Abstract
Although it has become routine to consider DNA in terms of its role as a carrier of genetic information, it is also an important contributor to the control of gene expression. This regulatory principle arises from its structural properties. DNA is maintained in an underwound state in most bacterial cells and this has important implications both for DNA storage in the nucleoid and for the expression of genetic information. Underwinding of the DNA through reduction in its linking number potentially imparts energy to the duplex that is available to drive DNA transactions, such as transcription, replication and recombination. The topological state of DNA also influences its affinity for some DNA binding proteins, especially in DNA sequences that have a high A + T base content. The underwinding of DNA by the ATP-dependent topoisomerase DNA gyrase creates a continuum between metabolic flux, DNA topology and gene expression that underpins the global response of the genome to changes in the intracellular and external environments. These connections describe a fundamental and generalised mechanism affecting global gene expression that underlies the specific control of transcription operating through conventional transcription factors. This mechanism also provides a basal level of control for genes acquired by horizontal DNA transfer, assisting microbial evolution, including the evolution of pathogenic bacteria.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA carries the genetic information needed to build and operate the cell. Although the expression of this information is controlled at multiple levels, regulation at the level of transcription initiation allows a gene to be controlled at the earliest stage of its expression. The literature is replete with examples of regulatory proteins that control transcription initiation and the history of the field is dominated by examples of protein-dependent control mechanisms, such as those controlling the life cycle of bacteriophage lambda or the expression of the lac operon (Lewis 2011; Oppenheim et al. 2005; Wilson et al. 2007). Research that has focused intensively on protein regulators for more than five decades has pushed the regulatory role of DNA into the background where, at best, it is regarded as contributing to gene control by providing cis-acting sites for the binding of regulatory proteins or through its possession of a structural flexibility that facilitates interactions between bound proteins via DNA looping (Schleif 2013; Semsey et al. 2005). This regulatory model, where DNA plays largely a passive role, is incomplete because it omits the active contribution that is made by DNA itself through its topological dynamism.
In most cells, DNA is found to have a deficit of helical turns and this underwound state places the DNA duplex under torsional stress (Bauer et al. 1980; Boles et al. 1990; Vinograd et al. 1965). This stress is a reflection of the energy content of the DNA and, in bacteria, the ultimate source of this energy is metabolic activity (Westerhoff and van Workum 1990; van Workum et al. 1996). The physical manifestation of its underwound state is seen in the adoption by DNA of a minimal energy conformation in which the helical axis writhes about itself; it can also be expressed through the opening of part of the DNA helix through a loss of base-pairing or by a combination of the two (Bauer et al. 1980). The writhing of the underwound duplex is described as negative supercoiling (Higgins and Vologodskii 2015). In a bacterium such as Escherichia coli, about half of the DNA supercoils are constrained by interaction with proteins (Bliska and Cozzarelli 1987; Pettijohn and Pfenninger 1980), but the energy in the unconstrained portion is available to drive DNA transactions (Booker et al. 2010). The constrained and unconstrained regions of the chromosome are unlikely to be fixed, displaying a dynamism that reflects changes in growth rate, growth phase and changes in the nature and sizes of the populations of DNA binding proteins that decorate the DNA.
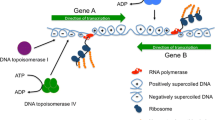
DNA gyrase is the bacterial topoisomerase that underwinds DNA, using energy from ATP hydrolysis to drive the reaction (Gellert et al. 1976; Higgins et al. 1978) (Fig 1). Gyrase activity is sensitive to the ratio of ATP to ADP in the cell and this is, in turn, a reflection of metabolic flux. Cells that are growing rapidly with a high metabolic flux have a higher ratio of ATP to ADP compared to metabolically quiescent cells (van Workum et al. 1996). The resulting enhancement in the negative supercoiling activity of DNA gyrase causes the DNA within rapidly growing bacteria to be more negatively supercoiled than the DNA of slow-growing or non-growing cells (Conter et al. 1997).
Transcription-induced DNA supercoiling resolved by DNA gyrase and DNA topoisomerase I. Genes A and B are transcribed convergently by RNA polymerase (red). The extent of each gene and its direction of transcription are shown by horizontal green arrows. RNA polymerases cannot rotate around the DNA as they track along the template and the template is also unable to rotate. This results in the creation and accumulation of overwound (positively supercoiled) DNA ahead of the polymerases and underwound (negatively supercoiled) DNA behind. The positive supercoils are removed by DNA gyrase in a reaction that is dependent on ATP hydrolysis. This dependency connects DNA gyrase activity to the metabolic status of the bacterium. The negative supercoils are removed by DNA topoisomerase I through an ATP-independent mechanism; the ATP-dependent DNA topoisomerase IV can also relax negative supercoils
Rapidly growing bacteria also support more transcription, especially of the genes and operons that express the translational machinery of the cell and, as we will see below, transcription also contributes to DNA topological change, albeit at a local level (Rovinskiy et al. 2012). It is also prudent to keep in mind that E. coli and related bacteria also possess DNA topoisomerase IV, an enzyme with a structure that is closely related to that of gyrase but which lacks the ability to negatively supercoil DNA (Kato et al. 1990). Topo IV is also ATP dependent and is the principal decatenase of the cell, responsible for the topological separation of daughter chromosomes at the end of genome replication; Topo IV also has the ability to relax negatively supercoiled DNA (Bates and Maxwell 2007; Zawadzki et al. 2015; Zechiedrich et al. 2000). Like DNA gyrase, Topo IV is sensitive to adenylylation by FicT toxins that interfere with ATP binding activity (Harms et al. 2015). Topo IV is not found in all bacteria. For example, Mycobacterium tuberculosis and M. smegmatis combine the decatenase activity of Topo IV with the negative supercoiling activity of gyrase in a single topoisomerase (Aubry et al. 2004; Jain and Nagaraja 2005).
Topo I, which is encoded by the topA gene, catalyses the relaxation of negatively supercoiled DNA in bacteria (Dekker et al. 2002) (Fig. 1). A related topoisomerase, Topo III (topB), contributes weak DNA relaxing activity but acts chiefly as a decatenase (Nurse et al. 2003; Perez-Cheeks et al. 2012). Drugs are available that inhibit gyrase and Topo IV. Of these, novobiocin is particularly useful because it interferes with ATP binding in a dose-dependent manner (Hardy and Cozzarelli 2003; Khodursky et al. 1995; Sugino et al. 1978). Experiments investigating the in vivo effects of the loss of Topo I and III have mainly relied on the use of strains with mutations in their respective genes, topA (Margolin et al. 1985) and topB (Perez-Cheeks et al. 2012).
Work with model bacteria has revealed a homeostatic control mechanism for DNA supercoiling in which feedback onto topoisomerase gene expression helps to keep the superhelical density of the genomic DNA within limits that are compatible with the survival of the organism (Menzel and Gellert 1983; Snoep et al. 2002). A similar control mechanism has been described in Mycobacterium spp. (Ahmed et al. 2016). At its simplest, homeostasis is achieved by inhibiting the promoters of the genes encoding DNA negative supercoiling activity by negative DNA supercoiling, and the promoters of genes encoding DNA relaxing activity by DNA relaxation. The topA gene, encoding DNA topoisomerase I, is stimulated by DNA negative supercoiling (Menzel and Gellert 1983). A promoter architecture in which the −35 and −10 elements have a sub-optimal spacing renders topA sensitive to supercoiling sensitive transcription: this sub-optimal spacing diminishes RNA polymerase binding efficiency but is offset by the DNA twist associated with negative supercoiling of the promoter DNA (Ahmed et al. 2016). In the case of DNA gyrase, the gyr gene promoters experience relaxation stimulated transcription (RST). In E. coli, RST sensing is intrinsic to the gyrA and gyrB promoter elements (Menzel and Gellert 1987; Straney et al. 1994; Liebart et al. 1989), whereas in Mycobacterium spp., it involves longer portions of the chromosome adjacent to the gyrBA operon promoter (Unniraman and Nagaraja 1999; Unniraman et al. 2002).
DNA supercoiling and transcription
DNA supercoiling has the potential to influence transcription initiation, elongation and termination (Chong et al. 2014; Kotlajich et al. 2015; Kravatskaya et al. 2013; Ma and Wang 2014a). Historically, much of the available information about the link between DNA topology and gene expression in bacteria has come from work with the Gram-negative organism Escherichia coli K-12 and its pathogenic relative Salmonella enterica serovar Typhimurium.
A leucine auxotrophic strain of Salmonella Typhimurium harbouring a promoter mutation called leu-500 in its leucine biosynthetic operon led to the identification of supX, a leu-500 suppressor, as the gene encoding DNA topoisomerase I (Friedman and Margolin 1968; Graf and Burns 1973; Margolin et al. 1985). DNA topoisomerase I is an enzyme that relaxes negative supercoils in DNA by raising the linking number of the molecule in steps of 1, making the linking number of the DNA less negative (Wang 1971). The discovery that loss of topoisomerase I expression resulted in a suppression of the leu-500 promoter defect led to speculation that this might be due to DNA in the cell being maintained generally in a more negatively supercoiled state, a state that promoted transcription initiation of the leuABCD operon (Richardson et al. 1984, 1988). This hypothesis is consistent with the observation that the leu-500 mutation results in a more G + C-rich Pribnow box at the leucine biosynthetic operon. Put simply, the more underwound state of the DNA in the supX mutant background might be expected to facilitate the formation of an open complex at the G + C-rich leu-500 promoter (Chen et al. 1992). Furthermore, treatment of a supX leu-500 strain with a gyrase-inhibiting antibiotic was reported to eliminate the suppressive effect of supX, in keeping with a role for elevated DNA supercoiling in leu-500 promoter activation (Pruss and Drlica 1985).
This simple model was shown to be incomplete following studies with derivatives of the supX strain that had topoisomerase one suppressor (tos) mutations and DNA negative supercoiling values close to those of the wild-type Salmonella strain (Richardson et al. 1984, 1988) (Fig. 2). These tos strains remained resolutely Leu+, showing that a simple correlation between leu-500 promoter activity and global levels of DNA supercoiling did not hold (Richardson et al. 1988). Instead, a clear correlation could be seen between leu-500 promoter activity and the absence of DNA topoisomerase I. How could this paradox be resolved?
The leu-500 paradox. The Gibbs free energy of supercoiling (ΔGSC) values (Y-axis), obtained using reporter plasmid supercoiling measurements, are represented for five strains of Salmonella Typhimurium by the five horizontal red lines (Richardson et al. 1984). The wild-type strain is shown on the left, followed by the four strains harbouring the leu-500 promoter mutation. The Leu phenotypes of the strains are given above the horizontal red lines as either Leu+ or Leu−. The presence (TopA+) or absence (TopA−) of active DNA topoisomerase I is indicated below the red line for each strain. The Leu− TopA+ strain (leu-500 strain 1) has a ΔGSC value that is identical to that of the wild type Salmonella Typhimurium strain with its full complement of functioning topoisomerases. The leu-500 strain 2 is Leu+ TopA− strain and has an elevated ΔGSC value because the loss of topoisomerase I results in a globally more negatively supercoiled genome. Strains with a Tos phenotype have topoisomerase one suppressor (tos) mutations. These mutations are typically defects in the negative supercoiling activity of gyrase that return ΔGSC to values that are close to wild type (leu-500 strain 3) or to a lower-than-wild-type value (leu-500 strain 4). If increased negative supercoiling suppresses the leu-500 mutation, then increasing this globally by elimination of the topA gene should create a Leu+ phenotype, and this is, indeed, observed (leu-500 strain 2). Paradoxically, no overall correlation exists between ΔGSC and Leu phenotype in the four strains. Instead, a Leu+ phenotype correlates with the absence of DNA topoisomerase I (TopA−)
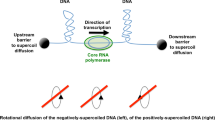
The solution to the leu-500 paradox came from applying the twin supercoiling domain model (Liu and Wang 1987) to the problem. Here, the movement of RNA or DNA polymerase along the DNA duplex introduces topological distortions ahead of and behind itself (Fig. 1). The DNA ahead of the moving polymerase becomes overwound (or positively supercoiled), while the DNA in the wake of the polymerase becomes underwound, or negatively supercoiled (Chong et al. 2014; Higgins 2014; Rahmouni and Wells 1992; Wu et al. 1988). Unless these zones of differentially supercoiled DNA are relaxed, the polymerase can become jammed on its DNA template (Ma et al. 2013). An escape route from this topological dilemma must now be found. The polymerase could rotate around the DNA, allowing the stress created by its tracking movement to be relieved, but this is thought to be unlikely in the crowded and viscous conditions found in the cytoplasm (Koster et al. 2010). The formation of R-loops where nascent RNA forms base pairs with the DNA template strand can also impede RNA polymerase’s freedom to rotate (Drolet 2006). Supercoils can dissipate rapidly by lateral diffusion along the DNA in vitro (van Loenhout et al. 2012), but in vivo, they can quickly be replaced by the next fast-moving polymerase or have their freedom to diffuse impeded by diffusion barriers in the chromosome, including those due to other moving polymerases and their associated local supercoiling domains (Leng and McMacken 2002) or by barriers erected by proteins capable of bridging different parts of a DNA molecule (Ding et al. 2014; Fulcrand et al. 2016). The local supercoiling created by the movement of the polymerases can be eliminated by the action of topoisomerases and this is probably the most common solution employed under physiological conditions, with DNA gyrase relaxing the overwound DNA ahead of the moving polymerase and DNA topoisomerase I relaxing the negatively supercoiled DNA behind (Koster et al. 2010) (Fig. 1). The ability of transcription or DNA replication units to act as engines of local DNA topological change introduces an important mechanism for communication along the chromosome (or other replicon) (El Hanafi and Bossi 2000; Ma and Wang 2014b; Naughton et al. 2013; Tsao et al. 1989; Zhi and Leng 2013). It may also represent part of the organising logic of the chromosome (Booker et al. 2010; Dorman 2013; Higgins 2014; Rovinskiy et al. 2012; Sobetzko 2016; Sobetzko et al. 2012).
Gene-to-gene communication involving local DNA supercoiling is an under-researched topic and some of the clearest examples have grown from investigations of possible solutions to the leu-500 paradox (Fig. 2). An early solution to the paradox posited a role for an (unidentified) upstream promoter in the creation of a local domain of negative supercoiling capable of driving a transition to an open transcription complex at the leu-500 promoter (Lilley and Higgins 1991). This model received strong support from artificial promoter–promoter communication systems established within heavily engineered recombinant plasmids (Chen et al. 1992). The solution to the leu-500 paradox came from an appreciation that it was not the global role of DNA supercoiling but the local action of topoisomerase I acting on locally generated negative supercoils at the leuO promoter that affects leu-500 promoter activity. Activation of the leu-500 promoter in its native location on the chromosome involves collaboration between transcriptionally generated negative supercoiling acting locally in cis via a promoter relay, together with transcription factors acting in trans (Fig. 3). The divergently oriented promoter of the ilvIH operon lies almost 2 kbp upstream of the leu-500 promoter. Transcription from P ilvIH activates the promoter of the leuO gene that is located immediately upstream of P leu-500 ; the divergently oriented P leuO and P leu-500 promoters are separated by a segment of A + T-rich DNA that is a binding and nucleation site for the H-NS nucleoid-associated protein (Fig. 3). The H-NS protein silences the P leu-500 promoter (or the wild type P leu promoter), something that the trans-acting LysR-like LeuO DNA binding protein prevents. LeuO does this by binding at the P leu-500 end of the A + T-rich DNA segment, insulating the P leu-500 promoter from encroachment by the transcription-silencing H-NS protein (Fig. 3). Unravelling the details of this complicated regulatory relay provided an important advance in our understanding of the role of DNA topology in setting the gene regulatory landscape in the genome (Fang and Wu 1998a; 1998b; Opel and Hatfield 2001; Rhee et al. 1999; Wu et al. 1995).
The ilvIH-leuO-leuABCD promoter relay. The segment of the Salmonella Typhimurium chromosome between ilvIH and leuABCD is shown. This genetic arrangement is conserved in E. coli. Activation of the leu-500 mutant promoter in a topA strain is achieved in cis by a promoter-to-promoter DNA supercoiling relay involving promoters P ilvIH , P leuO and P leu-500 (Wu et al. 1995). There is also a positive role for the LysR-like transcription factor, LeuO, encoded by the leuO gene. LeuO prevents encroachment by the H-NS nucleoid-associated transcription-silencing protein from the region of A + T-rich DNA into the promoter P leu-500 . Although the DNA sequence of the corresponding region in E. coli is different, the average A + T content is the same (Haughn et al. 1986) and the promoter relay is conserved in both species (Chen et al. 2005). Another DNA binding protein, the leucine-responsive regulatory protein, Lrp, contributes by stimulating transcription of P ilvIH (Wang et al. 1993)
Barrier proteins, DNA supercoiling and nucleoid structure
The intense investigation of the relationship between leu-500 promoter activity and DNA supercoiling revealed a role for the LeuO DNA binding protein as a structural element in the vicinity of the leuABCD operon (Chen and Wu 2005; Chen et al. 2003). The LeuO protein can form tetramers (Guadarrama et al. 2014) and LeuO has the ability to bridge together different parts of the same DNA molecule (Chen and Wu 2005). Closing loops of DNA in this way has the potential to create independently supercoiled domains and any DNA binding protein that has DNA–protein–DNA bridge-forming activity has the potential to create such domains. For example, the unrelated repressor proteins LacI and lambda CI have been shown to do this at, respectively, operator sites in the E. coli lac operon and in bacteriophage lambda (Ding et al. 2014; Fulcrand et al. 2016). The positioning of the bridges is determined by the locations of the binding sites for the DNA bridging proteins. LacI and lambda CI have relatively stringent binding site sequence requirements, unlike LeuO, which recognises a much more degenerate DNA sequence (Dillon and Dorman 2012; Shimada et al. 2011). Consequently, LeuO has been found to bind to scores of sites around the bacterial chromosome, giving it the potential to participate in the general organisation of the nucleoid. LeuO also contributes to nucleoid architecture through its ability to influence the DNA-dependent lateral spreading of the H-NS nucleoid-associated protein locally (Chen and Wu 2005) (Fig. 3) and globally (Dillon et al. 2012; Shimada et al. 2011).
The H-NS protein plays a major role in the global control of transcription in Gram-negative bacteria and alterations to its expression pattern have profound impacts on bacterial physiology (Dorman 2013; Fitzgerald et al. 2015; McGovern et al. 1994). This protein has a promiscuous relationship with the genome, binding to hundreds of sites that have little in common other than a high A + T base content and some intrinsic DNA flexibility (Dillon et al. 2010; Lucchini et al. 2006; Navarre et al. 2006; Oshima et al. 2006). It binds to nucleation sites on the DNA and then spreads laterally; its bridging capacity lends to it a Velcro-like quality that can zip together different segments of the chromosome (Bouffartigues et al. 2007; Dame et al. 2006; Maurer et al. 2009; Noom et al. 2007). DNA-dependent lateral polymerisation of H-NS can result in intrusion into gene promoters, resulting in transcription silencing, so limiting the advance of H-NS using appropriately placed barrier proteins provides a mechanism to prevent silencing of selected targets (Caramel and Schnetz 1998) (Fig. 3). Data from atomic force microscopy experiments indicate that the compaction associated with plectonemic wrapping of supercoiled DNA encourages DNA–H-NS–DNA bridging (Maurer et al. 2009). In principle, the protein could also bridge adjacent turns of toroidal DNA too.
An environmentally responsive global regulatory system
Variable DNA topology serves as the basis of a global regulatory system that allows the gene expression profile of the bacterium to vary in response to those environmental influences that cause DNA topology to change (Dorman 1991, 2006). Shifts in the ratio of ATP to ADP can arise from a very wide range of circumstances and, ultimately, this ratio serves as a powerful barometer of the health of the bacterial cell (Hsieh et al. 1991a, b; Meury and Kohiyama 1992). Experimental data demonstrating a response at the level of DNA supercoiling to variations in pH (Bang et al. 2002; Karem and Foster 1993; Quinn et al. 2014), temperature (Goldstein and Drlica 1984), osmotic pressure (Alice and Sanchez-Rivas 1997; Bordes et al. 2003; Cheung et al. 2003; Higgins et al. 1988; Meury and Kohiyama 1992; O’Byrne et al. 1992; Sheehan et al. 1992), oxygen supply (Bebbington and Williams 2001; Cameron et al. 2011, 2013; Cortassa and Aon 1993; Dixon et al. 1988; Dorman et al. 1988; Malkhosyan et al. 1991; Yamamoto and Droffner 1985), oxidative stress (Weinstein-Fischer et al. 2000), nutrition (Balke and Gralla 1987), growth phase (Bordes et al. 2003; Conter et al. 1997), intracellular growth (Ó Cróinín et al. 2006) and much more have been reported. A rapidly growing cell has different gene expression requirements to one that is in stationary phase and one that is undergoing a growth phase transition has yet another set of needs. Exploiting the topological state of the DNA in the chromosome as both a sensor and a gene-to-gene telegraph of physiological status allows the individual cell to adjust its transcription profile to ensure an optimal response to changing circumstances. Variations in the quality and quantity of the response between individual cells creates variety across a population of genetically identical bacterial cells that allows differences in relative competitive fitness to emerge that may prove useful in the survival of the population.
The relationship between DNA topology and gene expression is not unidirectional: transcription itself creates topological change in the DNA template, so environmental factors that influence transcription initiation, elongation or termination all have the potential to modulate the superhelicity of the DNA in the genome (Bohrer and Roberts 2016; Kouzine et al. 2008; Kotlajich et al. 2015; Ma and Wang 2014a).
Protein binding as a function of DNA topology
The conventional view of DNA binding by proteins emphasises the importance of the base sequence of the binding site. This is, indeed, important for binding by proteins that rely chiefly on a direct readout mechanism in which a domain of the protein, typically the helix–turn–helix motif of the DNA binding motif, searches for a match to the consensus binding site sequence for that protein. Proteins that rely on an indirect readout mechanism of site recognition are more sensitive to variations in DNA structure than simply the presence of a linear sequence of bases (Chiu et al. 2015; Zhou et al. 2013). H-NS appears to belong to this latter category, as shown by its sensitivity to DNA curvature and the presence of nucleotide sequence steps (e.g. TpA) that lend themselves to DNA flexibility (Bouffartigues et al. 2007; Lang et al. 2007). The promiscuous relationship of H-NS with DNA has been mentioned already. This is a feature that H-NS shares with many other proteins that bind to A + T-rich DNA, including proteins that bind using a winged helix–turn–helix motif (Brennan 1993; Dolan et al. 2011; Kenney 2002; Martínez-Hackert and Stock 1997). Here, the protein is interacting with both the major groove (where the DNA base sequence is accessible) and the minor groove (where a specific DNA sequence is not being sought). LeuO belongs to this category of proteins. As a LysR-like transcription factor, LeuO typically binds to regulatory sequences located between divergently transcribed genes, where it seeks a match to a very loose consensus binding site sequence: T-N11-A (Alanazi et al. 2013; Schell 1993; Sheehan and Dorman 1998) (Fig. 3). These very low stringency requirements for binding site matches make LeuO a useful antagonist for the transcription silencer, H-NS, with its preference for binding to A + T-rich DNA (Dillon et al. 2012). A predilection for A + T-rich DNA means that proteins like LeuO and other winged helix–turn–helix proteins must dock with DNA, where the minor groove is up to three times narrower than in more G + C-rich sequences (Oguey et al. 2010; Rohs et al. 2009). Supercoiling such DNA sequences to different levels is itself likely to impose restrictions on successful DNA binding. This has been tested in the case of OmpR, a response regulator protein that binds using a winged helix–turn–helix motif (Kenney 2002). Here, the binding of OmpR to the same DNA sequence was progressively impeded by the negative supercoiling of that sequence within a circular DNA molecule in vitro or on a negatively supercoiled chromosome in vivo (Cameron and Dorman 2012). Because they rely on DNA structure rather than the presence of a specific base sequence, indirect readout mechanisms of DNA binding offer a powerful means to drive the evolution of gene regulation (Xu et al. 2014). Proteins like OmpR, with low requirements for specific base sequences in their binding sites but which are sensitive to the superhelicity of the binding site DNA, are well placed to recruit new genes to their regulons (Quinn et al. 2014), including genes that arrive in the bacterial cell via horizontal gene transfer (Cordeiro et al. 2011).
DNA supercoiling, bacterial evolution and pathogenesis
Horizontal gene transfer plays an important role in the rapid evolution of bacterial species (Porwollik and McClelland 2003; Soucy et al. 2015; Touchon and Rocha 2016). Newly acquired genes have to be integrated not only into their new host physically, but also from a regulatory perspective (Dorman 2007; Higashi et al. 2016). Comprehensive information about the average superhelical density of DNA in different bacterial species is not available and the information that is in the literature has come from studies of just a few model organisms. However, it is clear that supercoiling set points can differ even between closely related examples, such as E. coli and Salmonella Typhimurium (Cameron et al. 2011; Champion and Higgins 2007; Quinn et al. 2014). Despite this, groups of genes that respond to changes in DNA supercoiling in one species may still do so in another; for example, the supercoiling-dependent promoter coupling that affects leuABCD expression in Salmonella Typhimurium is conserved in E. coli (Chen et al. 2005). Differences in supercoiling levels between species mean that an imported gene may acquire a pattern of expression in its new host that differs from its previous pattern, even if the gene is placed in an equivalent location of the new host’s genome. In this way, the gene may contribute in new ways to the life of the cell due to adjustments to its expression pattern, setting the novel gene–organism partnership on a new evolutionary trajectory.
A gene may not have to leave its original host cell to experience new patterns of DNA topological influence. Simply repositioning the gene on the single, circular chromosome of the bacterium may achieve this effect (Brambilla and Sclavi 2015; Fitzgerald et al. 2015; Gerganova et al. 2015). Bioinformatic analyses and experimental work have indicated that gene location can affect expression for reasons other than gene dosage (i.e. because a gene near the origin of chromosome replication doubles its copy number earlier in the cell cycle than one near the replication terminus) and that part of the underlying reason reflects differences in DNA supercoiling at different chromosomal sites (Bryant et al. 2014; Sobetzko et al. 2012).
Horizontal gene transfer has played an important role in the evolution of bacterial pathogens with many of their key virulence genes being located in genetic elements that were acquired in this way (Cotter and DiRita 2000; Groisman and Casadesús 2005; Porwollik and McClelland 2003). In Gram-negative pathogens, these laterally acquired genes are characterised by a higher A + T base content than the core genome, sensitivity to variations in DNA supercoiling in their expression and silencing by the H-NS nucleoid-associated protein (Dorman and Corcoran 2009).
Bacterial pathogenesis frequently involves the expression of specialist genes encoding factors that attach the bacterium to its host, that allow it to enter and survive within host cells and to evade the host defences (Hay and Zhu 2015). These genes are upregulated in response to physical and chemical signals that are encountered in the host environment. Other signals indicate that the bacterium has exited the host (Merrell et al. 2002). Experiments with many pathogens involved in a wide variety of infections have shown that these infection-relevant signals modulate the supercoiling of bacterial DNA, providing a background regulatory influence upon which the dedicated gene regulatory proteins operate. Examples have come from studies in Bordetella pertussis (Graeff-Wohlleben et al. 1995), Campylobacter jejuni (Dedieu et al. 2002), Chlamydia (Niehus et al. 2008), Dickeya dadantii (Hérault et al. 2014), E. coli (Beltrametti et al. 1999; Sánchez-Céspedes et al. 2015), Helicobacter pylori (Ye et al. 2007), Salmonella spp. (Bang et al. 2002; Galán and Curtiss 1990; Leclerc et al. 1998; Ó Cróinín et al. 2006; Webber et al. 2013), Shigella flexneri (Dorman et al. 1990), Vibrio cholerae (Parsot and Mekalanos 1992), Staphylococcus aureus (Fournier and Klier 2004; Schröder et al. 2014; Sheehan et al. 1992) and Yersinia enterocolitica (Rohde et al. 1994).
DNA relaxation is an important event in controlling the expression of genes that adjust Salmonella physiology to its host (Cameron and Dorman 2012; Ó Cróinín et al. 2006; Quinn et al. 2014). This bacterium is a facultative intracellular pathogen that can survive in the normally lethal environment of the macrophage in mammalian hosts. It does this by expressing clusters of horizontally acquired genes coding for an elaborate protein secretion machine and effector proteins that, once exported, modify the vacuole to render it harmless to the bacterium (Fass and Groisman 2009). This gene cluster is located within the SPI2 pathogenicity island and the transcription promoters in SPI2 become more active when the DNA template relaxes (Cameron and Dorman 2012; Quinn et al. 2014). In part, this is because the relaxed DNA conformation makes the promoters into better binding targets for the OmpR transcription factor, with its winged helix–turn–helix DNA-binding motif (Cameron and Dorman 2012). OmpR works in tandem with the SPI2-encoded SsrB response regulator to activate SPI2 promoters (Feng et al. 2004). Macrophage–vacuole-associated signals such as acid pH and low magnesium concentrations also serve to trigger these molecular events (Fass and Groisman 2009). Such additional information from the environment is likely to be essential in mounting an appropriate response at the level of specific gene expression activation or inhibition: a global event such as DNA relaxation or supercoiling is too general in its effects to represent a reliable regulatory circuit on its own.
This example from Salmonella pathogenesis and virulence gene control illustrates both the utility and the limitations of variable DNA topology as a regulatory principle in gene regulation. It has an impressive ‘reach’ because potentially every gene in the cell can be influenced by global adjustments to DNA superhelicity. It functions as a local actor, allowing topological disturbance caused by the transcription of one gene to influence its upstream and downstream neighbours. However, its effects are too generalised to make it useful as a regulator of transcription in response to specific signals from the external or the internal environment. Typically, this specificity is provided by proteins that sense and report individual changes to the chemical or physical environment. The role of DNA topology lies in facilitating or impeding the work of these specific protein actors, both individually and collectively, as it helps to integrate the many stimuli that the bacterium receives as it navigates its environment.
Concluding remarks
Genome-wide approaches to understanding gene expression patterns have become commonplace in modern molecular microbiology, providing us with the means to explore microbial cell biology at the interface between nucleoid structure and gene regulation. To derive the maximum benefit from this exploration, it will be important to appreciate the pervasive influence of variable DNA topology on gene expression. In this way, our research will not only advance the understanding of existing microbes, but also guide endeavours in synthetic biology that either seek to ‘rewire’ existing bacteria or produce synthetic ones. In addition, a view of gene regulation that is informed by an understanding of the importance of DNA topology will assist us in appreciating the (often unintended) consequences of exposing populations of bacterial cells to antimicrobial agents that target topoisomerases at concentrations that are too low to ensure lethality. Clinical practice may be improved and environmental contamination avoided through an understanding of the impact of drug treatments on DNA topology and, hence, on gene expression patterns in bacteria that are either in the patient or in the external environment.
References
Ahmed W, Menon S, Karthik PVDNB, Nagaraja V (2016) Autoregulation of topoisomerase I expression by supercoiling sensitive transcription. Nucleic Acids Res 44:1541–1552
Alanazi AM, Neidle EL, Momany C (2013) The DNA-binding domain of BenM reveals the structural basis for the recognition of a T-N11-A sequence motif by LysR-type transcriptional regulators. Acta Crystallogr D Biol Crystallogr 69:1995–2007
Alice AF, Sanchez-Rivas C (1997) DNA supercoiling and osmoresistance in Bacillus subtilis 168. Curr Microbiol 35:309–315
Aubry A, Pan XS, Fisher LM, Jarlier V, Cambau E (2004) Mycobacterium tuberculosis DNA gyrase: interaction with quinolones and correlation with antimycobacterial drug activity. Antimicrob Agents Chemother 48:1281–1288
Balke VL, Gralla JD (1987) Changes in the linking number of supercoiled DNA accompany growth transitions in Escherichia coli. J Bacteriol 169:4499–4506
Bang IS, Audia JP, Park YK, Foster JW (2002) Autoinduction of the ompR response regulator by acid shock and control of the Salmonella enterica acid tolerance response. Mol Microbiol 44:1235–1250
Bates AD, Maxwell A (2007) Energy coupling in type II topoisomerases: why do they hydrolyze ATP? Biochemistry 46:7929–7941
Bauer WR, Crick FHC, White JH (1980) Supercoiled DNA. Sci Am 243:100–113
Bebbington KJ, Williams HD (2001) A role for DNA supercoiling in the regulation of the cytochrome bd oxidase of Escherichia coli. Microbiology 147:591–598
Beltrametti F, Kresse AU, Guzmán CA (1999) Transcriptional regulation of the esp genes of enterohemorrhagic Escherichia coli. J Bacteriol 181:3409–3418
Bliska JB, Cozzarelli NR (1987) Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol 194:238–218
Bohrer CH, Roberts E (2016) A biophysical model of supercoiling dependent transcription predicts a structural aspect to gene regulation. BMC Biophys 9:2
Boles TC, White JH, Cozzarelli NR (1990) Structure of plectonemically supercoiled DNA. J Mol Biol 213:931–951
Booker BM, Deng S, Higgins NP (2010) DNA topology of highly transcribed operons in Salmonella enterica serovar Typhimurium. Mol Microbiol 78:1348–1364
Bordes P, Conter A, Morales V, Bouvier J, Kolb A, Gutierrez C (2003) DNA supercoiling contributes to disconnect sigmaS accumulation from sigmaS-dependent transcription in Escherichia coli. Mol Microbiol 48:561–571
Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S (2007) H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat Struct Mol Biol 14:441–448
Brambilla E, Sclavi B (2015) Gene regulation by H-NS as a function of growth conditions depends on chromosomal position in Escherichia coli. G3 (Bethesda) 5:605–614
Brennan RG (1993) The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell 74:773–776
Bryant JA, Sellars LE, Busby SJ, Lee DJ (2014) Chromosome position effects on gene expression in Escherichia coli K-12. Nucleic Acids Res 42:11383–11392
Cameron AD, Dorman CJ (2012) A fundamental regulatory mechanism operating through OmpR and DNA topology controls expression of Salmonella pathogenicity islands SPI-1 and SPI-2. PLoS Genet 8, e1002615
Cameron AD, Stoebel DM, Dorman CJ (2011) DNA supercoiling is differentially regulated by environmental factors and FIS in Escherichia coli and Salmonella enterica. Mol Microbiol 80:85–101
Cameron AD, Kröger C, Quinn HJ, Scally IK, Daly AJ, Kary SC, Dorman CJ (2013) Transmission of an oxygen availability signal at the Salmonella enterica serovar Typhimurium fis promoter. PLoS One 8, e84382
Caramel A, Schnetz K (1998) Lac and Lambda repressors relieve silencing of the Escherichia coli bgl promoter. Activation by alteration of a repressing nucleoprotein complex. J Mol Biol 284:875–883
Champion K, Higgins NP (2007) Growth rate toxicity phenotypes and homeostatic supercoil control differentiate Escherichia coli from Salmonella enterica serovar Typhimurium. J Bacteriol 189:5839–5849
Chen CC, Wu HY (2005) LeuO protein delimits the transcriptionally active and repressive domains on the bacterial chromosome. J Biol Chem 280:15111–15121
Chen D, Bowater R, Dorman CJ, Lilley DM (1992) Activity of a plasmid-borne leu-500 promoter depends on the transcription and translation of an adjacent gene. Proc Natl Acad Sci U S A 89:8784–8788
Chen CC, Ghole M, Majumder A, Wang Z, Chandana S, Wu HY (2003) LeuO-mediated transcriptional derepression. J Biol Chem 278:38094–38103
Chen CC, Chou MY, Huang CH, Majumder A, Wu HY (2005) A cis-spreading nucleoprotein filament is responsible for the gene silencing activity found in the promoter relay mechanism. J Biol Chem 280:5101–5112
Cheung KJ, Badarinarayana V, Selinger DW, Janse D, Church GM (2003) A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res 13:206–215
Chiu TP, Yang L, Zhou T, Main BJ, Parker SC, Nuzhdin SV, Tullius TD, Rohs R (2015) GBshape: a genome browser database for DNA shape annotations. Nucleic Acids Res 43:D103–D109
Chong S, Chen C, Ge H, Xie XS (2014) Mechanism of transcriptional bursting in bacteria. Cell 158:314–326
Conter A, Menchon C, Gutierrez C (1997) Role of DNA supercoiling and rpoS sigma factor in the osmotic and growth phase-dependent induction of the gene osmE of Escherichia coli K12. J Mol Biol 273:75–83
Cordeiro TN, Schmidt H, Madrid C, Juárez A, Bernadó P, Griesinger C, García J, Pons M (2011) Indirect DNA readout by an H-NS related protein: structure of the DNA complex of the C-terminal domain of Ler. PLoS Pathog 7, e1002380
Cortassa S, Aon MA (1993) Altered topoisomerase activities may be involved in the regulation of DNA supercoiling in aerobic-anaerobic transitions in Escherichia coli. Mol Cell Biochem 126:115–124
Cotter PA, DiRita VJ (2000) Bacterial virulence gene regulation: an evolutionary perspective. Annu Rev Microbiol 54:519–565
Dame RT, Noom MC, Wuite GJ (2006) Bacterial chromatin organization by H-NS protein unravelled using dual DNA manipulation. Nature 444:387–390
Dedieu L, Pagès JM, Bolla JM (2002) Environmental regulation of Campylobacter jejuni major outer membrane protein porin expression in Escherichia coli monitored by using green fluorescent protein. Appl Environ Microbiol 68:4209–4215
Dekker NH, Rybenkov VV, Duguet M, Crisona NJ, Cozzarelli NR, Bensimon D, Croquette V (2002) The mechanism of type IA topoisomerases. Proc Natl Acad Sci U S A 99:12126–12131
Dillon SC, Cameron AD, Hokamp K, Lucchini S, Hinton JC, Dorman CJ (2010) Genome-wide analysis of the H-NS and Sfh regulatory networks in Salmonella Typhimurium identifies a plasmid-encoded transcription silencing mechanism. Mol Microbiol 76:1250–1265
Dillon SC, Espinosa E, Hokamp K, Ussery DW, Casadesús J, Dorman CJ (2012) LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol Microbiol 85:1072–1089
Ding Y, Manzo C, Fulcrand G, Leng F, Dunlap D, Finzi L (2014) DNA supercoiling: a regulatory signal for the λ repressor. Proc Natl Acad Sci U S A 111:15402–15407
Dixon RA, Henderson NC, Austin S (1988) DNA supercoiling and aerobic regulation of transcription from the Klebsiella pneumoniae nifLA promoter. Nucleic Acids Res 16:9933–9946
Dolan KT, Duguid EM, He C (2011) Crystal structures of SlyA protein, a master virulence regulator of Salmonella, in free and DNA-bound states. J Biol Chem 286:22178–22185
Dorman CJ (1991) DNA supercoiling and environmental regulation of gene expression in pathogenic bacteria. Infect Immun 59:745–749
Dorman CJ (2006) DNA supercoiling and bacterial gene expression. Sci Prog 89:151–166
Dorman CJ (2007) H-NS, the genome sentinel. Nat Rev Microbiol 5:157–161
Dorman CJ (2013) Genome architecture and global gene regulation in bacteria: making progress towards a unified model? Nat Rev Microbiol 11:349–355
Dorman CJ, Corcoran CP (2009) Bacterial DNA topology and infectious disease. Nucleic Acids Res 37:672–678
Dorman CJ, Barr GC, Ní Bhriain N, Higgins CF (1988) DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J Bacteriol 170:2816–2826
Dorman CJ, Ni Bhriain N, Higgins CF (1990) DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature 344:789–792
Drolet M (2006) Growth inhibition mediated by excess negative supercoiling: the interplay between transcription elongation, R-loop formation and DNA topology. Mol Microbiol 59:723–730
El Hanafi D, Bossi L (2000) Activation and silencing of leu-500 promoter by transcription-induced DNA supercoiling in the Salmonella chromosome. Mol Microbiol 37:583–594
Fang M, Wu HY (1998a) A promoter relay mechanism for sequential gene activation. J Bacteriol 180:626–633
Fang M, Wu HY (1998b) Suppression of leu-500 mutation in topA+ Salmonella typhimurium strains. The promoter relay at work. J Biol Chem 273:29929–29934
Fass E, Groisman EA (2009) Control of Salmonella pathogenicity island-2 gene expression. Curr Opin Microbiol 12:199–204
Feng X, Walthers D, Oropeza R, Kenney LJ (2004) The response regulator SsrB activates transcription and binds to a region overlapping OmpR binding sites at Salmonella pathogenicity island 2. Mol Microbiol 54:823–835
Fitzgerald S, Dillon SC, Chao TC, Wiencko HL, Hokamp K, Cameron AD, Dorman CJ (2015) Re-engineering cellular physiology by rewiring high-level global regulatory genes. Sci Rep 5:17653. doi:10.1038/srep17653
Fournier B, Klier A (2004) Protein A gene expression is regulated by DNA supercoiling which is modified by the ArlS-ArlR two-component system of Staphylococcus aureus. Microbiology 150:3807–3819
Friedman SB, Margolin P (1968) Evidence for an altered operator specificity: catabolite repression control of the leucine operon in Salmonella typhimurium. J Bacteriol 95:2263–2269
Fulcrand G, Dages S, Zhi X, Chapagain P, Gerstman BS, Dunlap D, Leng F (2016) DNA supercoiling, a critical signal regulating the basal expression of the lac operon in Escherichia coli. Sci Rep 6:19243. doi:10.1038/srep19243
Galán JE, Curtiss R 3rd (1990) Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun 58:1879–1885
Gellert M, Mizuuchi K, O’Dea MH, Nash HA (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A 73:3872–3876
Gerganova V, Berger M, Zaldastanishvili E, Sobetzko P, Lafon C, Mourez M, Travers A, Muskhelishvili G (2015) Chromosomal position shift of a regulatory gene alters the bacterial phenotype. Nucleic Acids Res 43:8215–8226
Goldstein E, Drlica K (1984) Regulation of bacterial DNA supercoiling: plasmid linking numbers vary with growth temperature. Proc Natl Acad Sci U S A 81:4046–4050
Graeff-Wohlleben H, Deppisch H, Gross R (1995) Global regulatory mechanisms affect virulence gene expression in Bordetella pertussis. Mol Gen Genet 247:86–94
Graf LH Jr, Burns RO (1973) The supX-leu-500 mutations and expression of the leucine operon. Mol Gen Genet 126:291–301
Groisman EA, Casadesús J (2005) The origin and evolution of human pathogens. Mol Microbiol 56:1–7
Guadarrama C, Medrano-López A, Oropeza R, Hernández-Lucas I, Calva E (2014) The Salmonella enterica serovar Typhi LeuO global regulator forms tetramers: residues involved in oligomerization, DNA binding, and transcriptional regulation. J Bacteriol 196:2143–2154
Hardy CD, Cozzarelli NR (2003) Alteration of Escherichia coli topoisomerase IV to novobiocin resistance. Antimicrob Agents Chemother 47:941–947
Harms A, Stanger FV, Scheu PD, de Jong IG, Goepfert A, Glatter T, Gerdes K, Schirmer T, Dehio C (2015) Adenylylation of gyrase and Topo IV by FicT toxins disrupts bacterial DNA topology. Cell Rep 12:1497–1507
Haughn GW, Wessler SR, Gemmill RM, Calvo JM (1986) High A + T content conserved in DNA sequences upstream of leuABCD in Escherichia coli and Salmonella typhimurium. J Bacteriol 166:1113–1117
Hay AJ, Zhu J (2015) Host intestinal signal-promoted biofilm dispersal induces Vibrio cholerae colonization. Infect Immun 83:317–323
Hérault E, Reverchon S, Nasser W (2014) Role of the LysR-type transcriptional regulator PecT and DNA supercoiling in the thermoregulation of pel genes, the major virulence factors in Dickeya dadantii. Environ Microbiol 16:734–745
Higashi K, Tobe T, Kanai A, Uyar E, Ishikawa S, Suzuki Y, Ogasawara N, Kurokawa K, Oshima T (2016) H-NS facilitates sequence diversification of horizontally transferred DNAs during their integration in host chromosomes. PLoS Genet 12, e1005796
Higgins NP (2014) RNA polymerase: chromosome domain boundary maker and regulator of supercoil density. Curr Opin Microbiol 22:138–143
Higgins NP, Vologodskii AV (2015) Topological behavior of plasmid DNA. Microbiol Spectr 3(2). doi: 10.1128/microbiolspec.PLAS-0036-2014
Higgins NP, Peebles CL, Sugino A, Cozzarelli NR (1978) Purification of subunits of Escherichia coli DNA gyrase and reconstitution of enzymatic activity. Proc Natl Acad Sci U S A 75:1773–1777
Higgins CF, Dorman CJ, Stirling DA, Waddell L, Booth IR, May G, Bremer E (1988) A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell 52:569–584
Hsieh LS, Burger RM, Drlica K (1991a) Bacterial DNA supercoiling and [ATP]/[ADP]. Changes associated with a transition to anaerobic growth. J Mol Biol 219:443–450
Hsieh LS, Rouviere-Yaniv J, Drlica K (1991b) Bacterial DNA supercoiling and [ATP]/[ADP] ratio: changes associated with salt shock. J Bacteriol 173:3914–3917
Jain P, Nagaraja V (2005) An atypical type II topoisomerase from Mycobacterium smegmatis with positive supercoiling activity. Mol Microbiol 58:1392–1405
Karem K, Foster JW (1993) The influence of DNA topology on the environmental regulation of a pH-regulated locus in Salmonella typhimurium. Mol Microbiol 10:75–86
Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H (1990) New topoisomerase essential for chromosome segregation in E. coli. Cell 63:393–404
Kenney LJ (2002) Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol 5:135–141
Khodursky AB, Zechiedrich EL, Cozzarelli NR (1995) Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci U S A 92:11801–11805
Koster DA, Crut A, Shuman S, Bjornsti M-A, Dekker NH (2010) Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell 142:519–530
Kotlajich MV, Hron DR, Boudreau BA, Sun Z, Lyubchenko YL, Landick R (2015) Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. Elife 4 doi: 10.7554/eLife.04970
Kouzine F, Sanford S, Elisha-Feil Z, Levens D (2008) The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol 15:146–154
Kravatskaya GI, Chechetkin VR, Kravatsky YV, Tumanyan VG (2013) Structural attributes of nucleotide sequences in promoter regions of supercoiling-sensitive genes: how to relate microarray expression data with genomic sequences. Genomics 101:1–11
Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, Stella S, Babu MM, Travers A (2007) High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res 35:6330–6337
Leclerc GJ, Tartera C, Metcalf ES (1998) Environmental regulation of Salmonella typhi invasion-defective mutants. Infect Immun 66:682–691
Leng F, McMacken R (2002) Potent stimulation of transcription-coupled DNA supercoiling by sequence-specific DNA-binding proteins. Proc Natl Acad Sci U S A 99:9139–9144
Lewis M (2011) A tale of two repressors. J Mol Biol 409:14–27
Liebart JC, Paolozzi L, Camera MG, Pedrini AM, Ghelardini P (1989) The expression of the DNA ligase gene of Escherichia coli is stimulated by relaxation of chromosomal supercoiling. Mol Microbiol 3:269–273
Lilley DM, Higgins CF (1991) Local DNA topology and gene expression: the case of the leu-500 promoter. Mol Microbiol 5:779–783
Liu LF, Wang JC (1987) Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A 84:7024–7027
Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC (2006) H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog 2, e81
Ma J, Wang M (2014a) Interplay between DNA supercoiling and transcription elongation. Transcription 5, e28636
Ma J, Wang MD (2014b) RNA polymerase is a powerful torsional motor. Cell Cycle 13:337–338
Ma J, Bai L, Wang MD (2013) Transcription under torsion. Science 340:1580–1583
Malkhosyan SR, Panchenko YuA, Rekesh AN (1991) A physiological role for DNA supercoiling in the anaerobic regulation of colicin gene expression. Mol Gen Genet 225:342–345
Margolin P, Zumstein L, Sternglanz R, Wang JC (1985) The Escherichia coli supX locus is topA, the structural gene for DNA topoisomerase I. Proc Natl Acad Sci U S A 82:5437–5441
Martínez-Hackert E, Stock AM (1997) Structural relationships in the OmpR family of winged-helix transcription factors. J Mol Biol 269:301–312
Maurer S, Fritz J, Muskhelishvili G (2009) A systematic in vitro study of nucleoprotein complexes formed by bacterial nucleoid-associated proteins revealing novel types of DNA organization. J Mol Biol 387:1261–1276
McGovern V, Higgins NP, Chiz RS, Jaworski A (1994) H-NS over-expression induces an artificial stationary phase by silencing global transcription. Biochimie 76:1019–10129
Menzel R, Gellert M (1983) Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell 34:105–113
Menzel R, Gellert M (1987) Modulation of transcription by DNA supercoiling: a deletion analysis of the Escherichia coli gyrA and gyrB promoters. Proc Natl Acad Sci U S A 84:4185–4189
Merrell DS, Butler SM, Qadri F, Dolganov NA, Alam A, Cohen MB, Calderwood SB, Schoolnik GK, Camilli A (2002) Host-induced epidemic spread of the cholera bacterium. Nature 417:642–645
Meury J, Kohiyama M (1992) Potassium ions and changes in bacterial DNA supercoiling under osmotic stress. FEMS Microbiol Lett 78:159–164
Naughton C, Corless S, Gilbert N (2013) Divergent RNA transcription: a role in promoter unwinding? Transcription 4:162–166
Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC (2006) Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236–238
Niehus E, Cheng E, Tan M (2008) DNA supercoiling-dependent gene regulation in Chlamydia. J Bacteriol 190:6419–6427
Noom MC, Navarre WW, Oshima T, Wuite GJ, Dame RT (2007) H-NS promotes looped domain formation in the bacterial chromosome. Curr Biol 17:R913–R914
Nurse P, Levine C, Hassing H, Marians KJ (2003) Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J Biol Chem 278:8653–8660
Ó Cróinín T, Carroll RK, Kelly A, Dorman CJ (2006) Roles for DNA supercoiling and the Fis protein in modulating expression of virulence genes during intracellular growth of Salmonella enterica serovar Typhimurium. Mol Microbiol 62:869–882
O’Byrne CP, Ní Bhriain N, Dorman CJ (1992) The DNA supercoiling-sensitive expression of the Salmonella typhimurium his operon requires the his attenuator and is modulated by anaerobiosis and by osmolarity. Mol Microbiol 6:2467–2476
Oguey C, Foloppe N, Hartmann B (2010) Understanding the sequence-dependence of DNA groove dimensions: implications for DNA interactions. PLoS One 5, e15931
Opel ML, Hatfield GW (2001) DNA supercoiling-dependent transcriptional coupling between the divergently transcribed promoters of the ilvYC operon of Escherichia coli is proportional to promoter strengths and transcript lengths. Mol Microbiol 39:191–198
Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S (2005) Switches in bacteriophage lambda development. Annu Rev Genet 39:409–429
Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N (2006) Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res 13:141–153
Parsot C, Mekalanos JJ (1992) Structural analysis of the acfA and acfD genes of Vibrio cholerae: effects of DNA topology and transcriptional activators on expression. J Bacteriol 174:5211–5218
Perez-Cheeks BA, Lee C, Hayama R, Marians KJ (2012) A role for topoisomerase III in Escherichia coli chromosome segregation. Mol Microbiol 86:1007–1022
Pettijohn DE, Pfenninger O (1980) Supercoils in prokaryotic DNA restrained in vivo. Proc Natl Acad Sci U S A 77:1331–1335
Porwollik S, McClelland M (2003) Lateral gene transfer in Salmonella. Microbes Infect 5:977–989
Pruss GJ, Drlica K (1985) DNA supercoiling and suppression of the leu-500 promoter mutation. J Bacteriol 164:947–949
Quinn HJ, Cameron AD, Dorman CJ (2014) Bacterial regulon evolution: distinct responses and roles for the identical OmpR proteins of Salmonella Typhimurium and Escherichia coli in the acid stress response. PLoS Genet 10, e1004215
Rahmouni AR, Wells RD (1992) Direct evidence for the effect of transcription on local DNA supercoiling in vivo. J Mol Biol 223:131–144
Rhee KY, Opel M, Ito E, Hung Sp, Arfin SM, Hatfield GW (1999) Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc Natl Acad Sci U S A 96:14294–14299
Richardson SM, Higgins CF, Lilley DM (1984) The genetic control of DNA supercoiling in Salmonella typhimurium. EMBO J 3:1745–1752
Richardson SM, Higgins CF, Lilley DM (1988) DNA supercoiling and the leu-500 promoter mutation of Salmonella typhimurium. EMBO J 7:1863–1869
Rohde JR, Fox JM, Minnich SA (1994) Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol 12:187–199
Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B (2009) The role of DNA shape in protein-DNA recognition. Nature 461:1248–1253
Rovinskiy N, Agbleke AA, Chesnokova O, Pang Z, Higgins NP (2012) Rates of gyrase supercoiling and transcription elongation control supercoil density in a bacterial chromosome. PLoS Genet 8, e1002845
Sánchez-Céspedes J, Sáez-López E, Frimodt-Møller N, Vila J, Soto SM (2015) Effects of a mutation in the gyrA Gene on the virulence of uropathogenic Escherichia coli. Antimicrob Agents Chemother 59:4662–4668
Schell MA (1993) Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol 47:597–626
Schleif RF (2013) Modulation of DNA binding by gene-specific transcription factors. Biochemistry 52:6755–6765
Schröder W, Bernhardt J, Marincola G, Klein-Hitpass L, Herbig A, Krupp G, Nieselt K, Wolz C (2014) Altering gene expression by aminocoumarins: the role of DNA supercoiling in Staphylococcus aureus. BMC Genomics 15:291
Semsey S, Virnik K, Adhya S (2005) A gamut of loops: meandering DNA. Trends Biochem Sci 30:334–341
Sheehan BJ, Dorman CJ (1998) In vivo analysis of the interactions of the LysR-like regulator SpvR with the operator sequences of the spvA and spvR virulence genes of Salmonella typhimurium. Mol Microbiol 30:91–105
Sheehan BJ, Foster TJ, Dorman CJ, Park S, Stewart GS (1992) Osmotic and growth-phase dependent regulation of the eta gene of Staphylococcus aureus: a role for DNA supercoiling. Mol Gen Genet 232:49–57
Shimada T, Bridier A, Briandet R, Ishihama A (2011) Novel roles of LeuO in transcription regulation of E. coli genome: antagonistic interplay with the universal silencer H-NS. Mol Microbiol 82:378–397
Snoep JL, van der Weijden CC, Andersen HW, Westerhoff HV, Jensen PR (2002) DNA supercoiling in Escherichia coli is under tight and subtle homeostatic control, involving gene-expression and metabolic regulation of both topoisomerase I and DNA gyrase. Eur J Biochem 269:1662–1669
Sobetzko P (2016) Transcription-coupled DNA supercoiling dictates the chromosomal arrangement of bacterial genes. Nucleic Acids Res 44:1514–1524
Sobetzko P, Travers A, Muskhelishvili G (2012) Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc Natl Acad Sci U S A 109:E42–E50
Soucy SM, Huang J, Gogarten JP (2015) Horizontal gene transfer: building the web of life. Nat Rev Genet 16:472–482
Straney R, Krah R, Menzel R (1994) Mutations in the -10 TATAAT sequence of the gyrA promoter affect both promoter strength and sensitivity to DNA supercoiling. J Bacteriol 176:5999–6006
Sugino A, Higgins NP, Brown PO, Peebles CL, Cozzarelli NR (1978) Energy coupling in DNA gyrase and the mechanism of action of novobiocin. Proc Natl Acad Sci U S A 75:4838–4842
Touchon M, Rocha EP (2016) Coevolution of the organization and structure of prokaryotic genomes. Cold Spring Harb Perspect Biol 8:a018168
Tsao YP, Wu HY, Liu LF (1989) Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell 56:111–118
Unniraman S, Nagaraja V (1999) Regulation of DNA gyrase operon in Mycobacterium smegmatis: a distinct mechanism of relaxation stimulated transcription. Genes Cells 4:697–706
Unniraman S, Chatterji M, Nagaraja V (2002) DNA gyrase genes in Mycobacterium tuberculosis: a single operon driven by multiple promoters. J Bacteriol 184:5449–5456
van Loenhout MT, de Grunt MV, Dekker C (2012) Dynamics of DNA supercoils. Science 338:94–97
van Workum M, van Dooren SJ, Oldenburg N, Molenaar D, Jensen PR, Snoep JL, Westerhoff HV (1996) DNA supercoiling depends on the phosphorylation potential in Escherichia coli. Mol Microbiol 20:351–360
Vinograd J, Lebowitz J, Radloff R, Watson R, Laipis P (1965) The twisted circular form of polyoma viral DNA. Proc Natl Acad Sci U S A 53:1104–1111
Wang JC (1971) Interaction between DNA and an Escherichia coli protein omega. J Mol Biol 55:523–533
Wang Q, Sacco M, Ricca E, Lago CT, De Felice M, Calvo JM (1993) Organization of Lrp-binding sites upstream of ilvIH in Salmonella typhimurium. Mol Microbiol 7:883–891
Webber MA, Ricci V, Whitehead R, Patel M, Fookes M, Ivens A, Piddock LJ (2013) Clinically relevant mutant DNA gyrase alters supercoiling, changes the transcriptome, and confers multidrug resistance. MBio 4:e00273-13
Weinstein-Fischer D, Elgrably-Weiss M, Altuvia S (2000) Escherichia coli response to hydrogen peroxide: a role for DNA supercoiling, topoisomerase I and Fis. Mol Microbiol 35:1413–1420
Westerhoff HV, van Workum M (1990) Control of DNA structure and gene expression. Biomed Biochim Acta 49:839–853
Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS (2007) The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell Mol Life Sci 64:3–16
Wu HY, Shyy SH, Wang JC, Liu LF (1988) Transcription generates positively and negatively supercoiled domains in the template. Cell 53:433–440
Wu HY, Tan J, Fang M (1995) Long-range interaction between two promoters: activation of the leu-500 promoter by a distant upstream promoter. Cell 82:445–451
Xu X, Ben Imeddourene A, Zargarian L, Foloppe N, Mauffret O, Hartmann B (2014) NMR studies of DNA support the role of pre-existing minor groove variations in nucleosome indirect readout. Biochemistry 53:5601–5612
Yamamoto N, Droffner ML (1985) Mechanisms determining aerobic or anaerobic growth in the facultative anaerobe Salmonella typhimurium. Proc Natl Acad Sci U S A 82:2077–2081
Ye F, Brauer T, Niehus E, Drlica K, Josenhans C, Suerbaum S (2007) Flagellar and global gene regulation in Helicobacter pylori modulated by changes in DNA supercoiling. Int J Med Microbiol 297:65–81
Zawadzki P, Stracy M, Ginda K, Zawadzka K, Lesterlin C, Kapanidis AN, Sherratt DJ (2015) The localization and action of topoisomerase IV in Escherichia coli chromosome segregation is coordinated by the SMC complex, MukBEF. Cell Rep 13:2587–2596
Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR (2000) Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J Biol Chem 275:8103–8113
Zhi X, Leng F (2013) Dependence of transcription-coupled DNA supercoiling on promoter strength in Escherichia coli topoisomerase I deficient strains. Gene 514:82–90
Zhou T, Yang L, Lu Y, Dror I, Dantas Machado AC, Ghane T, Di Felice R, Rohs R (2013) DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res 41:W56–W62
Acknowledgements
This work was supported by Science Foundation Ireland Principal Investigator Award 13/IA/1875.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Charles J. Dorman declares that he has no conflict of interest.
Matthew J. Dorman declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
This article is a contribution to Special Issue “DNA Supercoiling” but has already been published in BREV, September 2016, Volume 8, Issue 3, pp 209–220, DOI 10.1007/s12551-016-0205-y.
Rights and permissions
About this article
Cite this article
Dorman, C.J., Dorman, M.J. DNA supercoiling is a fundamental regulatory principle in the control of bacterial gene expression. Biophys Rev 8 (Suppl 1), 89–100 (2016). https://doi.org/10.1007/s12551-016-0238-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-016-0238-2