Abstract
Plants being sessile are constantly exposed to several stresses, which involve different types of abiotic and biotic stress factors. Biotic stress in plants is caused by various living organisms called plant pathogens including bacteria, viruses, fungi and parasites. Among these pathogens, plant viruses cause severe damage to world agricultural productivity. The reason behind such widespread destruction caused by viruses is their ability to frequently evolve them through mutation and genetic recombination, to succeed over the unfavourable conditions. The virus infects both susceptible and tolerant/resistant plants by the similar and systematic manner but resistant/tolerant plants combat the virus spread and suppress the viral growth. When pathogen enters the plant system, diverse defense responses are initiated which are mediated by plant disease resistance genes (R genes) mediated resistance and hormone based signaling pathways which restrict the viral spread by initiating hypersensitive response. To further enhance our knowledge regarding resistance mechanisms, the virus infection pattern and interactions of virus within resistant and susceptible plants needs to be analysed. At present, most successful strategy involves deployment of crops possessing resistance/tolerance against viruses with the foremost interest of detecting genes associated with resistance or recovery. Among several plant viruses, ‘Geminiviruses’ are the most devastating. In this article we have provided a comprehensive overview of Tomato leaf curl New Delhi virus (ToLCNDV), a member of family Geminiviridae and the plant defense system initiated against this virus. The evaluation of ToLCNDV infection in a variety of hosts differing in their tolerance and identification of differentially expressed genes would be helpful in speculating the threats associated with similar begomoviral invasions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Solanaceae is economically the most important family and are highly divergent in regard to habitat and morphology [27]. Tomato is considered as protective food based upon the significant amount of nutrients it provides which are essential for human health. Tomato yield has been affected by more than 200 diseases caused by fungi, bacteria, viruses, and nematodes worldwide. It was reported to serve as a key host crop for 24 fungi, 7 bacteria, 10 viruses, 3 viroids, and multiple nematodes which indicate its significance as a model plant for studying the molecular mechanisms governing plant-pathogen interactions [20]. Viral diseases are major limiting factor in plant cultivation and extremely difficult to control or eradicate. About 136 viral species have been described to infect tomato crops, which is notably higher to any other vegetable crop. Among that Tomato yellow leaf curl virus (TYLCV; Begomovirus), Tomato spotted wilt virus (TSWV; Tospovirus) Pepino mosaic virus (PepMV; Potexvirus), Tomato mosaic virus (TMV; Tobamovirus) and Tomato torrado virus (ToTV; Torradovirus) are the most important emerging viruses causing severe diseases to tomato [32]. The members of Geminiviridae family have caused severe diseases and yield loss in tomato. The symptoms of virus infection in host plants include yellow mosaic, upward curling of leaf margin, vein swelling, decreased leaflet area, and plant stunting. Amongst the geminivirus infecting tomato, Tomato leaf curl New Delhi virus (ToLCNDV) has been reported to hinder the agricultural production in a broad geographical regions of the world comprising Middle and Far East, Europe, North Africa and predominant in northern India. In recent past, comprehensive research has been conducted related to virus genome, encoded proteins and their functions, genetic variability and plant-virus interactions. Depending on the plant-virus interaction, several strategies have been proposed to control/ reduce the virus spread. Still the sources of resistance against ToLCNDV are limited. Therefore, there is an immediate need to identify the mechanism that leads to the natural resistance in the wild relatives against ToLCNDV. This review highlights our current knowledge of ToLCNDV infection, interaction with host plants and significance of these associations.
Tomato leaf curl New Delhi virus
The family Geminiviridae consists of seven genera, namely, Becurtovirus, Begomovirus, Mastrevirus, Curtovirus, Eragrovirus, Topocuvirus and Turncurtovirus, which are characterized by single stranded circular deoxyribonucleic acid (ssDNA) genomes encapsidated in a geminate shape particle. Geminivirus has a small genome (2.5–3.0 kb), comprised of either one (monopartite) or two circular ssDNA molecules (bipartite) and are transmitted by specific insect vectors [31]. Amongst all the members, Begomoviruses cause a significant loss in yield of various economically important dicotyledonous crops and are transmitted through whitefly (Bemisia tabaci). It is the largest genus of Geminiviridae family containing 322 species. The members of this genus can be either monopartite or bipartite. They encode for 7–8 proteins on both virion and complementary strands. Depending on the phylogeny begomoviruses can be distinguished into two groups: Old World (OW) and New World (NW). These groups are genetically distinct as NW viruses entail bipartite genome, whereas the OW begomoviruses comprise either a monopartite along with satellite DNAs or bipartite genome [22]. Moreover, OW viruses are genetically more diverse and additionally have a conserved gene, AV2/V2 (pre-coat protein) in DNA-A which is not encoded by the NW begomoviruses [91]. As it has been reported that begomoviruses have higher tendency to recombination and attaining new DNA components, thus they are extremely vulnerable to evolutionary process, leading to emergence of new virulent strains. ToLCNDV is a bipartite begomovirus and is naturally transmitted via whitefly species (Bemisia tabaci; order: Hemiptera). Due to its association with severe diseases in economically and socially relevant crops, researchers have made tremendous progress to illustrate and understand the biology of these viruses.
Geographical distribution, host range and virus–vector interaction
ToLCNDV was recognized around 20 years ago [72] in solanaceous crops in India. Initially it was reported to infect tomato plants particularly in several countries of Asia [35, 64, 73]. Recently, it was identified to be distributed in areas of southern Italy [76], Tunisia [65], Iran [121] and Spain [45]. In India, ToLCNDV majorly affects tomato yield in Andhra Pradesh, Delhi, Gujarat, Haryana, Maharashtra, Punjab, Uttar Pradesh, and West Bengal.
The host of ToLCNDV are mainly the members of Solanaceae family such as tomato causing Tomato leaf curl disease (ToLCD) [123], potato causing potato apical leaf curl disease [13], eggplant causing yellow mosaic disease [84], pepper [35] and tobacco [1]. However, recently it was also reported to infect cucurbit crops in Murai, Spain [76, 105, 110]. ToLCNDV strains have been isolated from cucumber, bottle gourd and muskmelon in Thailand, bitter guard in Pakistan, zucchini squash in Spain and Italy [45], Luffa, pumpkin and ash gourd (Benincasa hispida) in India. ToLCNDV also infects plants belonging to Malvaceae, Fabaceae, Phyllanthaceae, and Papaveraceae families. In Malvaceae family it causes yellow vein mosaic virus in okra and cotton leaf curl disease in cotton. In Fabaceae it infects soybean (Glycine max), opium poppy (P. somniferum) from Papaveraceae and sweet leaf (S. androgynous) from Phyllanthaceae family. The infection by ToLCNDV is evidenced by abnormality in tomato, such as stunted or dwarfed growth, leaflets are curled upwards and inwards, slightly chlorotic and yellowish in texture and are stiff, thicker than normal, crumpling, and mosaic/mottling symptoms [104]. ToLCNDV is naturally transmitted via whitefly species (Bemisia tabaci). Bemisia tabaci exists in tropical and subtropical regions and is a major threat to agronomically important plants. Bemisia tabaci consists of cryptic species complex and 37 discrete species of whiteflies have been recognized based on the DNA markers (mitochondrial cytochrome oxidase I; mtCOI) [17] and these species vary in range of infecting hosts, insecticide resistance, and their ability to transmit virus. Middle East-Asia Minor 1 (MEAM-1) (previously known as B biotype), Asian 1 and Asia II species of B. tabaci have been reported to transmit the virus worldwide. Contrastingly, Mediterranean Q1 (MED Q1) species were observed to cause ToLCNDV epidemics specifically in southern Spain in various crops viz tomato, zucchini, and melon. Initially, through its stylet, insect acquires or and inoculate the virus from the plant phloem and the acquisition access period (AAP) and inoculation access period (IAP) varies from 10 to 60 min. In case of B. tabaci infecting tomato plants the AAP and IAP was 30 min [11]. It transmits virus in a persistent, circulative manner with a latent period of 8–21 h during which the virus circulates from digestive tract of insect to the salivary gland from where with the help of stylet viruses are transmitted with the saliva into the plant. This translocation of ToLCNDV is facilitated by a protein termed as midgut protein. Since, ToLCNDV has been reported to infect crops other than Solanaceae family and the information for B. tabaci-ToLCNDV interactions is limited, thus the significance of host specificity of B. tabaci and ToLCNDV during infection need to be studied.
Genome organization and function of virus-encoded proteins
ToLCNDV genome consists of two genomic components; DNA-A which encodes for six ORFs i.e., AV1/CP; coat protein and AV2; pre-coat protein, AC1/Rep; replication initiation protein, AC2/TrAP; transcription activator protein, AC3/REn; replication enhancer and AC4; pathogenesis related protein. On the other hand, DNA-B has two ORFs which encodes for BV1/NSP, nuclear shuttle protein and BC1/MP, movement protein [22]. Apart from mono- or bi-partite genome, presence of an additional molecule i.e., satellite DNA has also been reported in geminiviruses. Amongst the bipartite ToLCNDV 44% were found to be associated with betasatellites [47].
Virus-encoded proteins
Coat Protein (AV1/CP) is the only structural protein of geminiviruses and forms twinned virion structure required to encapsidate the new viral genome. It has the ability to bind ssDNA and dsDNA and regulates viral replication by down-regulating the nicking and closing activity of Rep [10, 60]. It accumulates in the nucleolus and facilitates viral DNA shuttle from nucleus to cytoplasm in monopartite begomovirus. In bipartite viruses, CP has been reported to specifically function in the nuclear shuttling in the mutant virus of nuclear shuttle protein [37]. It is a late expressing gene and the major determinant in viral transmission. In contrast to CP, AV2 protein promotes Rep activities by enhancing its nicking activity [90]. In tomato plants infected with AC2 mutant clones of Tomato leaf curl virus (ToLCV) and ToLCNDV, reduction in the viral DNA accumulation was reported [71].
Rep protein is highly conserved and it is a prerequisite for initiation of RCR [53]. It has domains for nicking, DNA-binding, oligomerization and ATPase activity. During RCR and transcription of viral genome, it recognizes conserved nonanucleotide TAATATT↓AC within a hairpin loop of the plus-strand and initiates DNA cleavage/ligation process [24]. Rep also plays an essential role in activating the host replication machinery by interacting with the plant cell cycle regulatory elements. Rep interacts with retinoblastoma-related protein (RBR) [29] and activates E2F-family transcription factors which in turn removes the cell cycle arrest. Apart from this, Rep interacts with replication factor C (RFC) complex and minichromosome maintenance complex component 3 (MCM3) and proliferating cell nuclear antigen (PCNA) to aid the host DNA replication event [3, 5, 58]. In transgene-based approach, considering its critical roles in the genome replication and interactions with multiple proteins, Rep is a potential target for the generation of geminivirus resistant plant.
Moreover, AC2 and AC3 are transcribed together to produce a dicistronic messenger RNA (mRNA) and their promoter sequence is encoded within the coding sequence of Rep gene [102]. Initially, AC2 was identified as a transcription activator which interacts with DNA-binding protein PEAPOD2 (PPD2), and binds onto the CP promoter to activate its transcription [51]. Further, it was revealed that AC2 acts as the suppressor of silencing activity, both Post-transcriptional gene silencing (PTGS) and Transcriptional gene silencing (TGS). AC3 enhances the viral DNA replication in association with AC1 and PCNA. AC3 interplay with Rep and enhances its ATPase activity, thus facilitating the unwinding of viral DNA [79]. In tomato, REn has been reported to interact with NAC1 (NAM, ATAF, and CUC1) which assist in viral DNA replication [98] and increase the viral titre.
Simultaneously, geminiviruses encode a small protein AC4 that completely overlaps AC1 transcript. AC4 has been evidenced to be engaged in virus movement, virus replication and as a suppressor of PTGS mechanism of host plant. In Curtovirus, (Beet severe curly top virus; BSCTV) disruption of C4 did not affect the virus replication within the infected N. benthamiana leaf but inhibited the spread of virus in the new emerging leaves [46]. In N. benthamiana, C4 was found to bind with ssDNA and dsDNA of virus and help in its movement [109]. Simultaneously, it was reported that C4 induced the expression level of cell-cycle related genes such as PCNA, Cyclin-dependent kinases (CDKs) and cyclins (cyc1, cyc2, and cyc2b), and down-regulated the expression of CDK inhibitor (ICK1) and the retinoblastoma-related protein (RBR1), resulting in virus replication, accumulation and symptom development [77].
Likewise, AC5 ORF has been annotated in various geminiviruses. It is encoded by downstream region of AC3 and covers a portion of coding region of CP. Its functions may vary amongst different viruses. In Mungbean yellow mosaic India virus (MYMIV), AC5 facilitates viral infection and movement, whereas in Tomato Chlorotic Mottle Virus (ToCMoV), Watermelon Chlorotic Stunt Virus (WmCSV) and Tomato Leaf Deformation Virus (TLDV), AC5 is non-essential for viral-induced pathogenesis [63]. Further, the DNA-B encoded Nuclear shuttle protein (BV1/NSP) and Movement protein (BC1, MP) are prerequisite for movement of virus. NSP is required for shuttling of newly synthesized viral DNA from nucleus to cytoplasm. The plant acetyltransferase (AtNSI; Arabidopsis thaliana nuclear shuttle protein interactor) associates with NSP and assists in nuclear export of viral DNA [62]. NSP acts as pathogenicity determinant by interacting and inhibiting the NSP-interacting kinases (NIKs)-mediated antiviral responses and increases the susceptibility of plants to geminivirus infection [23]. Additionally, MP actively binds with NSP-viral DNA complex and participates in cell-to-cell movement of virus through plasmodesmata [26]. Further NSP, MP, viral DNA and Histone 3 form a complex which aid in cell-to-cell trafficking of the viral genome [124]. It interacts with plant protein such as Arabidopsis regulator of endocytosis, synaptotagmin (SYTA) and heat shock cognate 70 kDa protein (Hsc70), which facilitates the viral DNA movement between the cells. Further, MPs also act as virulence factor of bipartite begomoviruses and mutation at its 3′ region has been linked with symptom development [26].
ToLCNDV multiplies by a specific mechanism known as rolling circle replication (RCR) within the plant nucleus utilizing the host proteins such as primase and polymerase to produce a dsDNA replicative form (RF) of viral genome [31]. DNA-A and DNA-B are transcribed in bi-directional manner and are separated by a non-coding region termed as common region (CR) or Intergenic region (IR) of 180 to 200 nt. This region consists of origin of replication (Ori) required for rolling circle replication and promoter elements such as TATA boxes and sequences recognized by various transcription factors including ‘AG’ motif and G-box [31]. IR comprises a conserved hairpin motif which encodes a consensus AT-rich nonanucleotide sequence, 5’-TAATATTAC-3’, in the loop where Rep-mediated nicking initiates RCR. IR consists of repeated upstream iterative sequence motifs known as ‘iterons’ for binding of Rep proteins. The region between repeats of iteron and stem loop is called as origin of replication or ori.
Suppressors of plant defense mechanism
Apart from assisting in virus replication and movement, virus proteins have evolved to act as suppressors of plant defense mechanism, thus inhibiting the response of plant against viral infection. Against DNA viruses, RNA interference (RNAi)-mediated silencing machinery is the major defense mechanism. Several virus encoded proteins have been reported to play key roles in suppressing host defences specifically targeting the silencing machinery. Initially, AC2 was found to act as the suppressor of silencing activity, both PTGS and TGS. AC2 interact with Adenosine kinase (ADK), enzyme required to generate S-adenosyl methionine (SAM) and inhibit the methylation cycle [117]. Further, it attenuates PTGS by inhibiting the activity of RNA dependent RNA polymerase 6 (RDR6), Argonaute1 (AGO1) [49] and calmodulin like protein (rgs-CaM) [122] proteins which are required for siRNA biogenesis thus making the host plant susceptible for virus infection. Besides AC2, AV2 was hypothesized to function in suppressing the PTGS mediated defense mechanism in host plant [126]. It was demonstrated that AV2 interacts with SGS3 which prevents conversion of ssRNA to dsRNA, thus inhibits the production of siRNAs [25]. AV2 also suppresses the TGS in plants but the mechanism is still not elucidated [116]. Another study highlighted the role of AC3 in controlling the virus-induced gene [78] but the exact mechanism is still unknown. Further, in begomoviruses, AC4 acts as a suppressor of silencing mechanism by binding the single stranded siRNA and miRNA, thus inhibiting the cleavage of target mRNA and inducing the viral spread [14]. Additionally, AC5 represses both PTGS and TGS of host plant. It interacts with ssRNA to inhibit the PTGS machinery. It was hypothesized that AC5 suppresses chromatin methylation by reducing the expression of Domains Rearranged Methyltransferase-2 (DRM2; CpHpH cytosine methyltransferase) in N. benthamiana [55]. Recent information suggests the role of Rep in TGS [87]. Simultaneously, Rep associates with SUMO-conjugating enzyme 1 (SCE1) and modifies the sumoylation pattern of host factors to create conditions suitable for viral infection [12]. Thus, by targeting these virus protein through transgenic approach, resistance can be developed against ToLCNDV.
Involvement of host factors during ToLCNDV–plant interactions
Our restricted information of host gene response during virus infection in plant is the major constrain for future studies. Several tomato cultivars have been derived by introgression of resistance against tomato leaf curl disease from wild Solanum species such as S. pimpinellifolium, S. peruvianum, S. chilense, S. habrochaites, S. chmielewskii and S. pinnellii [83, 104].
Various approaches have been exploited to establish the host gene-expression modulation during geminivirus infection [4, 50, 69, 96]. Differentially expressed genes such as those encoding ubiquitin-conjugating enzyme, receptor-like protein kinase, cell cycle proteins, hormone and its regulation, disease resistance genes and various antioxidants were identified during the interaction of ToLCNDV with tomato and potato plants [40, 69, 96]. Apart from this, many QTLs have been identified to be associated with tolerance/resistance against ToLCNDV [92]. Thus, well established resistant/tolerant hosts and comprehensive analysis of differentially expressed host genes may contribute in the discovery of resistance response against geminiviruses. In this regard, functional characterization of accessible or reported datasets of proteome and transcriptome of various plant-geminivirus interaction may assist in revealing the key regulatory component of resistance.
The ubiquitin-proteasome system
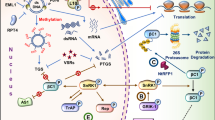
The 26SP, a multi-subunit complex, has ATP-dependent proteolytic function, which is required for the degradation of ubiquitinated intracellular proteins. Ubiquitin proteasome systems (UPS) have appeared as a new subject in plant-microbe interactions [101]. Role of ubiquitin and proteasome during plant-pathogen interaction were extensively reviewed [19, 94]. However, limited information is available defining the role of these networks in geminivirus-plant interaction. Transcript profiling in tolerant tomato plant upon ToLCNDV infection has shown induction of various UPS gene [96]. It suggests operation of a possible virus resistance pathway mediated or controlled by ubiquitin proteasomal degradation (Fig. 1). Apart from UPS, the individual subunit of this multimeric complex is implicated in the diverse function. SlRPT4, a component of 19S regulatory particle (RP) was identified to possess viral DNA binding properties, thus inhibiting the binding of SlRNA PolII on IR which in turn regulates the ToLCNDV genome transcription (Fig. 1). Simultaneously, SlRPT4 restricts the virus spread through the activation of HR and PCD, and thus contributed in the inhibition of ToLCNDV pathogenesis and progression, leading to reduced level of corresponding viral genes [93]. Furthermore, a comprehensive study is required to decipher the mechanism of other UPS components for improved understanding of plant-pathogen interactions.
Schematic representation of Tomato leaf curl New Delhi virus–plant host interaction. Virus entry into host cell initiates defense machinery and induces changes in gene expression level. It includes protein degradation, RNAi- mediated resistance to ToLCNDV, activation of Transcription factor and resistance genes
Resistance genes
Plants are a great source of nutrients and therefore are infected by several pathogens. They have developed different strategies which include avoidance (structural barriers), as well as suppression (by several secondary metabolites) and active defense (by antimicrobial molecules, such as degradative enzymes and phytoalexins) against pathogens. Plants’ response to pathogen often relies on the network of Resistance proteins (R proteins) [44]. These R genes can identify specific effectors and trigger the ‘hypersensitive response (HR)’, which is the key reaction of resistance mechanism in many plant-pathosystems. The HR induces programmed cell death (PCD), which controls several physiological and development specific processes in plants [33]. Till date several R genes have been reported in tomato to be involved in pathogen recognition [89]. Table 1 recapitulates the known R genes in plants against viruses.
In tomato, two NB-ARC-LRR R genes have been identified to regulate the viral defense viz; Sw5 and Tm2 [106]. Various studies have identified only five loci that are linked to viral resistance: Ty-1/Ty-3 and Ty-4 from S. chilense, Ty-2 from S. habrochaites and Ty-5 from S. peruvianum [2, 36, 42, 43, 52]. Amongst these Ty-1 and Ty-3 were found to encode RDRs that are associated with amplification of RNA silencing mechanism [113]. Besides this, the promoter region of TYLCV was found to be hypermethylated in Ty-1 expressing tomatoes. The Ty-5 locus is a recessive resistance QTL and associated with NAC1 marker [2]. It encodes for messenger RNA surveillance factor Pelo gene called Pelota and is required during recycling phase of protein biosynthesis (Fig. 1) [52]. A large number of resistance genes have been introgressed in tomato cultivars plants from wild tomato [66, 83, 104]. Through pyramiding of Ty-2 and Ty-3 in tomato cultivar via breeding, increased resistance was observed against ToLCNDV [83].
Recently, defense related genes nucleotide-binding site and leucine-rich repeat (NBS-LRR) were found to be differently expression in tolerant (Capsicum annuum and S. lycopersicum) and susceptible (Nicotiana benthamiana and N. tabacum) plants upon imposition of ToLCNDV infection [50]. Interestingly, NIK1 (NSP-interacting kinase 1) which consists of a leucine-rich repeat receptor-like kinase (LRR-RLK) domain, was identified to interact with Nuclear shuttle protein of begomoviruses (Fig. 1) [97]. Further, the interaction between NIK1-NSP initiates complete inhibition of translational machinery; regulated via RPL10 and L10-interacting MYB domain-containing protein (LIMYB) leading to antiviral defense in plants [125]. Due to accelerated evolution in genome of geminiviruses and variation in the host range, R gene responsible for tolerance/resistance against ToLCNDV need to be identified from the wild type species. These predicted R genes can be acclimated to develop defense mechanism in plants through transgenic approach or traditional pyramiding approach.
Transcription factors (TFs)
The imposition of biotic stress triggers transcriptional reprogramming in plants which regulates the metabolic and phenotypic changes and leads to defense response. Various transcription factors have been reported to be associated with plant immunity against plant viruses [111]. Transcription factors might directly or indirectly interact with signalling molecules to initiate defense response by regulating the downstream signaling networks. In a study on Nicotiana benthamiana it was observed that WRK1-WRY3 and MYB1 regulate the expression of N-gene required to initiate defense against begomoviruses [57]. Further, it was revealed that additionally N-gene associates with Squamosa promoter binding protein (SBP)-domain TF (NbSPL6). NbSPL6 initiates the transcription of defense related genes specifically RPS4-mediated immunity response against viruses (Fig. 1) [74].
In a recent study, membrane bound NAC transcription factors, SlNACMTF3 and SlNACMTF 8, were found to be differentially expressed upon ToLCNDV infection in tomato cv. Pusa Ruby. These membrane bound transcription factors were revealed to regulate the expression of biotic stress-related genes such as Senescence associated protein, Elicitor responsive protein 1, Pathogen related protein 1b (PR1b), hence elucidating the key role of these TFs in regulating biotic stress response [9]. Similarly, SlWRKY16 was demonstrated to regulate the expression of Tornado1 (SlTRN1) upon salicyclic acid treatment. SlTRN1 is essential for cell expansion and vein formation, thus elevation in the level of its transcripts during ToLCNDV might be related to the appearance of elevated symptoms (Fig. 1) [61]. Since, TFs play vital role in governing the alterations in defense pathway, it is essential to identify the transcription factors regulating the plant defense responses against viruses.
Role of RNA interference mechanism during geminivirus-plant interaction
In plants, RNA interference (RNAi) inhibits gene expression using three approaches: by degradation of transcripts (PTGS); inhibiting translation of mRNAs, or by promoting methylation of targeted DNA fragment affecting transcription of genes (TGS) [99]. PTGS and TGS are correlative since both the mechanisms require the development of small interfering RNAs (siRNAs). PTGS is induced when siRNA produced is complementary to the coding region of the target gene and entails sequence-specific RNA degradation. Contrastingly, siRNA complementary to the target gene promoter region initiates TGS, which causes chromatin remodeling through methylation [21, 95]. In case of geminivirus, these siRNAs are produced during virus replication by the action of host RDRs, or by transcription of inverted repeats or converged promoters [75, 99, 107]. Virus-derived siRNAs have been extensively studied during various plant-virus interactions (Fig. 1) [16, 85]. The siRNA-mediated gene silencing has been shown to be involved in the recovery from the specific virus infection in plants [86, 88, 95, 120]. Further, this phenomenon has been used to develop tolerance against ToLCNDV through transgenic approach [103, 115].
PTGS as antiviral defense
Upon virus infection in plants, RNA silencing is the innate antiviral defense response, during which dsRNAs derived from virus replication are recognized as the pathogen-associated molecular patterns (PAMPs), activating the pattern-recognition receptors (PRRs); DCL enzymes leading to synthesis of vsiRNA [127]. The increased accumulation of vsiRNAs during viral infections establishes the function of RNA silencing in antiviral defense. In a study against ToLCNDV in tomato, a naturally tolerant cultivar of tomato, namely H-88-78-1, was identified which has reduced infectivity and virus titre at 21 dpi in contrast to a susceptible cv. Punjab Chhuhara [96]. A crucial role of antivirus activity of siRNA was established, as in cv. H-88-78-1 less abundance of virus genome was correlated with a relatively higher level of vsiRNAs production [96]. Recently, it was depicted that the variation in tolerance is attributed to the differential level of siRNAs production [50]. A significant variation was observed in the transcript level of genes associated with the gene silencing machinery (RDR6, AGO1 and SGS3). Besides, PTGS another mechanism of RNA silencing is Transcriptional Gene Silencing (TGS). Its mechanism and role in virus resistance is discussed below.
Role of epigenetics in regulating the geminiviruses infection
Epigenetic is a vital mechanism for development and stress associated phenotype of a system. It is related to heritable but concurrently reversible alterations in the expression of genes that are not due to DNA sequence variation [8]. It is a crucial mechanism which regulates the gene transcription, and thus gene expression by determining the binding of transcription regulators to DNA [21, 99]. In eukaryotic cell, it involves variations in the chromatin structure by some of the chemical modulations of DNA through methylation of cytosine residues (DNA methylation) or by posttranslational modifications of proteins associated with DNA (histones). DNA methylation involves contracting processes; addition and removal of a methyl group at C5 (5th carbon) of pyrimidine ring of cytosine. Two types of DNA methyltransferases are identified in plants, for methylation maintenance; Chromomethylase (CMT; methylation maintenance), Methyltransferase (MET; maintenance of methylation), Kryptonite (KYP; methylation maintenance and H3K9 methylation) and de novo methylation; Domains Rearranged Methyltransferase (DRM; de novo methylation) [118]. Contrastingly, repressor of silencing1 (ROS1), demeter-like2 (DML2), and DML3 are DNA glycosylases that actively mediate DNA demethylation via a base excision repair process [28]. This DNA methylation confers gene silencing and plays crucial roles in plant development and defense against viruses, transposons, and transgenes [75, 99].
Further, this epigenetic pathway involves siRNAs; 23–24 nt long) directing epigenetic changes termed as RNA-directed DNA methylation (RdDM). These siRNAs are homologous to promoter regions of a gene and stimulate de novo cytosine methylation in the promoter region [99, 118]. Synthesis of these siRNAs relies on RNAi machinery core proteins: Dicer (DCL3), DNA dependent RNA polymerase (RNA PolIV/V), and Argonaute (AGO4). The biogenesis of heterochromatic siRNA requires transcription by DNA-dependent RNA polymerase PolIV (RNA PolIV) and ssRNAs thus produced are converted to dsRNA by RNA dependent RNA polymerase (RDR2). These dsRNAs are diced by DCL3 to produce 24 nt siRNAs. The guide strand incorporates into AGO4/ AGO6 containing RITS complex, which then enters the RNA PolV-mediated pathway of de novo DNA methylation. PolV transcripts function as a scaffold RNA which interacts with the siRNA associated with AGO4/6. This siRNA bound AGO4/6 associates with chromatin modifiers such as DRM2, which catalyzes de novo methylation at the siRNA-targeted site [30]. Apart from antivirus defense RdDM mechanism guides chromatin modifications and silences transposons in plants.
TGS-mediated resistance to ToLCNDV
In plants, transcripts of virus genome trigger the RNA silencing machinery [80]. Apart from viral transcript degradation, several reports suggest that plants employ DNA methylation to instigate defense against geminiviruses. The dsRNA generated during viral infection can initiate sRNAs directed de novo DNA methylation via RdDM pathway resulting in transcriptional silencing of viruses [18, 39, 120]. RdDM entails virus genome modification by de novo methylation at cytosine-residue in virus DNA and associated histones and thus suppresses the transcription and/or replication of virus [18, 85, 88, 120]. Against ToLCNDV, RdDM has been reported to initiate methylation in the coding (AC1/Rep) and promoter regions (IR). Moreover, significant variation in expression of genes for cytosine or histone methyltransferase, methyl cycle enzymes, and DCL proteins and components of RdDM pathways was observed in the tolerant cv. as compare to susceptible cv. (Fig. 1) [95]. This study provides support to the hypothesis that tolerant/resistant plants utilize small RNA-directed methylation as one of the defense strategy against geminiviruses, thus suppressing the transcription of virus genome. Further, role of different virus components and plant factors liable for defense mechanism needs to be explored.
MicroRNA as antiviral defense
During ToLCNDV infection, regulation of gene expression at transcription level has been quite well explained [50, 96]. However, recently miRNAs have emerged as an important class of regulators which controls the expression of both host and viral genes at post-transcriptional levels (Fig. 1) [68, 70, 82]. MiRNAs are non-coding RNAs of 19 to 24 nt that govern gene expression at post-transcriptional level by binding and initiating the degradation of its complementary target mRNAs, thus inhibiting its translation.
The role of miRNAs in regulating biotic stress response in plants is being investigated. Specific set of miRNAs has been found to differentially express during virus infections in different plant-virus interactions [48, 82]. During begomovirus infection, expression pattern of diverse miRNA was predicted to be altered. The expression of miR159 was up-regulated during ToLCNDV infection. It was predicted to target MYB transcription factor, which binds to the promoter region of LEAFY gene (LFY) which is broadly expressed throughout the transition from vegetative to reproductive phase. Thus, during ToLCNDV infection enhanced expression of miR159 was related to the symptom (leaf curling) development [70]. Similarly, other miRNAs such as miR156, miR164, miR166, miR160/167 and miR170, which modifies the expression of key development related transcription factors, such as SPL (Squamosa Promoter Binding Protein), NAC, HD-Zip (Homeodomain-leucine zipper), and ARFs (Auxin responsive factor), Scare-crow like genes respectively, were observed to have differential expression pattern during ToLCNDV infection (Fig. 1). These studies propose that the virus infection specifically regulate a group of miRNAs which leads to the symptom development. In addition to this, in several studies significant difference was observed in expression of miRNA targeting the defense related genes. In tomato, miRNAs were identified which were predicted to target functional R genes viz, sly-miR6022 (21 nt; targets Hcr9), sly-miR6023 (22nt; targets Hcr9) and miR6024 and miRNA 6026 (22 nt; targets Tm2) and sly-miR6027 (22 nt; targets Sw5) [56]. Thus, indicating that miRNAs have a crucial role in regulating the role of R genes triggered resistance responses.
Furthermore, few host-miRNAs have been revealed to directly bind the viral transcripts. In silico analysis predicted several tomato miRNAs/miRNAs* (such as miR1918, miR156b*, miR159, miR164a, miR169b*, miR169d, miR171d, miR172b* miR166a* and miR319) having significant affinity to associate with ToLCV genome or encoded transcripts [68]. Although, numerous plant miRNAs have been classified/predicted to have a crucial role during virus infection but the precise function of the most is still unidentified. Although various studies have been executed on diverse plant species unveiling the significance of miRNAs during virus infection, no comprehensive study has identified and elucidated the precise role of virus infection-related miRNAs in tomato crop.
Regulation of virus infection through contrived resistance
Geminiviruses are controllers of host cellular machinery and encode several viral suppressors of RNA silencing (VSRs). Further, due to the viral genome evolution, breeding and transgenic approaches have not induced stable resistant lines. Recently, RNAi silencing based genetic engineering approaches have been utilized as effective defense strategy. The artificial miRNA (amiRNA)-based transgenics approach has been used to decipher the gene silencing attribute of miRNAs. Subsequently, this antiviral therapy has been used to generate plants producing amiRNAs corresponding to pre-coat and coat protein transcripts of ToLCNDV evolve resistance in tomato [115]. Apart from amiRNAs, artificial transacting siRNAs regulating the VSRs has been considered an effective approach [104]. Conversely, in case of combined infection by multiple virus these RNAi-based approaches would not be sufficient to overcome virus infection. Recently, invented CRISPR/Cas9 technology (Clustered regularly interspaced short palindromic repeats/ CRISPR-associated9) aiming the virus genomic regions has emerged as a novel tool to engineer resistance against geminiviruses [6, 41, 123]. This approach has been activated against Rep, CP and promoter region (IR) of Tomato yellow leaf curl virus (TYLCV), Beet curly top virus (BCTV), and Merremia mosaic virus (MeMV). This approach can be used to target essential machineries of virus replication and transcription. It could be postulated that Cas9/sgRNA module associates with virus genome, specifically origin of replication and inhibits the binding of Rep proteins, thus regulating the virus accumulation. Notoriously, the robustness of CRISPR/Cas9 technology needs to be verified in field conditions. In the upcoming decade the CRISPR/Cas9 technology along with NGS would be applicable to establish the disease resistance against ToLCNDV by generating varieties of staple crops.
Conclusion and future prospects
Excessive ToLCNDV-induced yield losses have reported worldwide in most of the agriculturally important staple crops. The spread of ToLCNDV, its management strategies and the future prospects to regulate the spread of ToLCNDV are depicted in Fig. 2. Initially, the virus control strategies involved management of B. tabaci via use of pesticides [54]. However, notably new strains of viruses are emerging due to recombination and mutations in the genome of ssDNA viruses. Comprehensive research is requisite to establish the molecular basis and mechanistic details of evolution of ToLCNDV genome and the mechanism by which ToLCNDV associates with different viruses and satellites.
Further, identification of plant defense mechanism evolved against these viruses in natural genotypes varying in their tolerance/resistance against viruses needs to be explored. Simultaneously, an approach concerning apperception of R gene responsible for defense against ToLCNDV needs to be proposed to develop resistant/tolerant plants through transgenic or traditional pyramiding strategies. However, our effort in finding the appropriate genes/or pathways involved in plant defense against ToLCNDV is reduced due to the inadequate well-characterized, compatible host model suitable for transcriptome profiling studies.
Apart from genes such as R genes, Ubiquitin proteasomal genes and Transcription factors, small RNAs (sRNAs) have also been demonstrated as a source of plant defense against ssDNA viruses. NGS can be utilized to decipher the repertoire of sRNAs, aiding in understanding the perspective of host defense and viral counter-defense measures. In particular, identification of plant miRNAs is difficult due to their small size (18–24 nt) and multiple occurrences in the genome. Recent advancements in high throughput sequencing technology have led to the discovery of species-specific miRNAs in many plant species in addition to conserved miRNAs during biotic stress [48, 82]. Further, illustration and identification of additional miRNAs is required during ToLCNDV infection and the machinery involved for defense through miRNA is a prerequisite to understand the molecular mechanism related to regulation of viral infection through variation in expression of miRNA targets.
During infection, viruses activate the transcription of host genes (like those involved in cell-cycle and signaling), which are required for viral DNA replication. These results validate that geminivirus are the models for studying the mechanism of genome methylation and pathways regulating cytosine methylation in plants. Further, due to the inability to attain field resistance in transgenic tomato, application of high-throughput technologies and reverse genetic approaches, like virus-induced gene silencing (VIGS) [57] and RNAi would be beneficial to develop defense mechanism against ToLCNDV. Lately, upon targeting ToLCNDV genes (AC1, AC2, AC4, AV1 and AV) [100, 115] a significant decrease in the virus titre was witnessed.
Concurrently, based on the role of ssDNA viruses in modification of host machinery for replication and transcription its application in gemone editing is under process. Advancement in this aspect can be done based on advancement in the geminivirus-plant interactions. Imminently, geminivirus-based tools will be established to gain insight of the plant machinery.
References
Amari K, Gonzalez-Ibeas D, Gomez P, Sempere RN, Sanchez-Pina MA, Aranda MA, et al. Tomato torrado virus is transmitted by Bemisia tabaci and infects pepper and eggplant in addition to tomato. Plant Dis. 2008;92:1139.
Anbinder I, Reuveni M, Azari R, Paran I, Nahon S, Shlomo H, et al. Molecular dissection of tomato leaf curl virus resistance in tomato line TY172 derived from Solanum peruvianum. Theor Appl Genet. 2009;119:519–30.
Arguello-Astorga G, Lopez-Ochoa L, Kong LJ, Orozco BM, Settlage SB, Hanley-Bowdoin L. A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma-related protein. J Virol. 2004;78:4817–26.
Ascencio-Ibanez JT, Sozzani R, Lee TJ, Chu TM, Wolfinger RD, Cella R, et al. Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 2008;148:436–54.
Bagewadi B, Chen S, Lal SK, Choudhury NR, Mukherjee SK. PCNA interacts with Indian mung bean yellow mosaic virus Rep and downregulates Rep activity. J Virol. 2004;78:11890–903.
Baltes NJ, Hummel AW, Konecna E, Cegan R, Bruns AN, Bisaro DM, et al. Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat Plants. 2015;1:15145.
Bendahmane A, Kanyuka K, Baulcombe DC. The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell. 1999;11:781–91.
Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12.
Bhattacharjee P, Das R, Mandal A, Kundu P. Functional characterization of tomato membrane-bound NAC transcription factors. Plant Mol Biol. 2017;93:511–32.
Briddon RW, Pinner MS, Stanley J, Markham PG. Geminivirus coat protein gene replacement alters insect specificity. Virology. 1990;177:85–94.
Butter NS, Rataul HS. The virus vector relationship of the tomato leaf curl virus (TLCV) and its vector Bemisia tabaci Gennadius (Homoptera: Aleyrodidae.). Phytoparasitica. 1977;5:173–86.
Castillo AG, Kong LJ, Hanley-Bowdoin L, Bejarano ER. Interaction between a geminivirus replication protein and the plant sumoylation system. J Virol. 2004;78:2758–69.
Chandel RS, Banyal DK, Singh BP, Malik K, Lakra BS. Integrated management of whitefly, Bemisia tabaci (Gennadius) and potato apical leaf curl virus in India. Potato Res. 2010;53:129–39.
Chellappan P, Vanitharani R, Fauquet CM. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc Natl Acad Sci USA. 2005;102:10381–6.
Cooley MB, Pathirana S, Wu H-J, Kachroo P, Klessig DF. Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell. 2000;12:663–76.
Dalakouras A, Dadami E, Wassenegger M. Viroid-induced DNA methylation in plants. Biomol Concepts. 2013;4:557–65.
De Barro PJ, Ahmed MZ. Genetic networking of the Bemisia tabaci cryptic species complex reveals pattern of biological invasions. PLoS ONE. 2011;6:e25579.
Deuschle K, Kepp G, Jeske H. Differential methylation of the circular DNA in geminiviral minichromosomes. Virology. 2016;499:243–58.
Dielen AS, Badaoui S, Candresse T, German-Retana S. The ubiquitin/26S proteasome system in plant-pathogen interactions: a never-ending hide-and-seek game. Mol Plant Pathol. 2010;11:293–308.
Emmanuel E, Levy AA. Tomato mutants as tools for functional genomics. Curr Opin Plant Biol. 2002;5:112–7.
Espinas NA, Saze H, Saijo Y. Epigenetic control of defense signaling and priming in plants. Front Plant Sci. 2016;11:1201.
Fauquet CM, Briddon RW, Brown JK, Moriones E, Stanley J, Zerbini FM, et al. Geminivirus strain demarcation and nomenclature. Arch Virol. 2008;153:783–821.
Fontes EPB, Santos AA, Luz DF, Waclawovsky AJ, Chory J. The geminivirus nuclear shuttle protein is a virulence factor that suppresses transmembrane receptor kinase activity. Genes Dev. 2004;18:2545–56.
Fontes EPB, Gladfelter HJ, Schaffer RL, Petty IT, Hanley-Bowdoin L. Geminivirus replication origins have a modular organization. Plant Cell. 1994;6:405–16.
Fukunaga R, Doudna JA. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009;28:545–55.
Gafni Y, Epel BL. The role of host and viral proteins in intra- and inter-cellular trafficking of geminiviruses. Physiol Mol Plant Pathol. 2002;60:231–41.
Gebhardt C. The historical role of species from the Solanaceae plant family in genetic research. Theor Appl Genet. 2016;129:2281–94.
Gehring M, Reik M, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90.
Gutierrez C. DNA replication and cell cycle in plants: learning from geminiviruses. EMBO J. 2000;19:792–9.
Haag JR, Pikaard CS. Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol. 2011;12:483–92.
Hanley-Bowdoin L, Bejarano ER, Robertson D, Mansoor S. Geminiviruses: masters at redirecting and reprogramming plant processes. Nat Rev Microbiol. 2013;11:777–88.
Hanssen IM, Lapidot M, Thomma BP. Emerging viral diseases of tomato crops. Mol Plant-Microbe Int. 2010;23:539–48.
Hatsugai N, Iwasaki S, Tamura K, Kondo M, Fuji K, Ogasawara K, et al. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009;23:2496–506.
Hayes AJ, Jeong SC, Gore MA, Yu YG, Buss GR, Tolin SA, et al. Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics. 2004;166:493–503.
Hussain M, Mansoor S, Iram S, Zafar Y, Briddon RW. First report of Tomato leaf curl New Delhi virus affecting chilli pepper in Pakistan. Plant Pathol. 2004. doi:10.1111/j.1365-3059.2004.01073.x.
Hutton SF, Scott JW, Schuster DJ. Recessive resistance to Tomato yellow leaf curl virus from the tomato cultivar Tyking is located in the same region as Ty-5 on chromosome 4. HortScience. 2012;47:324–7.
Ingham DJ, Pascal E, Lazarowitz SG. Both bipartite geminivirus movement proteins define viral host range, but only BL1 determines viral pathogenicity. Virology. 1995;207:191–204.
Ishibashi K, Mawatari N, Miyashita S, Kishino H, Mashi T, Ishikawa M. Coevolution and hierarchical interactions of Tomato mosaic virus and the resistance gene Tm-1. PLoS Pathog. 2012;8:e1002975.
Jackel JN, Storer JM, Coursey T, Bisaro DM. Arabidopsis RNA polymerases IV and V are required to establish H3K9 methylation, but not cytosine methylation, on Geminivirus chromatin. J Virol. 2016;90:7529–40.
Jeevalatha A, Siddappa S, Kumar A, Kaundal P, Guleria A, Sharma S, et al. An insight into differentially regulated genes in resistant and susceptible genotypes of potato in response to tomato leaf curl New Delhi virus-[potato] infection. Virus Res. 2017;15:22–33.
Ji X, Zhang H, Zhang Y, Wang Y, Gao C. Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat Plants. 2015;1:15144.
Ji Y, Scott JW, Schuster DJ, Maxwell DP. Molecular mapping of Ty-4, a new Tomato Yellow Leaf Curl Virus resistance locus on chromosome 3 of tomato. Hortic Sci. 2009;134:281–8.
Ji Y, Schuster DJ, Scott JW. Ty-3, a begomovirus resistance locus near the Tomato yellow leaf curl virus resistance locus Ty-1 on chromosome 6 of tomato. Mol Breed. 2007;20:271–84.
Jones JD, Dangle JL. The plant immune system. Nature. 2006;444:323–9.
Juarez M, Tovar R, Fiallo-Olive E, Aranda MA, Gosalvez B, Castillo P, et al. First detection of Tomato leaf curl new Delhi virus infecting zucchini in Spain. Plant Dis. 2014;98:857.
Jupin I, De Kouchkovsky F, Jouanneau F, Gronenborn B. Movement of tomato yellow leaf curl geminivirus (TYLCV): involvement of the protein encoded by ORF C4. Virology. 1994;204:82–90.
Jyothsna PQ, Haq QMI, Singh P, Sumiya KV, Praveen S, Rawat R, et al. Infection of tomato leaf curl New Delhi virus (ToLCNDV), a bipartite begomovirus with betasatellites, results in enhanced level of helper virus components and antagonistic interaction between DNA B and betasatellites. Appl Microbiol Biotechnol. 2013;97:5457–71.
Khraiwesha B, Zhua JK, Zhuc J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta. 2012;1819:137–48.
Kumar V, Mishra SK, Rahman J, Taneja J, Sundaresan G, Mishra NS, et al. Mungbean yellow mosaic Indian virus encoded AC2 protein suppresses RNA silencing by inhibiting Arabidopsis RDR6 and AGO1 activities. Virology. 2015;486:158–72.
Kushwaha N, Singh AK, Basu S, Chakraborty S. Differential response of diverse solanaceous hosts to tomato leaf curl New Delhi virus infection indicates coordinated action of NBS-LRR and RNAi-mediated host defense. Arch Virol. 2015;160:1499–509.
Lacatus G, Sunter G. Functional analysis of bipartite begomovirus coat protein promoter sequences. Virology. 2008;376:79–89.
Lapidot M, Karniel U, Gelbart D, Fogel D, Evenor D, Kutsher Y, et al. A novel route controlling begomovirus resistance by the messenger RNA surveillance factor pelota. PLoS Genet. 2015;11:e1005538.
Laufs J, Jupin I, David C, Schumacher S, Heyraud-Nitschke F, Gronenborn B. Geminivirus replication: genetic and biochemical characterization of Rep protein function, a review. Biochimie. 1995;77:765–73.
Legg PJ, Shirima R, Tajebe SL, Guastella D, Boniface S, Jeremiah S, et al. Biology and management of Bemisia whitefly vectors of cassava virus pandemics in Africa. Pest Manag Sci. 2014;70:1446–53.
Li F, Xu X, Huang C, Gu Z, Cao L, Hu T, et al. The AC5 protein encoded by Mungbean yellow mosaic India virus is a pathogenicity determinant that suppresses RNA silencing-based antiviral defenses. New Phytol. 2015;208:555–69.
Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, et al. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA. 2012;109:1790–5.
Liu Y, Schiff M, Dinesh-Kumar SP. Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J. 2004;38:800–9.
Luque A, Sanz-Burgos AP, Ramirez-Parra E, Castellano MM, Gutierrez C. Interaction of geminivirus Rep protein with replication factor C and its potential role during geminivirus DNA replication. Virology. 2002;302:83–94.
Maiti S, Paul S, Pal A. Isolation, characterization, and structure analysis of a non-TIR-NBS-LRR encoding candidate gene from MYMIV-resistant Vigna mungo. Mol Biotechnol. 2012;52:217–33.
Malik PS, Kumar V, Bagewadi B, Mukherjee SK. Interaction between coat protein and replication initiation protein of Mung bean yellow mosaic India virus might lead to control of viral DNA replication. Virology. 2005;337:273–83.
Mandal A, Sarkar D, Kundub S, Kundu P. Mechanism of regulation of tomato TRN1 gene expression in late infection with tomato leaf curl New Delhi virus (ToLCNDV). Plant Sci. 2015;241:221–37.
McGarry RC, Barron YD, Carvalho MF, Hill JE, Gold D, Cheung E, et al. A novel Arabidopsis acetyltransferase interacts with the geminivirus movement protein NSP. Plant Cell. 2003;15:1605–18.
Melgarejo TA, Kon T, Rojas MR, Paz-Carrasco L, Zerbini FM, Gilbertson RL. Characterization of a new world monopartite begomovirus causing leaf curl disease of tomato in Ecuador and Peru reveals a new direction in geminivirus evolution. J Virol. 2013;87:5397–413.
Mizutani T, Daryono BS, Ikegami M, Natsuaki KT. First report of Tomato leaf curl New Delhi virus infecting cucumber in Central Java, Indonesia. Plant Dis. 2001;95:1485.
Mnari-Hattab M, Zammouri S, Belkadhi MS, Bellon Dona D, Ben Nahia E, Hajlaoui MR. First report of Tomato leaf curl New Delhi virus infecting cucurbits in Tunisia. New Dis Rep. 2015;31:21.
Momotaz A, Scott JW, Schuster DJ. Identification of quantitative trait loci conferring resistance to Bemisia tabaci in an F2 population of Solanum lycopersicum x Solanum habrochaites accession LA1777. J Am Soc Hortic Sci. 2010;135:134–42.
Moury B, Verdin E. Viruses of pepper crops in the mediterranean basin: a remarkable stasis. Adv Virus Res. 2012;84:127–62.
Naqvi AR, Choudhury NR, Mukherjee SK. Mohd. Rizwanul Haq Q. In silico analysis reveals that several tomato microRNA/microRNA* sequences exhibit propensity to bind to tomato leaf curl virus (ToLCV) associated genomes and most of their encoded open reading frames (ORFs). Plant Physiol Biochem. 2011;49:13–7.
Naqvi AR, Sarwat M, Pradhan B, Choudhury NR, Haq QM, Mukherjee SK. Differential expression analyses of host genes involved in systemic infection of Tomato leaf curl New Delhi virus (ToLCNDV). Virus Res. 2011;160:395–9.
Naqvi AR, Haq QM, Mukherjee SK. MicroRNA profiling of tomato leaf curl New Delhi virus (ToLCNDV) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol J. 2010;7:281.
Padidam M, Beachy RN, Fauquet CM. The role of AV2 (“precoat”) and coat protein in viral replication and movement in tomato leaf curl geminivirus. Virology. 1996;224:390–404.
Padidam M, Beachy RN, Fauquet CM. Tomato leaf curl geminivirus from India has a bipartite genome and coat protein is not essential for infectivity. J GenVirol. 1995;76:25–35.
Padidam M, Beachy RN, Fauquet CM. Classification and identification of Geminiviruses using sequence comparisons. J Gen Virol. 1995;76:249–63.
Padmanabhan MS, Ma S, Burch-Smith TM, Czymmek K, Huijser P, Dinesh Kumar SP. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 2013;9:e1003235.
Pandey G, Sharma N, Sahu PP, Prasad M. Chromatin-based epigenetic regulation of plant abiotic stress response. Curr Genom. 2016;17:490–8.
Panno S, Iacono G, Davino M, Marchione S, Zappardo V, Bella P, et al. First report of Tomato leaf curl New Delhi virus affecting zucchini squash in an important horticultural area of southern Italy. New Dis Rep. 2016;33:6.
Park J, Hwang HS, Buckley KJ, Park JB, Auh CK, Kim DG, et al. C4 protein of Beet severe curly top virus is a pathomorphogenetic factor in Arabidopsis. Plant Cell Rep. 2010;29:1377–89.
Pasumarthy KK, Mukherjee SK, Choudhury NR. The presence of tomato leaf curl Kerala virus AC3 protein enhances viral DNA replication and modulates virus induced gene-silencing mechanism in tomato plants. Virol J. 2011;8:178.
Pasumarthy KK, Choudhury NR, Mukherjee SK. Tomato leaf curl Kerala virus (ToLCKeV) AC3 protein forms a higher order oligomer and enhances ATPase activity of replication initiator protein (Rep/AC1). Virol J. 2010;7:128.
Patil BL, Fauquet CM. Studies on differential behavior of cassava mosaic geminivirus DNA components, symptom recovery patterns, and their siRNA profiles. Virus Genes. 2015;50:474–86.
Pelham J. Resistance in tomato to Tobacco mosaic virus. Euphytica. 1996;15:258.
Pradhana B, Naqvi AR, Saraf S, Mukherjee SK, Deya N. Prediction and characterization of Tomato leaf curl New Delhi virus (ToLCNDV) responsive novel microRNAs in Solanum lycopersicum. Virus Res. 2015;2:183–95.
Prasanna H, Sinha D, Rai G, Krishna R, Kashyap SP, Singh N, et al. Pyramiding Ty-2 and Ty-3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Pathol. 2015;64:256–64.
Pratap D, Kashikar AR, Mukherjee SK. Molecular characterization and infectivity of a Tomato leaf curl New Delhi virus variant associated with newly emerging yellow mosaic disease of eggplant in India. Virol J. 2011;8:305.
Raja P, Jackel JN, Li S, Heard IM, Bisaro DM. Arabidopsis double-stranded RNA binding protein DRB3 participates in methylation-mediated defense against geminiviruses. J Virol. 2014;88:2611–22.
Rajeswaran R, Seguina J, Chabannesc M, Duroyc PO, Laboureauc N, Farinellib L, et al. Evasion of short interfering RNA-directed antiviral silencing in Musa acuminata persistently infected with six distinct banana streak pararetroviruses. J Virol. 2014;88:11516–28.
Rodríguez-Negrete EA, Lozano-Duran R, Piedra-Aguilera A, Cruzado L, Bejarano ER, Castillo AG. Geminivirus Rep protein interferes with the plant DNA methylation machinery and suppresses transcriptional gene Silencing. New Phytol. 2013;199:464–75.
Rodriguez-Negrete EA, Carrillo-Tripp J, Rivera-Bustamante RF. RNA silencing against Geminivirus: complementary action of posttranscriptional gene silencing and transcriptional gene silencing in host recovery. J Virol. 2009;83:1332–40.
Ronde DD, Butterbach P, Kormelink R. Dominant resistance against plant viruses. Front Plant Sci. 2014;5:307.
Rouhibakhsh A, Choudhury NR, Mukherjee SK, Malathi VG. Enhanced nicking activity of Rep in presence of pre-coat protein of Mungbean yellow mosaic India virus. Virus Genes. 2012;44:356–61.
Rybicki EP. A phylogenetic and evolutionary justification for three genera of Geminiviridae. Arch Virol. 1997;139:49–77.
Saez C, Esteras C, Martinez C, Ferriol M, Dhillon NPS, Lopez C, et al. Resistance to tomato leaf curl New Delhi virus in melon is controlled by a major QTL located in chromosome 11. Plant Cell Rep. 2017; doi:10.1007/s00299-017-2175.
Sahu PP, Sharma N, Puranik S, Chakraborty S, Prasad M. Tomato 26S Proteasome subunit RPT4a regulates ToLCNDV transcription and activates hypersensitive response in tomato. Sci Rep. 2016;6:270–8.
Sahu PP, Sharma N, Puranik S, Muthamilarasan M, Prasad M. Involvement of host regulatory pathways during geminivirus infection: a novel platform for generating durable resistance. Funct Integr Genom. 2014;14:47–58.
Sahu PP, Sharma N, Puranik S, Prasad M. Post-transcriptional and epigenetic arms of RNA silencing: a defense machinery of naturally tolerant tomato plant against Tomato Leaf Curl New Delhi Virus. Plant Mol Biol Rep. 2014;32:1015–29.
Sahu PP, Rai NK, Chakraborty S, Singh M, Ramesh B, Chattopadhyay D, et al. Tomato cultivar tolerant to Tomato leaf curl New Delhi virus infection induces virus-specific short interfering RNA accumulation and defence-associated host gene expression. Mol Plant Pathol. 2010;11:531–44.
Santos AA, Lopes KVG, Apfata JAC, Fontes EPB. NSP-interacting kinase, NIK: a transducer of plant defence signalling. J Exp Bot. 2010;61:3839–45.
Selth LA, Dogra SC, Rasheed MS, Healy H, Randles JW, Rezaian MA. A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell. 2005;17:311–25.
Sharma N, Sahu PP, Puranik S, Prasad M. Recent advances in plant-virus interaction with emphasis on small interfering RNAs (siRNAs). Mol Biotechnol. 2013;55:63–77.
Sharma VK, Basu S, Chakraborty S. RNAi mediated broad spectrum transgenic resistance to chilli-infecting begomoviruses. Plant Cell Rep. 2015;34:1389–99.
Shen Q, Hu T, Bao M, Cao L, Zhang H, Song F, et al. Tobacco RING E3 Ligase NtRFP1 mediates ubiquitination and proteasomal degradation of a Geminivirus-Encoded βC1. Mol Plant. 2016;9:911–25.
Shivaprasad PV, Akbergenov R, Trinks D, Rajeswaran R, Veluthambi K, Hohn T, Pooggin MM. Promoters, transcripts, and regulatory proteins of Mungbean yellow mosaic geminivirus. J Virol. 2005;79:8149–63.
Singh A, Taneja J, Dasgupta I, Mukherjee SK. Development of plants resistant to tomato geminiviruses using artificial trans-acting small interfering RNA. Mol Plant Pathol. 2015;16:724–34.
Singh RK, Rai N, Singh M, Saha S, Singh SN. Detection of tomato leaf curl virus resistance and inheritance in tomato (Solanum lycopersicum L.). J Agric Sci Camb. 2015;153:78–89.
Sorab SS, Karim S, Varma A, Azhar EI, Mandal B, Abuzenadah AM, et al. Factors affecting sap transmission of Tomato leaf curl New Delhi begomovirus infecting sponge gourd in India. Phytoparasitica. 2013;41:591–2.
Spassova MI, Prins TW, Folkertsma RT, Klein-Lankhorst RM, Hille J, Goldbach RW, et al. The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol Breed. 2001;7:151–61.
Szittya G, Burgyan J. RNA interference-mediated intrinsic antiviral immunity in plants. Curr Top Microbiol Immunol. 2013;371:153–81.
Takahashi H, Suzuki M, Natsuaki K, Shigyo T, Hino K, Teraoka T, et al. Mapping the virus and host genes involved in the resistance response in Cucumber mosaic virus infected Arabidopsis thaliana. Plant Cell Physiol. 2001;42:340–7.
Teng K, Chen H, Lai J, Zhang Z, Fang Y, Xia R, et al. Involvement of C4 protein of beet severe curly top virus (Family Geminiviridae) in virus movement. PLoS ONE. 2010;5:e11280.
Tiwari AK, Sharma PK, Khan MS, Snehi SK, Raj SK, Rao GP. Molecular detection and identification of Tomato leaf curl New Delhi virus isolate causing yellow mosaic disease in bitter gourd (Momordica charantia), a medicinally important plant in India. Med Plants. 2010;2:117–23.
Tsuda K, Somssich IE. Transcriptional networks in plant immunity. New Phytol. 2015;206:932–47.
Vallejos CE, Astua-Monge G, Jones V, Plyler TR, Sakiyama NS, Mackenzie SA. Genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics. 2006;172:1229–42.
Verlaan MG, Hutton SF, Ibrahem RM, Kormelink R, Visser RGF, Scott JW, et al. The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 2013;9:e1003399.
Vidal S, Cabrera H, Andersson RA, Fredriksson A, Valkonen JPT. Potato gene Y-1 is an N gene homolog that confers cell death upon infection with Potato virus Y. Mol Plant Microbe Interact. 2002;15:717–27.
Vu TV, Choudhury NR, Mukherjee SK. Transgenic tomato plants expressing artificial microRNAs for silencing the pre-coat and coat proteins of a begomovirus, Tomato leaf curl New Delhi virus, show tolerance to virus infection. Virus Res. 2013;172:35–45.
Wang B, Li F, Huang C, Yang X, Qian Y, Xie Y, et al. V2 of tomato yellow leaf curl virus can suppress methylation-mediated transcriptional gene silencing in plants. J Gen Virol. 2014;95:225–30.
Wang H, Hao L, Shung CY, Sunter G, Bisaro DM. Adenosine kinase is inactivated by geminivirus AL2 and L2 proteins. Plant Cell. 2003;15:3020–32.
Wendte JM, Pikaard CS. The RNAs of RNA-directed DNA methylation. Biochem Biophys Acta. 2016;1860:140–8.
Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr C, Baker B. The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell. 1994;78:1101–15.
Yadav RK, Chattopadhyay D. Enhanced viral intergenic region specific siRNA accumulation and DNA methylation correlates with resistance against a geminivirus. Mol Plant-Microbe Interact. 2011;24:1189–97.
Yazdani-Khameneh S, Aboutorabi S, Shoori M, Aghazadeh A, Jahanshahi P, Golnaraghi A, et al. Natural occurrence of tomato leaf curl New Delhi virus in Iranian cucurbit crops. Plant Pathol J. 2016;32:201–8.
Yong CH, Lacatus G, Sunter G. Geminivirus AL2 protein induces expression of, and interacts with, a calmodulin-like gene, an endogenous regulator of gene silencing. Virology. 2014;460:108–18.
Zaidi S, Martin DP, Amin I, Farooq M, Mansoor S. Tomato leaf curl New Delhi virus: a widespread bipartite begomovirus in the territory of monopartite begomoviruses. Mol Plant Pathol. 2016;18:901–11.
ZhouY Rojas MR, Park MR, Seo YS, Lucas WJ, Gilbertson RL. Histone H3 interacts and colocalizes with the nuclear shuttle protein and the movement protein of a geminivirus. J Virol. 2011;85:11821–32.
Zorzatto C, Machado JB, Lopes KVG, Nascimento KJT, Pereira WA, Brustolini OJB, et al. NIK1-mediated translation suppression functions as a plant antiviral immunity mechanism. Nature. 2015;520:679–82.
Zrachya A, Glick E, Levy Y, Arazi T, Citovsky V, Gafni Y. Suppressor of RNA silencing encoded by Tomato yellow leaf curl virus-Israel. Virology. 2007;358:159–65.
Zvereva AS, Pooggin MM. Silencing and innate immunity in plant defense against viral and non-viral pathogens. Viruses. 2012;4:2578–97.
Acknowledgements
The authors’ work in the area of plant-virus interaction was supported by the core grant of National Institute of Plant Genome Research (NIPGR), New Delhi. N.S. and M.P. acknowledges the award of Senior Research Fellowship and TATA Innovation Fellowship (BT/HRD/35/01/02/2017) from Department of Biotechnology, Govt. of India, India, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Additional information

In Honour of Prof AK Sharma, the Founder and Editor-in-Chief of the Nucleus.
Rights and permissions
About this article
Cite this article
Sharma, N., Prasad, M. An insight into plant–Tomato leaf curl New Delhi virus interaction. Nucleus 60, 335–348 (2017). https://doi.org/10.1007/s13237-017-0224-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-017-0224-5