each primer set according to the PCR assay conditions. PCR analysis of the genomic DNA of V. alginolyticus, V. harveyi, and V. parahaemolyticus indicated fragment sizes of 185 bp, 396 bp and 805 bp, respectively. A band at 596 bp appeared when using the universal primers for the 16S rRNA gene in all three Vibrio species.
Multiplex PCR was performed with multiple samples and primers (Fig. 4). Three Vibrio species with all primer sets obtained clear 396-bp, 185-bp, 805-bp, and 596-bp bands (Fig. 4a, lane 1). V. vulnificus and V. mimicus control samples were inserted with all primer sets (harv-F, harv-R, algi-F, algi-R, para-F, para-R, universal-F, and universal-R), but only a 596-bp band was observed for the 16S rRNA gene to detect common bacteria (Fig. 4b, lanes 1 and 2). Additional multiplex PCR was performed on unknown Vibrio species along with identification by direct sequencing, and the same results were obtained (Supplementary Fig. 1).
Sensitivity test by PCR to determine the template concentration detection limit of V. alginolyticus (a), Vibrio harveyi (b), and Vibrio parahaemolyticus (c). Each lane represents sample concentration as follows; lane M—100-bp marker; lane 1—3 ng; lane 2—1 ng; lane 3—0.3 ng; lane 4—0.1 ng; lane 5—0.03 ng; lane 6—0.01 ng; 7—0.003 ng. PCR products were analysed quantitatively using the Image-J program
Sensitivity test for PCR
PCR was performed to determine how many nanograms of template could be recognized. PCR detected up to 0.1 ng in V. alginolyticus, 0.03 ng in V. harveyi, and 0.003 ng in V. parahaemolyticus (Fig. 1).
Phylogenetic analysis
The phylogenetic tree for the 16S rRNA gene and YeaD was constructed with MEGA 7 software (Fig. 2). Sequences for phylogenetic tree contained approximately 1000 bp from the 16S rRNA gene and YeaD. All samples were registered in NCBI and recognized by designed primers using primer-BLAST. For the 16S rRNA gene, it was difficult to classify V. parahaemolyticus ATCC 17802 or V. alginolyticus ATCC 33787, K09K1, and ZJ-T from other species. In contrast, for YeaD, the sequence difference between species of V. alginolyticus, V. harveyi, and V. parahaemolyticus was remarkable, indicating that three species samples showed clear separation in phylogeny tree.
Discussion
Multiplex PCR is useful for rapid and accurate measurement if reliable primers are used (Wei et al. 2014). Multiplex PCR can detect template concentrations as low as 0.1 ng in Vibrio alginolyticus, 0.03 ng in V. harveyi, 0.003 ng in V. parahaemolyticus. The copy numbers of a template can be calculated through nanograms of DNA considering the 5.2 million bp of Vibrio chromosomes and 650 daltons per base pair (Staroscik 2004; Danna et al. 1973). 0.1 ng means 17,800 templates, 0.03 ng means 5340 templates, and 0.003 ng means 534 templates. Since Vibrio has a generation time of 27 min in the glucose medium and 19 min in the LB medium, 2.15 × 29 Vibrio species in the LB medium and 4.20 × 26 Vibrio species in the glucose medium are obtained in 1 colony after 10 h of culture (Stokke et al. 2011). Therefore, a single colony is fully identifiable when extracted from Litopenaeus vannamei. In preceding experiments, V. harveyi, V. alginolyticus, and V. parahaemolyticus were extracted and cultured from L. vannamei showing vibriosis symptoms. The Vibrio species is extractable from stomach, hepatopancreas, haemolymph, muscle, and gut of L. vannamei (Sirirustananun et al. 2011; Yatip et al. 2018; Ananda et al. 2017). In the case of V. alginolyticus, 6 × 106 cfu shrimp-1 was obtained in the ventral sinus of L. vannamei on a challenge test (Sirirustananun et al. 2011).
To be reliable, the primers must recognize all strains of the same species without detecting other species. Bioinformatics analysis by primer-BLAST in NCBI supported the reliability of our primers (Supplementary Fig. 3). No Vibrio species were identified other than the target species among all NCBI strains using the designed-primers. Only a few other Vibrio species were searched but not identified because they differed by more than five nucleotides from the primers. In addition, all but one of the 17 V. parahaemolyticus, 11 V. alginolyticus, and 3 V. harveyi strains registered in NCBI were detected, except for 1 V. alginolyticus. However, the 1 V. alginolyticus (strain K08M4) is registered as V. splendidus in the BioProject section of NCBI. Thus, except for the confounding strain, 100% matching was achieved. This suggests that the designed primers are reliable and induce low mutation rate.
For YeaD, there was a distinct sequence difference between all Vibrio species used for comparison (Supplementary Fig. 2). However, YeaD gene sequences of the same species were well-conserved. Although YeaD is the essential gene involved in galactose metabolism, it remains unclear how YeaD functions using different sequences for each species (You et al. 2010). We initially attempted to construct primers for gapA. Although the three species were distinguished from each other using gapA, it was more convenient to construct primers for YeaD to compare all Vibrio species than gapA. Primers for YeaD clearly identified the different species. Stability of YeaD gene suggests to be genetic marker to identify gram-negative bacteria.
Litopenaeus vannamei is predominantly related to V. harveyi, V. alginolyticus, and V. parahaemolyticus based on previous infection rates and association studies. In the past, Anguirre-Guzmán et al., selected V. harveyi, V. alginolyticus, V. parahaemolyticus, and V. penaeicida as potential pathogenic Vibrio species of L. vannamei (Aguirre-Guzmán et al. 2001). In Ecuador and Mexico research, 90 isolates of diseased L. vannamei were found to have 42 Vibrio harveyi (46.7%), 14 V. alginolyticus (15.6%), and 8 V. parahaemolyticus (8.9%). 4 P. damselae (4.4%), 1 V. mimicus (1.1%) were also found in small amounts (Johannessen et al. 1999). In India, V. parahaemolyticus (83.4%) was found in the hepatopancreas of infected L. vannamei without identification of V. alginolyticus and V. harveyi (Shanmugasundaram et al. 2015). In shrimp culture ponds of Sri Lanka, V. parahaemolyticus (55.5%), V. alginolyticus (27.7%), V. damsela (5.6%), V. anguillarum (5.6%), unidentified Vibrio (5.6%) were found without identification of V. harveyi (Heenatigala and Fernando 2016). To find out association between L. vannamei and Vibrio species in research, we counted the number of research. The number of research associated with Vibrio and L. vannamei were 262 in PubMed. Among them, 45 papers related to V. harveyi (17.2%), 106 papers related to V. alginolyticus (40.5%), and 80 papers related to V. parahaemolyticus (30.5%). There were also 15 papaers of V. anguillarum (5.7%), 11 papers of V. campbellii (4.2%), 4 papers of V. penaeicida (1.5%). So, V. harveyi, V. alginolyticus and V. parahaemolyticus are major pathogens and high association with L. vannamei.
In addition to Vibrio harveyi, V. alginolyticus and V. parahaemolyticus, the other Vibrio species also exist in diseased Litopenaeus vannamei. Fourteen Vibrio species are reported to be infected in cultured shrimp; Vibrio harveyi, V. splendidus, V. parahaemolyticus, V. alginolyticus, V. anguillarum, V. vulnificus, V. campbellii, V. fischeri, V. damsella, V. pelagicus, V. orientalis, V. ordalii, V. menditerrani, V. logei (Annam 2015). According to a infect rate research in Ecuador and Mexico, the other Vibrio species can be detected with a probability of 28.9%. 16S rRNA gene-based primers only detect the other Vibrio species without identification. If 16S rRNA gene-based primers detect bacteria, direct sequencing or other PCR techniques can identify the undefined Vibrio species.
References
Aftabuddin S, Roman WU, Hasan CK, Ahmed M, Rahman H, Siddique MAM (2017) First incidence of loose-shell syndrome disease in the giant tiger shrimp Penaeus monodon from the brackish water ponds in Bangladesh. J Appl Anim Res 46:1–8
Aguirre-Guzmán G, Vázquez-Juárez R, Ascencio F (2001) Differences in the susceptibility of American white shrimp larval substages (Litopenaeus vannamei) to four Vibrio species. J Invertebr Pathol 78:215–219
Ananda RR, Sridhar R, Balachandran C, Palanisammi A, Ramesh S, Nagarajan K (2017) Pathogenicity profile of Vibrio parahaemolyticus in farmed Pacific white shrimp, Penaeus vannamei. Fish Shellfish Immunol 67:368–381
Annam RA (2015) Analysis of engine test and emission test of seaweed biodiesel for sustainable energy. J Chem Pharm Res 7(2):755–760
Biju VN, Gunalan B (2016) Prevalence of Vibrio infection in Penaeus (Litopenaeus) vannamei farms. Int J Sci Invent Today 5:485–493
Castroverde CDM, Luis BBS, Monsalud RG, Hedreyda CT (2006) Differential detection of vibrios pathogenic to shrimp by multiplex PCR. J Gen Appl Microbiol 52:273–280
Chittori S, Simanshu DK, Savithri HS, Murthy MRN (2007) Structure of the putative mutarotase YeaD from Salmonella typhimurium: structural comparison with galactose mutarotases. Biol Crystallogr 63:197–205
Danna KJ, Sack GH, Nathans D (1973) Studies of Simian virus 40 DNA. VII. A cleavage map of the SV40 genome. J Mol Biol 78:363–376
FAO (2016) The state of world fisheries and aquaculture. Food and Agriculture Organization of the United Nations, Rome
Goarant C, Reynaud Y, Ansquer D, de Decker S, Merien F (2007) Sequence polymorphism-based identification and quantification of Vibrio nigripulchritudo at the species and subspecies level targeting an emerging pathogen for cultured shrimp in New Caledonia. J Microbiol Methods 70:30–38
Gomez-Gil B, Tron-Mayen L, Roque AF, Turnbull J, Inglis V, Guerra-Flores AL (1998) Species of Vibrio isolated from hepatopancreas, haemolymph and digestive tract of a population of healthy juvenile Penaeus vannamei. Aquaculture 163:1–9
Gomez-Gil B, Soto-Rodriguez S, Garcia-Gasca A, Roque A, Vazquez-Juaarez R, Thompson FL, Swings J (2004) Molecular identification of Vibrio harveyi-related isolates associated with diseased aquatic organisms. Microbiology 150:1769–1777
Haldar S, Chatterjee S, Asakura M, Vijayakumaran M, Yamasaki S (2007) Isolation of Vibrio parahaemolyticus and Vibrio cholerae (Non-O1 and O139) from moribund shrimp (Penaeus monodon) and experimental challenge study against post larvae and juveniles. Ann Microbiol 57:55–60
Halder S, Neogi SB, Kogure K, Chatterjee S, Chowdhury N, Hinenoya A, Asakura M, Yamasaki S (2009) Development of a haemolysin gene-based multiplex PCR for simultaneous detection of Vibrio campbellii, Vibrio harveyi. and Vibrio parahaemolyticus. Lett Appl Microbiol 50:146–152
Heenatigala PPM, Fernando MUL (2016) Occurrence of bacteria species responsible for vibriosis in shrimp pond culture systems in Sri Lanka and assessment of the suitable control measures. Sri Lanka J Aquat Sci 21:1–17
Hossain MT, Kim EY, Kim YR, Kim DG, Kong IS (2012) Development of a groEL gene–based species specific multiplex polymerase chain reaction assay for simultaneous detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus. J Appl Microbiol 114:448–456
Jayasree L, Janakiram P, Madhavi R (2006) Characterization of Vibrio spp. Associated with diseased shrimp from culture ponds of Andhra Pradesh. J World Aquac Soc 37:523–532
Jung SH, Choi HS, Do JW, Kim SM, Kwon MG, Seo JS, Jee YH, Kim SR, Cho YR, Kim JD, Park MA, Jee BY, Cho MY, Kim JW (2012) Monitoring of bacteria and parasites in cultured olive flounder, black rockfish, red sea bream and shrimp during summer period in Korea from 2007 to 2011. J Fish Pathol 25:231–241
Johannessen J-Ar, Olsen B, Olaisen J (1999) Aspects of innovation theory based on knowledge-management. Int J Inf Manage 19(2):121–139
Kaplinski L, Andreson R, Puurand T, Remm M (2004) MultiPLX: automatic grouping and evaluation of PCR primers. Bioinform Appl Note 21:1701–1702
Kinne RK (1993) The role of organic osmolytes in osmoregulation: from bacteria to mammals. J Exp Zool Suppl 265(4):346–355
Kim HJ, Ryu JO, Lee SY, Kim ES, Kim HY (2015) Multiplex PCR for detection of the Vibrio genus and five pathogenic Vibrio species with primer sets designed using comparative genomics. BioMed Cent Microbiol. https://doi.org/10.1186/s12866-015-0577-3
Kim JK, Lee JB, Huh YR, Jang HA, Kim CH, Yoo JW, Lee BL (2015) Burkholderia gut symbionts enhance the innate immunity of host Riptortus pedestris. Dev Comp Immunol 53(1):265–269
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Evolut Genet Anal 33:1870–1874
Lan D, Lin B, Xiong X, Xiaonong Y, Li J (2016) Identification and characteristics analysis of toll-like receptors family genes in yak. Genes Genom 38:429–438
Lee TW, Delongchamp RR, Kim WK, Reis RJS (2016) Use of p-value plots to diagnose and remedy problems with statistical analysis of microarray data. Genes Genom 38:45–52
Lu X, Luan S, Cao B, Meng X, Sui J, Dai P, Luo K, Shi X, Hao D, Han G, Kong J (2017) Estimation of genetic parameters and genotype-by-environment interactions related to acute ammonia stress in Pacific white shrimp (Litopenaeus vannamei) juveniles at two different salinity levels. PLoS ONE. https://doi.org/10.1371/journal.pone.0173835
Mašková J (2011) Supercoiling (supercoiling of E. Coli). http://www.wikilectures.eu/w/File:Supercoiling.jpg#filelinks
Min JR, Na K, Chong HJ, Jeong HS (2015) Bactericidal efficacy of a monopersulfate compound against Vibrio harveyi and toxicity to Litopenaeus vannamei. Korean J Fish Aquat Sci 48:5 661–667
Pinto AD, Ciccarese G, Tantillo G, Catalano D, Forte VT (2005) A collagenase-targeted multiplex PCR assay for identification of Vibrio alginolyticus, Vibrio cholerae, and Vibrio parahaemolyticus. J Food Prot 68:150–153
Pourmozaffar S, Hajimoradloo A, Miandare HK (2017) Dietary effect of apple cider vinegar and propionic acid on immune related transcriptional responses and growth performance in white shrimp, Litopenaeus vannamei. Fish Shellfish Immunol 60:65–71
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sawabe T, Ogura Y, Matsumura Y, Feng G, Amin AKMR, Mino S, Nakagawa S, Sawabe T, Kumar R, Fukui Y, Satomi M, Matsushima R, Thompson FL, Gomez-Gil B, Christen R, Maruyama F, Kurokawa K, Hayashi T (2013) Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front Microbiol 4:414, 1–14
Shanmugasundaram S, Mayavu P, Manikandarajan M, Suriya A, Anbarasu ER (2015) Isolation and identification of Vibrio sp. in the Hepatopancreas of cultured white pacific shrimp (Litopenaeus vannamei). Int Lett Nat Sci 46:52–59
Sirirustananun N, Chen JC, Lin YC, Yeh ST, Liou CH, Chen LL, Sim SS, Chiew SL (2011) Dietary administration of a Gracilaria tenuistipitata extract enhances the immune response and resistance against Vibrio alginolyticus and white spot syndrome virus in the white shrimp Litopenaeus vannamei. Fish Shellfish Immunol 31:848–855
Soto-Rodriguez SA, Gomez-Gil B, Lozano R (2010) ‘Bright-red’ syndrome in Pacific white shrimp Litopenaeus vannamei is caused by Vibrio harveyi. Dis Aquat Org 92:11–19
Staroscik A (2004) Calculator for determining the number of copies of a template. http://www.uri.edu/research/gsc/resources/cndna.html
Stokke C, Waldminghaus T, Skarstad K (2011) Replication patterns and organization of replication forks in Vibrio cholerae. Microbiology 157:695–708
Sun K, Hu YH, Zhang XH, Bai FF, Sun L (2009) Identification of vhhP2, a novel genetic marker of Vibrio harveyi, and its application in the quick detection of V. harveyi from animal specimens and environmental samples. J Appl Microbiol 107:1251–1257
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple; sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid Res 22:4673–4680
Vallone PM, Butler JM (2004) AutoDimer: a screening tool for primer-dimer and hairpin structures. Biotechniques 37:226–231
Vandenberghe J, Verdonck L, Robles-Arozarena R, Rivera G, Bolland A, Balladares M, Gomez-Gil B, Calderon J, Sorgeloos P, Swings J (1999) Vibrios associated with Litopenaeus vannamei larvae, postlarvae, broodstock, and hatchery probionts. Appl Environ Microbiol 65:2592–2597
Wang Z, Shi X, Sun L, Bai Y, Zhang D, Tang B (2017a) Evolution of mitochondrial energy metabolism genes associated with hydrothermal vent adaption of Alvinocaridid shrimps. Genes & Genomics 39(12):1367–1376
Wang Z, Wang B, Chen G, Lu Y, Jian J, Wu Z (2017b) Identification and comparative analysis of the pearl oyster Pinctada fucata hemocytes microRNAs in response to Vibrio alginolyticus infection. Genes & Genomics 39(10):1069–1081
Wei S, Zhao H, Xian Y, Hussain MA, Wu X (2014) Multiplex PCR assays for the detection of Vibrio alginolyticus, Vibrio parahaemolyticus, Vibrio vulnificus, and Vibrio cholerae with an internal amplification control. Diagn Microbiol Infect Dis 79:115–118
Xue M, Wu L, He Y, Liang H, Wen C (2018) Biases during DNA extraction affect characterization of the microbiota associated with larvae of the Pacific white shrimp. Peer J 6:e5257
Yatip P, Nitin CTD, Flegel TW, Soowannayan C (2018) Extract from the fermented soybean product Natto inhibits Vibrio biofilm formation and reduces shrimp mortality from Vibrio harveyi infection. Fish Shellfish Immunol 72:348–355
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13:134
You W, Qiu X, Zhang Y, Ma J, Gao Y, Zhang X, Niu L, Teng M (2010) Crystallization and preliminary Xray diffraction analysis of the putative aldose 1-epimerase YeaD from Escherichia coli. Struct Biol Cryst Commun 66:951–953
Zhao C, Fan S, Qiu L (2018) Identification of MicroRNAs and Their Target Genes Associated with Ovarian Development in Black Tiger Shrimp (Penaeus monodon) Using High-Throughput Sequencing. Scientific Reports 8(1):11602
Zhang Y, Han Z, Gao T, Shi H (2018) Genetic structure analysis of mantis shrimp Oratosquilla oratoria based on mitochondrial DNA control region sequence. Genes & Genomics 40(9):1001–1009
Acknowledgements
This research was a part of the project titled “Omics based on fishery disease control technology development and industrialization (20150242),” funded by the Ministry of Oceans and Fisheries, Korea.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, YJ., Jo, A., Kim, SW. et al. Multiplex PCR using YeaD and 16S rRNA gene to identify major pathogens in vibriosis of Litopenaeus vannamei. Genes Genom 41, 35–42 (2019). https://doi.org/10.1007/s13258-018-0736-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-018-0736-7