Abstract
Oecidiobranchus (Isopoda, Asellota) is distinctive among asellote isopod genera in that most of its known species occur in the Nordic Seas and the Arctic Ocean. Some of these species are known only from a few specimens (i.e., poorly known). We used a combined morphological and genetic approach to evaluate the diversity of Oecidiobranchus species in this region. On the basis of genetics, at least three species were recognized, representing Oecidiobranchus cf. nanseni, Oecidiobranchus cf. plebejum, and a third, probably undescribed species. Oecidiobranchus cf. plebejum was found at several locations to the north of the large Greenland-Iceland-Faeroe Ridge, while O. cf. nanseni occurred on both sides of the ridge; temporal or spatial changes during and after the last ice age may have contributed to the genetic differences of populations on each side of the ridge. The wide distribution of the genus in the Nordic Seas and the Arctic Ocean suggests that the genus has been present there for an extensive period.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Recent genetic studies have revealed high genetic diversity and even cryptic species among many morphologically well-defined species of deep-sea benthic invertebrates (e.g., Raupach et al. 2007; Brix et al. 2014b; Gubili et al. 2017). The evolutionary forces promoting speciation in the deep sea are, however, still poorly known, but are probably both of ecological and evolutionary (historical) causes. The latter include barriers to species distributions, which are generally poorly understood in the deep sea. Rex and Etter (2010) list distance, currents, topography, oxygen levels, and vicariance in their evaluation of potential isolating barriers. Bober et al. (2017) for instance attested reduced gene flow across hadal trenches for deep-sea benthic isopods.
The Greenland-Iceland-Faeroe Ridge (GIF Ridge), with its complex hydrography, is one of the most pronounced distributional barriers in the Atlantic Ocean (Svavarsson et al. 1993), ranging across the Atlantic Ocean from east to west with a maximum saddle depth of 840 m (Hansen and Østerhus 2000), whereas between Iceland and the Faeroe Islands, the maximum saddle depths are 420 m (close to Iceland) and 480 m (close to the Faeroes), and in the Denmark Strait (between Iceland and Greenland), the maximum saddle depth is 620 m (Hansen and Østerhus 2000). The GIF Ridge separates the abyssal basins of the North Atlantic proper from the sub-Arctic and Arctic basins to the north. Several primary water masses have been defined in the region (Stefánsson 1962; Logemann et al. 2013) whose temperatures range from − 0.9 to 12 °C, with low and stable temperatures in deeper water, versus a considerable temperature range in shallow waters. In addition, the region is a very important component of the Atlantic meridional overturning circulation. Modified North Atlantic Water (MNAW, > 7.0 °C) flows northwards into the Greenland Sea, cools, and sinks, and colder water masses (< 0.5 °C, e.g., Norwegian Sea Deep Water (NSDW); Norwegian Sea Arctic Intermediate Water (NSAIW)) overflow the ridge from north to south via the ridge’s deep channels.
The deep regions on either side of the GIF Ridge have experienced very different ecological and evolutionary conditions throughout the last several million years (see Dahl et al. (1976) for the Nordic Seas), currently resulting in an extremely cold environment (temperatures < 0 °C) and a very low faunal diversity to the north of the ridge, whereas deep waters south of the ridge generally exhibit temperatures above 2 °C and considerably higher faunal diversity (Svavarsson et al. 1990; Svavarsson 1997; Stuart and Rex 2009; Rex and Etter 2010; Oug et al. 2017).
Many species reach their distributional limits at the GIF Ridge (isopod crustaceans: Negoescu and Svavarsson 1997; Brix and Svavarsson 2010; Brökeland and Svavarsson 2017; amphipod crustaceans: Weisshappel 2000, 2001; polychaete annelids: Parapar et al. 2014); however, several well-defined benthic species apparently exist on both sides of this extensive barrier and occur accordingly over a wide range of abiotic factors, such as temperature (e.g., the crustacean isopod Haploniscus bicuspis G.O. Sars, 1877 from − 0.86 to 7.11 °C, Brökeland and Svavarsson 2017). The intraspecific genetic diversity of species occurring on both sides of the ridge is still unknown.
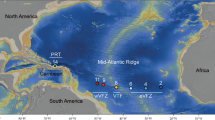
Oecidiobranchus Hessler, 1970 (Isopoda, Desmosomatidae) is a small eurybathic genus with only five known species, of which one occurs at abyssal depths off Australia (Brix 2006); the remaining four occur in the northern part of the North Atlantic Ocean, the Nordic Seas, and the Arctic Ocean (Fig. 1a) (Hansen 1916; Hessler 1970; Just 1980; Malyutina and Kussakin 1996a; Brix and Svavarsson 2010): Oecidiobranchus plebejum (Hansen 1916), also reported by Hessler (1970) from the Bermuda transect, O. nanseni Just, 1980 known from the Nansen Ridge north of Svalbard, and O. polare Gurjanova, 1946 and O. glacialis Malyutina and Kussakin, 1996a known from the Russian side of the Arctic (Fig. 1a) (Hansen 1916; Gurjanova 1946; Just 1980; Malyutina and Kussakin 1996a). While O. polare and O. glaciale occur in shallow, cold waters (Kussakin 1999), O. nanseni and O. plebejum are pronounced Arctic deep-water species. Oecidiobrancus nanseni was collected during the FRAM Drift-Ice Expedition from 83° 40′ N at depths of 2300 m at the Nansen Ridge north of Svalbard (Just 1980), at between 794 and 3709 m in the Nordic Seas, and between 850 and 3920 m in the Arctic Ocean (Svavarsson 1988), while the type material of O. plebejum originates from 66° 23′ N and 67° 29′ N, at depths between 1330 and 1620 m (Hansen 1916). All the above locations exhibit temperatures below zero.
a Distribution map of Oecidiobranchus. Oecidiobranchus plebejum collected (red circles), OTU1; O. cf. plebejum (dark red circles); O. nanseni collected (blue squares); OTU2, O. cf. nanseni (dark blue squares); O. polare (yellow triangle); O. glacialis (green pentagon); the type locality of O. plebejum and O. nanseni is marked in species colors at the stations according to Just (1980: FRAM I station 18) and Hansen (1916: Ingolf station 102). b Distribution of Oecidiobranchus nanseni in BIOICE RP sled samples according to Brix and Svavarsson (2010)
Oecidiobranchus is unique among asellote isopod genera in having such a large proportion of its known species in the Nordic Seas and the Arctic Ocean. Moreover, some of these species are quite common within this region. For instance, from all 239 RP (Rothlisberg and Pearcy 1977) epibenthic sled samples from the BIOICE expeditions (1991–2004), O. nanseni was the fourth most frequently occurring species, occurring at 55 stations (Brix and Svavarsson 2010; Fig. 1b). Oecidiobranchus nanseni extends deeper in the north than in the south and occurred in all seven local water masses across wide bathymetric and temperature ranges (north 209–1558 m, south 317–2215 m, − 0.86 to 7.12 °C). Oecidiobranchus nanseni occurs in sympatry with O. plebejum in circum-Icelandic waters, and was also found in sympatry on the two IceAGE (Icelandic Animals: Genetics and Ecology, 2011–2012) cruises conducted in these waters thus far (Fig. 1a). According to Kussakin (1999), both species occur sympatrically on the Pacific side of the Arctic Ocean as well. The only genera with similar distributions are Cryodesma Svavarsson, 1988 consisting of only two species, both occurring in the Nordic Seas and the Arctic Ocean (Svavarsson 1988; Malyutina and Kussakin 1996b), and Nymphodora Kaiser, 2009, with only a single species, N. fletcheri (Paul and George, 1975), endemic to the Arctic Ocean (Kaiser 2009). Aside from these, the asellote isopod fauna of the Nordic Seas and the Arctic Ocean consists mainly of families of less predominantly deep-sea distribution, like the desmosomatids, whereas predominantly deep-sea families are poorly represented or even absent (Svavarsson et al. 1993). Oecidiobranchus has, with its shallow and deep-water species, apparently adapted well to Arctic conditions and therefore may have been present in these regions for an extensive period.
Some species of Oecidiobranchus are still poorly known and need to be re-evaluated. Here, we evaluate the morphological and genetic diversity of Oecidiobranchus species in the GIF Ridge region. This paper provides a detailed look at all Oecidibranchus specimens collected. We describe herein the pattern of genetic diversity detected in Oecidiobranchus and its implications for the geographical distribution and species delimitation of Oecidibranchus species in the waters of the GIF Ridge.
Materials and methods
Sampling
Specimens for both molecular and morphological analysis were sampled during the recent IceAGE expeditions 1 (M85/3 on board RV Meteor in 2011) and 2 (POS456 on board RV Poseidon in 2013) (Brix et al. 2011; Brix 2013); these specimens were collected and treated as described in Brix et al. (2014a, b) and Riehl et al. (2014).
Molecular methods
Two mitochondrial markers were sequenced for genetic analysis: a roughly 400 bp portion of the small ribosomal subunit (16S) and a roughly 650 bp portion of cytochrome c oxidase subunit I (COI). Detailed protocols for DNA extraction, PCR, and sequencing are described in Riehl et al. (2014); briefly, for COI, the primers LCO-1490/HCO-2198 (Folmer et al. 1994) were used at 45 °C (first 5 cycles) and 50 °C (remaining 35 cycles) annealing temperature, and for 16S, the primers 16S-SF and 16S-SR (Tsang et al. 2009) were used at 48 °C annealing temperature. Sequences were edited in Geneious v.10 (Drummond et al. 2011). The alignment of 16S was performed with the online MAFFT server v7 (Katoh and Standley 2013), and ambiguously aligned portions were removed using the online Gblocks server (Talavera and Castresana 2007), employing all three criteria for less-stringent selection. The COI alignment was performed on DNA codons using the Clustal X algorithm (Larkin et al. 2007) in BioEdit (written by Tom Hall, Ibis Theraputics). All alignments were edited for consistency by hand, and the ends were trimmed to avoid large blocks of gaps. Haplotype networks were computed in popART (Leigh and Bryant 2015) using the TCS algorithm and converted to geographical haplotype maps by hand.
Species were delimited separately for each locus using three “discovery” methods (sensu Carstens et al. 2013): the ABGD algorithm (automated barcode gap discovery, Puillandre et al. 2011), GMYC (general mixed Yule coalescent, Pons et al. 2006), and mPTP (multiple threshold PTP, Kapli et al. 2016). The ABGD method was performed using the online version (http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html) on Kimura two-parameter (K2P, Kimura 1980) corrected pairwise distances and 20 algorithm steps. The GMYC and mPTP analyses require ultrametric input trees, which were computed in BEAST2 v2.4.6 (Bouckaert et al. 2014) using a four-category gamma-distributed model of sequence mutation. For 16S, the GTR model was employed, whereas for COI, the HKY model was employed. Strict clocks and Yule tree priors were used for both markers. All gamma priors were replaced with default lognormal priors. Convergence of the runs was assessed with Tracer v1.6 (Rambaut and Drummond 2014) to choose a burn-in such that all ESSs were at least 200. The trees were produced and annotated with Bayesian posterior probabilities (PP) using TreeAnnotator in the BEAST2 package, with sequences of the related genus Torwolia from the South Atlantic used to root the trees. The GMYC analysis was performed in R with the multiple threshold option. The mPTP analysis was performed with one million steps after 20,000 burn-in steps and three replicate runs.
All data are stored in the Barcode of Life Database (BoLD) project OECID, which contains all available data and is made publically available via GenBank submission. The BIN system in BoLD compares newly submitted sequences with all already available sequences in BoLD clustering them according to their molecular divergence using clustering algorithms. Each cluster receives a unique and specific BIN (barcode identity number).
Morphological methods
All specimens used for the molecular approach described above were determined to species level on board the vessels during the IceAGE expeditions, and the determinations were later verified at the German Centre for Marine Biodiversity Research (DZMB) using a Leica MZ 12.5 dissection microscope and the original descriptions as reference (Hansen 1916; Gurjanova 1946; Hessler 1970; Just 1980; Malyutina and Kussakin 1996a) and were compared to the type specimens loaned from different museum collections (see list below). All voucher specimens are stored at the Center of Natural History, Hamburg (CeNak) (see Table 1). Three voucher specimens (IA2Desm07, IA2Desm09, and IDesm186) were used for confocal laser scanning microscopy (CLSM) and stained with Congo Red. The method was adapted from Michels and Büntzow (2010) and further established by Kihara and Arbizu (2012), Brix et al. (2014b), and Bober et al. (2017). The specimens were scanned in dorsal and lateral view using a Leica DM2500 with a Leica TCS SPE at a resolution of 2480 × 2480 pixels with a ×10 lens. The chitinous exoskeleton parts were excited by 532-nm laser light and detected with a bandpass filter set to 539–670 nm. Furthermore, the 405 and 488-nm laser lines were used with emission filters set to 420–480 and ≥ 490 nm, respectively (Michels and Gorb 2012). The resulting image stacks were further processed in Fiji (Schneider et al. 2012; Schindelin et al. 2012) and finalized in Adobe Photoshop CS5.
Abbreviations used in this study
Morphology: A1 = antennula; Mxp = maxilliped; Op = operculum; PI–PVII = pereopods I–VII; Plt = pleotelson; Prn1–7 = pereonites 1–7; ZMH = Zoological Museum, Hamburg; ZMUC or NHMD = Zoological Museum, University of Copenhagen; AM = Australian Museum. Topography: GIF Ridge = Greenland-Iceland-Faeroe Ridge. Water masses: Modified North Atlantic Waters (MNAW; 7.0–8.5 °C, salinity 35.10–35.30), Labrador Sea Water (LSW; 3–4 °C, salinity 34.90–34.95), Iceland Sea Overflow Water (ISOW; 2–3 °C, salinity 34.85–35.00); Modified East Icelandic Water (MEIW; 1–3 °C, salinity 34.70–34.90), Norwegian Sea Deep Water (NSDW; less than − 0.5 °C, salinity less than − 34.40), Norwegian Sea Arctic Intermediate Water (NSAIW; − 0.5–0.5 °C, salinity 34.85–34.90), and Arctic/Polar Water (A/PW; 0–2 °C, salinity 34.30–34.90).
Comparative material examined
Type material
ZMUC CRU-7810, female, adult, Oecidiobranchus plebejum (Hansen, 1916), lectotype
ZMUC CRU-9698, male, adult, Oecidiobranchus plebejum (Hansen, 1916), paralectotype
NHMD-155588, female, juvenile, Oecidiobranchus plebejum (Hansen, 1916), paralectotype
ZMUC-CRU-7485, female, adult, Oecidiobranchus nanseni (Just, 1980), holotype
ZMUC-CRU-7486, 2 specimens, preparatory females, Oecidiobranchus nanseni (Just, 1980), paratypes
Non-type material
NHMD-155589 Oecidiobranchus plebejum (Hansen, 1916), 6 specimens
NHMD-155591 Oecidiobranchus plebejum (Hansen, 1916), 1 specimen
AM P.59200 Oecidiobranchus plebejum (Hansen, 1916), 25 specimens
AM P.59201 Oecidiobranchus plebejum (Hansen, 1916), 36 specimens
AM P.65390 Oecidiobranchus plebejum (Hansen, 1916), 3 specimens
AM P.65391 Oecidiobranchus plebejum (Hansen, 1916), 24 specimens
AM P.65768 Oecidiobranchus sp., 3 specimens
AM P.65769 Oecidiobranchus sp., 19 specimens
AM P.65770 Oecidiobranchus sp., 2 specimens
Results
From the IceAGE collection of 26 specimens of Oecidiobranchus, 26 sequences of 16S and 21 of COI were obtained (Table 1). New sequences were deposited in GenBank under accession numbers MG831391–MG831411 (COI), and MG895871–MG895896 (16S), and final alignments were deposited in TreeBASE (treebase.org). Table 2 contains the alignment length, nucleotide composition and diversity, number of variable sites, and ABGD-determined pairwise distance threshold for each marker. All species delimitation methods detected the same three putative species (operational taxonomic units (OTUs)) at both loci, although mPTP detected a 49% probability that two specimens (i.e., IDesm075 and IDesm077) belonged to a fourth OTU (Figs. 2, 3, and 4). Analysis on the BoLD platform also determined a different BIN for these two specimens.
Ultrametric trees and species delimitations for both markers. The red ABGD lines mark the boundary between intraspecific nodes (closer to the tips) and interspecific nodes (closer to the root), given as Kimura two-parameter p-distances (K2P). Geographical position is marked by abbreviations: Denmark Strait (DEN), Norwegian Channel (NCH), Irminger Basin (IRM), Faroe-Iceland Ridge (FIR), Norwegian Basin (NOR). The clades are marked with the corresponding OTUs
Haplotype network for 16S (a). Circle size is proportional to the number of individuals sampled with that haplotype, and color denotes sample origin (abbreviations are as in Fig. 2), with the small black circle indicating an unsampled haplotype required to connect the network. Numbers next to branches indicate the number of mutational steps between haplotypes. Haplotype map for 16S (b). Circle size is proportional to the number of individuals sampled at each station, and color denotes 16S haplotype. The first number of stations labels is the IceAGE cruise number, followed by the station ID
Morphologically, OTU1 (Fig. 5a–c) most closely resembles Oecidiobranchus plebejum, while OTU2 (Fig. 5d, e) resembles more O. nanseni. OTU3 was found only at IceAGE2 stations close to the Faeroe Islands and with only two specimens (one male and one female, the latter highly damaged). This seems to be a third species, probably a species unknown to science. In the continuous sorting process, more specimens became available and will be used for a morphological study of this putative new species. At present, OTU3 relies on a damaged specimen and a male insufficient for species description.
CLSM of the three specimens in dorsal and lateral view representing the two main OTUs. a OTU1, O. cf. plebejum, male specimen, field ID IA2Desm09, lateral view. b OTU1, O. cf. plebejum, female specimen, field ID IA2Desm07, lateral view. c OTU1, O. cf. plebejum, female specimen, field ID IA2Desm07, dorsal view. d OTU2, O. cf. nanseni, ovigerous female specimen, field ID IDesm186, lateral view. e OTU2, O. cf. nanseni, ovigerous female specimen, field ID IDesm186, dorsal view. Scale bar = 0.1 mm
The three delimited species showed different, but partially overlapping geographic distributions, with OTU1 (O. cf. plebejum) found only north of the GIF Ridge, OTU2 (O. cf. nanseni) found both south and north of the GIF Ridge (i.e., circum-Icelandic), and OTU3 restricted to the northern part of the Iceland-Faeroe Ridge (IFR); if OTU4 is valid, currently available data would indicate a distribution only south of the ridge (16S, Fig. 4; COI, Fig. 5).
The species Oecidiobranchus plebejum and O. nanseni appear accordingly to be separate species, but probably closely related because they are difficult to distinguish morphologically (see Table 3, see also Svavarsson 1988). The distingushing morphological characters vary with developmental stage and due to sexual dimorphism. Specimens are frequently damaged (appendages broken off), making the defining length/width ratios not always determinable. Hessler (1970) described the first body leg (pereopod, PI) in detail for O. plebejum, but whether he used Hansen’s (1916) original material for his drawings or his own material from the Gay Head Bermuda transect is unclear. Re-examination of the lectotype revealed that all appendages of the lectotype, except the right pereopod I, were broken off. Just (1980) showed for O. nanseni a detailed drawing only for the claw seta, propodus, and dactylus of pereopod I, but not the whole pereopod I. Pereopod II was not drawn in detail (only shown in situ drawing of the specimen). If the type material is to be used for species identification, illustration of an intact paralectotype from the Hansen (1916) material from Ingolf station 102 and redescription of necessary appendages from the specimens Just (1980) designated as types would be necessary.
Discussion
As discussed above, Oecidiobranchus appears to be rare among desmosomatids in that most of its species are concentrated in the Nordic Seas and Arctic Ocean; however, the strength of this assertion rests on uncertain species boundaries and a paucity of data. According to available data at the time, Brix and Svavarsson (2010) identified all Oecidiobranchus specimens in BIOICE samples as O. nanseni morphologically, resulting in this species having a wide distribution on the GIF Ridge and occurring both south and north of the ridge. However, the validity of these specimens as a single single species was in doubt, due to the wide temperature tolerance implied by this distribution (Brix and Savavarsson pers. obs.; Pedro Martinez unpublished species distribution models from the Brix and Svavarsson (2010) data). Indeed, the genetic and morphological analyses herein suggest at least three species in the region (O. cf. nanseni, O. cf. plebejum, and one previously unknown species), but also that O. cf. nanseni occurred on both sides of the ridge.
Morphological determination of Oecidiobranchus species relies on minor differences (mostly length to width ratios, see Table 3). In the case of O. plebejum and O. nanseni, O. plebejum is in general “more robust” while O. nanseni is “more slender” as stated by Just (1980). Both species are superficially very similar, and visualizing differences between them requires a close look under the microscope. As the length-to-width ratios vary by developmental stage and are also influenced by sexual dimorphism, a morphometric approach measuring a significant number of specimens genetically assigned to one species would deliver a clearer picture. Characters of O. nanseni and O. plebejum indicating differences in morphology were shown by Just (1980) and are summarized in Table 3 with focus on characters visible without dissection. The characters that allow the clearest differentiation of species are only visible after dissection (setation of Mxp, two slender distal setae at Op or Op without setae).
The relatively small number of specimens analyzed per OTU presents, however, a challenge to accurately describe the geographical range of the OTUs, indicating the need to analyze more material. Likewise, the absence in IceAGE material of O. cf. plebejum south of the ridge may imply that Hessler’s O. plebejum from the Bermuda transect is a different species, or simply that greater numbers of specimens are needed to describe species distributions with sufficient confidence.
Several isopod species are known to occur in deep waters both south and north of the GIF Ridge despite considerable differences in the benthic temperatures (Negoescu and Svavarsson 1997; Brix and Svavarsson 2010; Brökeland and Svavarsson 2017). Whereas the deeper parts of the world oceans generally exhibit low temperatures (between 2 and 3 °C), the temperatures of the deeper Nordic Seas and the Arctic Ocean typically exhibit temperatures below zero (e.g., Norwegian Sea Deep Water, less than − 0.5 °C, Hansen and Østerhus 2000). A species occurring in deep waters both south and north of the GIF Ridge would therefore have to adjust to temperatures both above and below zero. Whether such an adjustment is physiologically demanding to these species or not is unclear; other species are known to overcome this problem by producing ice-binding proteins (Duman 2015).
The greater oceanographic mixing and topographical complexity associated with the Greenland-Iceland Ridge in the Denmark Strait is likely also associated with higher genetic diversity, which may favor speciation processes. The genetically diverse (comparatively speaking) stations in the Denmark Strait occur at slope depths, placing them in a region of sharp transition between shallow and deep waters; this depth gradient could increase genetic differentiation and decrease connectivity as has been reported in the Northwest Atlantic (Jennings et al. 2013), possibly due to stenobathic isolation. Distributions of Oecidiobranchus species do appear to be influenced by depth, or a variable correlated with depth: OTU3 (potentially a new species) was found only in shallow depths on the ridge, OTU2 (O. cf. nanseni) was found in the deep basins north and south of the ridge, and OTU1 (O. cf. plebejum) attained a broader bathymetric distribution but only north of the ridge. Similar patterns have been observed in another isopod species, Chelator insignis (Hansen, 1916) (Brix et al. 2014a). Furthermore, the GIF Ridge has undergone extensive environmental changes in its history, particularly during the most recent ice age, which ended about 15 ka BP. During the Last Glacial Maximum (LGM, 28.1–22.8 cal. ka BP; Patton et al. 2017), the extensive Icelandic ice sheet extended to the continental shelf break (around 300-m depth, Patton et al. 2017), considerably decreasing (potentially by as much as 100 km) the already narrow, ~ 300-km strait between Iceland and Greenland. The large ice sheets of Greenland and Iceland may have greatly influenced water exchange and interactions between the Nordic Seas and the North Atlantic during the LGM. Significant changes occurred in the currents of the Denmark Strait during the last 10,600 cal years BP, with large changes in the assemblages of the foraminiferans (Perner et al. 2016). Several isopod species feed extensively on foraminiferans (Guðmundsson et al. 2000), although little is known of feeding in Oecidiobranchus. A changing environment and concomitant changes in the community structure of foraminiferans may, however, have shaped the genetic patterns seen in O. nanseni (i.e., OTU2) across the GIF Ridge, indicating a temporal or spatial separation of the populations.
References
Bober S, Riehl T, Henne S, Brandt A (2017) New Macrostylidae (Isopoda) from the Northwest Pacific Basin described by means of integrative taxonomy with reference to geographical barriers in the abyss. Zool J Linnean Soc 1–53. https://doi.org/10.1093/zoolinnean/zlx042
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ, Prlic A (2014) BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput Biol 10(4):e1003537. https://doi.org/10.1371/journal.pcbi.1003537
Brix S (2006) A new genus and new species of Desmosomatidae (Crustacea: Isopoda: Asellota) from the deep sea of south-eastern Australia. Mem Mus Vic 63(2):175–205
Brix S (2013) IceAGE - Icelandic marine Animals: Genetics and Ecology, Cruise No. POS456, IceAGE2, 20.07.2013 – 04.08.2013, Kiel (Germany) - Reykjavik (Iceland). Report published 2013 via Deutsche Zentrum für Marine Biodiversitätsforschung, Senckenberg am Meer
Brix S, Meißner K, Stransky B, Halanych KM, Jennings RM, Kocot KM, Svavarsson J (2014a) The IceAGE project— a follow up of BIOICE. Polish Polar Res 35:141–150
Brix S, Riehl T, Leese F (2011) First genetic data for species of the genus Haploniscus Richardson, 1908 (Isopoda: Asellota: Haploniscidae) from neighbouring deep-sea basins in the South Atlantic. Zootaxa 2838:79–84
Brix S, Svavarsson J (2010) Distribution and diversity of desmosomatid and nannoniscid isopods (Crustacea) on the Greenland-Iceland-Faeroe Ridge. Polar Biol 33:515–530
Brix S, Svavarsson J, Leese F (2014b) A multi-gene analysis reveals multiple highly divergent lineages of the isopod Chelator insignis (Hansen, 1916) south of Iceland. Polish Polar Res 35:225–242. https://doi.org/10.2478/popore-2014-0015
Brökeland W, Svavarsson J (2017) Distribution of haploniscids (Isopoda, Asellota, Haploniscidae) in Icelandic waters, with description of Haploniscus astraphes n. sp. from the Iceland basin and the Southeast Atlantic. Zootaxa 4231:301–326
Carstens BC, Pelletier TA, Reid NM, Satler JD (2013) How to fail at species delimitation. Mol Ecol 22(17):4369–4383
Dahl E, Laubier L, Sibuet M, Strömberg J-O (1976) Some quantitative results on benthic communities of the deep Norwegian Sea. Astarte 9:61–79
Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A (2011) Geneious v.5.4, Available from http://www.geneious.com
Duman JG (2015) Animal ice-binding (antifreeze) proteins and glycolipids: an overview with emphasis on physiological function. J Exp Biol 218:1846–1855. https://doi.org/10.1242/jeb.116905
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3(5):294–299
Gubili C, Ross E, Billett DSM, Yool A, Tsairidis C, Ruhl HA, Rogacheva A, Masson D, Tyler PA, Hauton C (2017) Species diversity in the cryptic abyssal holothurian Psychropotes longicauda (Echinodermata). Deep-Sea Res II 137:288–296
Guðmundsson G, von Schmalensee M, Svavarsson J (2000) Are foraminifers (Protozoa) important food for small isopods (Crustacea) in the deep-sea? Deep-Sea Res 47:2093–2109
Gurjanova E (1946) New species of Isopoda and Amphipoda from the Arctic Ocean. Compendium of results, Drifting Expedition, Icebreaker “Cedov”, 1937–1940. Moscow 3:272–297
Hansen HJ (1916) Crustacea Malacostraca: the order Isopoda. Dan Ingolf Exped 3:1–262
Hansen B, Østerhus S (2000) North Atlantic–Nordic Seas exchanges. Prog Oceanogr 45(2):109–208. https://doi.org/10.1016/S0079-6611(99)00052-X
Hessler RR (1970) The Desmosomatidae (Isopoda, Asellota) of the Gay Head-Bermuda Transect. Bull Scripps Inst Oceanogr 15:1–185
Jennings RM, Etter RJ, Ficarra L (2013) Population differentiation and species formation in the deep sea: the potential role of environmental gradients and depth. PLoS One. https://doi.org/10.1371/journal.pone.0077594
Just J (1980) Polar Sea abyssal and deep bathyal Isopoda (Crustacea). Steenstrupia 6(14):197–230
Kaiser S (2009) Nymphodora gen. nov., a new genus of Nannoniscidae Hansen, 1916 (Isopoda, Asellota, Janiroidea) from the high Arctic. Zootaxa 2096:371–380
Kapli T, Lutteropp S, Zhang J, Kobert K, Pavlidis P, Stamatakis A, Flouri T (2016) Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 33(11):1630–1638
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30(4):772–780. https://doi.org/10.1093/molbev/mst010
Kihara TC, Arbizu PM (2012) Three new species of Cerviniella Smirnov, 1946 (Copepoda: Harpacticoida) from the Arctic. Zootaxa 3345:1–33
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kussakin OG (1999) Marine and brackish-water isopod crustaceans (Isopoda) of cold and temperate waters of the northern hemisphere. Vol. 3, Suborder Asellota, part 2. Families Joeropsididae, Nannoniscidae, Desmosomatidae, Macrostylidae. Opredeliteli po Faune, Izdavaemye Zoologicheskim Muzeem Akademii Nauk 169:1–384 (in Russian)
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Leigh JW, Bryant D (2015) popart: full-feature software for haplotype network construction. Methods Ecol Evol. https://doi.org/10.1111/2041-210X.12410
Logemann K, Olafsson J, Snorrason A, Valdimarsson H, Marteinsdottir G (2013) The circulation of Icelandic waters—a modelling study. Ocean Sci 9:931–955. https://doi.org/10.5194/os-9-931-2013
Malyutina MV, Kussakin OG (1996a) Addition to the Polar Sea bathyal and abyssal isopoda (Crustacea). Part 1. Anthuridea, Valvifera, Asellota (Ischnomesidae, Macrostylidae, Nannoniscidae). Zoosystem Rossica 4:49–62
Malyutina MV, Kussakin OG (1996b) Additions to the Polar Sea bathyal and abyssal Isopoda (Crustacea, Malacostraca), Part 2. Asellota, Desmosomatidae. Zoosystem Rossica 4:239–260
Michels J, Büntzow M (2010) Assessment of Congo red as a fluorescence marker for the exoskeleton of small crustaceans and the cuticle of polychaetes. J Microsc 238:95–101
Michels J, Gorb SN (2012) Detailed three-dimensional visualization of resilin in the exoskeleton of arthropods using confocal laser scanning microscopy. J Microsc 245:1–16
Negoescu I, Svavarsson J (1997) Anthurideans (Crustacea, Isopoda) from the North Atlantic and the Arctic Ocean. Sarsia 82:159–202
Oug E, Bakken T, Kongsrud JA, Alvestad T (2017) Polychaetous annelids in the deep Nordic Seas: strong bathymetric gradients, low diversity and underdeveloped taxonomy. Deep-Sea Res II 137:102–112. https://doi.org/10.1016/j.dsr2.2016.06.016
Parapar J, Helgason GV, Jirkov I, Moreira J (2014) Diversity and taxonomy of Ampharetidae (Polychaeta) from Icelandic waters. Polish Polar Res 35:311–340. https://doi.org/10.2478/popore-2014-0019
Patton H, Hubbard A, Bradwell T, Schomacker A (2017) The configuration, sensitivity and rapid retreat of the Late Weichselian Icelandic ice sheet. Earth Sci Rev 166:223–245. https://doi.org/10.1016/j.earscirev.2017.02.001
Perner K, Jennings AE, Moros M, Andrews JT, Wacker L (2016) Interaction between warm Atlantic-sourced waters and the East Greenland Current in northern Denmark Strait (68 degrees N) during the last 10 600 cal a BP. J Quaternary Sci 31:472–483. https://doi.org/10.1002/jqs.2872
Pons J, Barraclough T, Gomez-Zurita J, Cardoso A, Duran D, Hazell S et al (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol 55(4):595–609. https://doi.org/10.1080/10635150600852011
Puillandre N, Lambert A, Brouillet S, Achaz G (2011) ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol 21(8):1864–1877. https://doi.org/10.1111/j.1365-294X.2011.05239.x
Rambaut A, Drummond AJ. (2014) Tracer v1.6 (http://tree.bio.ed.ac.uk/software/tracer)
Raupach MJ, Malyutina M, Brandt A, Wägele J-W (2007) Molecular data reveal a highly diverse species flock within the munnopsoid deep-sea isopod Betamorpha fusiformis (Barnard, 1920) (Crustacea : Isopoda : Asellota) in the Southern Ocean. Deep-Sea Res II 54:1820–1830. https://doi.org/10.1016/j.dsr2.2007.07.009
Rex M, Etter RJ (2010) Deep-sea biodiversity. Harvard University Press, Cambridge, 354 pp
Riehl T, Brenke N, Brix S, Driskell A, Kaiser S, Brandt A (2014) Field and laboratory methods for DNA studies on deep-sea isopod crustaceans. Polish Polar Res 35:205–226. https://doi.org/10.2478/popore-2014-0018
Rothlisberg PC, Pearcy WG (1977) An epibenthic sampler used to study the ontogeny of vertical migration of Pandalus jordani (Decapoda, Caridea). Fish Bull 74:994–997
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682
Schneider CA, Rasband WS, Eliceiri KW et al (2012) NIH image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Stefánsson U (1962) North Icelandic waters. Rit Fiskid 3:1–269
Stuart CT, Rex MA (2009) Bathymetric patterns of deep-sea gastropod species diversity in 10 basins of the Atlantic Ocean and Norwegian Sea. Mar Ecol 30:164–180. https://doi.org/10.1111/j.1439-0485.2008.00269.x
Svavarsson J (1988) Desmosomatidae (Isopoda, Asellota) from bathyal and abyssal depths in the Norwegian, Greenland, and North Polar Seas. Sarsia 73:1–32
Svavarsson J (1997) Diversity of isopods (Crustacea), new data from the Arctic and Atlantic Oceans. Biodivers Conserv 6:1571–1579
Svavarsson J, Brattegard T, Strömberg J-O (1990) Distribution and diversity patterns of asellote isopods (Crustacea, Isopoda) in the deep Norwegian and Greenland Seas. Prog Oceanogr 24:297–310
Svavarsson J, Strömberg JO, Brattegard T (1993) The deep-sea asellote (Isopoda, Crustacea) fauna of the Northern Seas: species composition, distributional patterns and origin. J Biogeogr 20:537–555
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56(4):564–577. https://doi.org/10.1080/10635150701472164
Tsang LM, Chan BKK, Shih F-L, Chu KH, Chen AC (2009) Host-associated speciation in the coral barnacle Wanella milleporae (Cirripedia: Pyrgomatidae) inhabiting the Millepora coral. Mol Ecol 18:1463–1475
Weisshappel JB (2000) Distribution and diversity of the hyperbenthic amphipod family Eusiridae in the different seas around the Greenland-Iceland-Faeroe-Ridge. Sarsia 85:227–236
Weisshappel JB (2001) Distribution and diversity of the hyperbenthic amphipod family Calliopiidae in the different seas around the Greenland-Iceland-Faeroe-Ridge. Sarsia 86:143–151
Acknowledgements
We would like to thank the crews of RV Metor and RV Poseidon for their important help and contribution. We wish to thank all pickers and sorters during the IceAGE expeditions for providing a unique set of specimens. Without the help of Karen Jeskulke, Sarah Schnurr, and Andrea Ormos, working so hard on producing high-quality PCR products at the Smithsonian, the whole work would have been much slower. We thank Anna M. Jażdżewska for help with the data and two anonymous reviewers whose suggestions improved the manuscript.
Funding
This study was supported by the German Science Foundation under grant no. BR3843/6-1. This grant supported the travels of Robert Jennings and Saskia Brix within the American-German cooperation in the frame of the IceAGE project.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed by the authors.
Field study
Permits and approval of field or observational studies have been obtained by the authors, if applicable.
Additional information
Communicated by K. Halanych
This article is part of the Topical Collection on Biodiversity of Icelandic Waters by Karin Meißner, Saskia Brix, Ken M. Halanych and Anna Jazdzewska.
Rights and permissions
About this article
Cite this article
Jennings, R.M., Brix, S., Bober, S. et al. More diverse than expected: distributional patterns of Oecidiobranchus Hessler, 1970 (Isopoda, Asellota) on the Greenland-Iceland-Faeroe Ridge based on molecular markers. Mar Biodiv 48, 845–857 (2018). https://doi.org/10.1007/s12526-018-0857-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12526-018-0857-x