Abstract
A systematic study on the lack of dissimilatory nitrate reductase (NAR) properties in Halomonas strains had been reported so far. The effects of different factors on Halomonas sp. B01 NAR activity were investigated. The salt tolerance of NAR was characterized. The denitrification process under high salt conditions was reported. Halomonas sp. B01 expressed membrane-bound NAR under induced culture by nitrate. The optimum pH of the enzyme reaction system was 8, and the optimum temperature was 30 °C. The mRNA expression abundance of narH in NAR encoding gene was highest in the 60 g/L NaCl inducing matrix. The NaCl concentration of optimum growth and induction of NAR were both 60 g/L. The ectoine added to the NAR vitro enzyme reaction system could maintain NAR activity under high NaCl concentration. In the range of 0–60 g/L NaCl, the NAR activity was stable at 17.7 (± 0.3) U/mg. The denitrification was performed by Halomonas sp. B01 at 60 g/L NaCl, and the denitrification rate reached 97.1% at 24 h. This study reveals for the first time the NAR properties of Halomonas strains, which provides a theoretical and technical basis for the nitrogen removal of high-salt nitrogenous wastewater using this strain.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Halomonas sp. is a class of moderately halophilic bacteria (de la Haba et al. 2014). Some strains of the genus are reportedly capable of nitrification, denitrification or simultaneous heterotrophic nitrification, and aerobic denitrification (known as SND) (Berendes et al. 1996; Mormile et al. 1999; Guo et al. 2013; Wang et al. 2017). Te Wang reported that Halomonas sp. B01 can simultaneously tolerate high concentrations of NaCl and NH4+-N and efficiently perform SND. In a nitrogen (N) removal solution, which contains 60 g/L NaCl and 4000 mg/L NH4+-N, SND was performed over 180 h under optimal conditions; the residual total inorganic N concentration was 21.7 mg/L, and the N removal rate reached 99.2% (Wang et al. 2017). Halomonas N removal strains are important for the purification of high-salinity nitrogenous wastewater from leather processing, marine aquaculture, synthetic ammonia, landfills, and nickel battery production.
The process of microbial N removal is generally NH4+ → NO2− → NO3− → NO2− → NO→N2O → N2, in which NO3− → NO2− is catalyzed by dissimilatory nitrate reductase (Jetten et al. 1997). This enzyme is a rate-limiting enzyme in a series of enzymes involved in the entire N removal process. The properties and activity of this enzyme have a great influence on the N removal efficiency (Barber et al. 2002). There are two kinds of dissimilatory nitrate reductases expressed by bacteria: one is membrane-bound nitrate reductase (NAR), while the other is periplasmic nitrate reductase (NAP) (Argandoña et al. 2006). NAR is widely found in E. coli and has been well-studied (Zumft 1997). NAR consists of three subunits (α, β, and γ) and several cofactors and participates in the production of the transmembrane proton gradient, which is combined with nitrate reduction. Studies have shown that subunits α and β are located in the cytoplasm, while subunit γ is located in the membrane and connects subunits α and β to the cytoplasmic side of the intima (Morozkina and Zvyagilskaya 2007). NAP has 2 subunits: large subunit NapA and small subunit NapB. NapA is a catalytic subunit, and NapB contains cytochrome c (Arnoux et al. 2003). It is generally believed that NAR performs nitrate respiration under anaerobic and hypoxic conditions. NAR is sensitive to oxygen molecules and plays a major role in the biological N removal of anaerobic denitrification (Zumft 1997; Stolz and Basu 2002). NAP is not sensitive to oxygen molecules; therefore, its physiological function may be related to aerobic denitrification.
Llamas reported on the NAR of Halomonas maura (in regard to the NAR of Halomonas). RT-PCR analysis showed that the narGHJI operon was expressed under anaerobic conditions when nitrate could be used as an electron acceptor. This membrane-bound nitrate reductase is the only enzyme responsible for anaerobic respiration in H. Maura (Llamas et al. 2006). Te Wang reported on the gene cloning and structure analysis of the NAR in Halomonas sp. B01, which was isolated from the sediment of a saltern pool and could remove N by SND (Wang et al. 2017). The study on the properties and salt tolerance is of Halomonas strain NAR of great significance to the related research and the technological progress of N removal of high-salinity nitrogenous wastewater by the Halomonas strain.
The type identification of Halomonas sp. B01 nitrate reductase was carried out in this paper. The effects of different factors on the NAR activity of this strain were investigated. The salt tolerance of NAR was characterized. The denitrification process of this strain under high salt conditions was reported.
Materials and methods
Materials
Strain: Halomonas sp. B01 was isolated, screened, and identified by the author’s previous study (16S rRNA sequence GenBank No. KJ778559) (Wang et al. 2017), and the strain was deposited in the China Center for Type Culture Collection (CCTCC) under the accession No. CCTCC AB 2014335.
Reagents: Ectoine (1, 4, 5, 6-tetrahydro-2-methyl-4-pyrimidinecarboxylic acid) standard was purchased from Biomol GmbH (Germany). TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa Code. 9767), TaKaRa PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa Code. RR047), SYBR Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa Code. RR820) purchased from Takara Biotechnology (Dalian) Co., Ltd. BCA Protein Concentration Assay Kit (PC0020) purchased from Beijing Solarbio Science & Technology Co., Ltd.
Medium
LB medium (g/L): peptone 10, yeast extract powder 5, NaCl 60. The medium was autoclaved at 121 °C for 20 min.
Growth medium (g/L): glucose 20, (NH4)2SO4 10, yeast extract powder 0.5, K2HPO4·3H2O 9, KH2PO4 3, MgSO4·7H2O 0.4, MnSO4·H2O 0.01, NaCl 60. pH 7.2. The medium was autoclaved at 121 °C for 20 min.
Inducing medium (g/L): glucose 30, KNO3 0.4 (N element 50 mg/L), yeast extract powder 0.5, K2HPO4·3H2O 0.3, KH2PO4 0.1, MgSO4·7H2O 0.4, MnSO4·H2O 0.01, NaCl 60. pH 7.2. The medium was autoclaved at 121 °C for 20 min.
Denitrifying medium (g/L): glucose 30, KNO3 0.7 (N element 100 mg/L), K2HPO4·3H2O 9, KH2PO4 3, MgSO4·7H2O 0.4, MnSO4·H2O 0.01, NaCl 60. pH 7.2. The medium was autoclaved at 121 °C for 20 min.
Strain culture
The strains were cultivated in 5 mL LB medium at 30 °C and 120 rpm in a rotary shaker for 24 h. Then 1% of the cultures were inoculated in shake flasks (300 mL) containing 30 mL growth medium at 30 °C and 120 rpm in a rotary shaker for 36 h. The cells were collected by centrifugation (at 4 °C and 14,000×g for 15 min) and transferred to the inducing medium at 30 °C and 120 rpm for 36 h.
Determination of enzyme activity
A crude extract of each enzyme prepared for this study was used for enzyme activity assays. The cytoplasmic enzyme (including the periplasmic enzyme) was prepared in 20 mL of the cell culture medium, which was also used to determine enzyme activity, that was centrifuged at 14,000×g for 15 min at 4 °C, and the supernatant was discarded. Then, 100 mM pH 7.2 phosphate buffer was added to the centrifuged pellet to resuspend it. A freeze-thaw cycle (at − 20 °C ≥ 2 h and at 30 °C for 30 min.) was repeated 4 times. Ultrasound was used to disrupt cells in an ice bath (sonicated at 400 W for 3 s and stopped for 3 s) continuously through 30 cycles to obtain a solution of disrupted cells. The disrupted-cell solution was centrifuged at 14,000×g for 15 min at 4 °C, the supernatant was collected, and the sample was used for cytoplasmic and periplasmic enzyme assays. Cell membrane debris was in the centrifuged pellet. The membrane-bound enzyme was prepared as follows: Dodecyl-β-D-maltoside is an alkyl glycoside nonionic surfactant, which promotes the disintegration of lipid membranes to release membrane proteins, and provides a hydrophobic environment for membrane proteins in the state of membrane removal in solution. It maintains and protects the hydrophobic transmembrane structure of membrane proteins, thereby maintaining the structure and function of membrane proteins. Therefore dodecyl-β-D-maltoside is often used to extract proteins from cell membranes. In this paper, dodecyl-β-D-maltoside was used to dissolve the cytoplasmic membrane and release the membrane-bound proteins. The disrupted-cell centrifuge pellet, as described above, was resuspended in 100 mM phosphate buffer (pH 7.2) containing 1.0% dodecyl-β-D-maltoside, incubated for 1 h at 4 °C in the dark, and centrifuged at 14,000×g for 15 min at 4 °C. Then, the supernatant was collected, and the sample was used for the cell membrane-bound enzyme assay (Zhang et al. 2015). The total protein concentration of the enzyme solution to be determined was assayed as follows: Protein was determined using the BCA Protein Concentration Assay Kit (PC0020, Solarbio, Beijing, China). The amount of the enzyme solution to be determined was based on the total protein (mg).
The NAR activity was assayed as follows: NAR reduced NO3− to NO2− in the presence of reduced coenzyme (Barber et al. 2002). One unit of NAR activity (U) is the amount of NAR that reduces 1 μmol NO3− per min at the reaction substrate of KNO3 (0.5 mmol/L), pH 7.2, and 30 °C under the specified reaction conditions.
Determination of inorganic N concentration
NH4+-N was determined by Nessler’s reagent method (APHA 1999). NO2−-N was determinated by diazotization-coupling reaction method (APHA 1999). NO3−-N was determinated by zinc-cadmium reduction method (Sun et al. 2013). The water used for the determination was nitrogen-free water.
Determination of ectoine concentration
The sample for determining ectoine was prepared by the ethanol extraction method (Zhang et al. 2009). Ectoine concentration was determined using high-performance liquid chromatography (HPLC) (Zhang et al. 2009).
Method of RT-PCR
RT-PCR was performed by SYBR Green I fluorescent dye method (Marino et al. 2003). The 16S rDNA sequence primers of Halomonas sp. B01 were 16S rDNA universal primers for bacterial, which were F1-ACATCCTGCGAACTTGTGAGAG and R1-CCGCTGGCAAATAA GGACA. The narH primers of Halomonas sp. B01 were F2-TCGAGTTCGACGGCATCCT and R2-AACACCACGGGCTCCTCTT. The nucleotide sequence of Halomonas sp. B01 narH had been registered on GenBank (KT792958). The total RNA of samples was extracted using TaKaRa MiniBEST Universal RNA Extraction Kit (TaKaRa Code. 9767). RT-PCR was performed using TaKaRa PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa Code. RR047) and SYBR Premix Ex Taq™ II (Tli RNaseH Plus) (TaKaRa Code. RR820) in apparatus of TaKaRa PCR Thermal Cycler Dice™ (TaKaRa Code.TP600) and TaKaRa PCR Thermal Cycler Dice™ Real Time System (TaKaRa Code. TP800). RT-PCR results were processed using Delta-delta Ct method. Expression abundance of the mRNA of narH was expressed by 2−△△Ct. Experimental samples were prepared by extracting the total mRNA A260/280 ≥ 2.0. The amplification curves of the experimental samples were all S-type curves. The melting curves of the specific primer amplification were all single peaks. The standard deviation of the mRNA expression abundance of parallel samples was less than or equal to 0.3%. The mRNA of 16s rDNA was used as an internal standard.
Culture of the denitrifier under aerobic condition
The strains were cultivated in 5 mL LB medium at 30 °C and 120 rpm in a rotary shaker for 24 h. Then 1% of the cultures were inoculated in shake flasks (300 mL) containing 30 mL denitrifying medium at 30 °C and 120 rpm in a rotary shaker for 24 h.
For all of the above measurements, three parallel samples were measured. The average of the three measurements with standard deviation values is presented in the results section.
Results and discussion
Type analysis and identification of Halomonas sp. B01 nitrate reductase
Dissimilatory nitrate reductase of bacterial expression has two types: NAR and NAP. The main difference between these two types of enzymes is that NAR is a membrane-bound enzymes and NAP is a periplasmic enzyme (Argandoña et al. 2006). A low concentration of sodium azide inhibits NAR activity, while NAP activity is not sensitive to low concentration sodium azide. NAR can use reductive coenzyme I (NADH) as an electron donor to reduce chlorate and NAP cannot (Bell et al. 1990; Carter et al. 1995). The above three points are the main basis for identifying the type of dissimilatory nitrate reductase (NAR or NAP) of bacterial expression.
Nitrate reductase of Halomonas sp. B01 is membrane-bound enzymes
There are three possible forms of nitrate reductase in Halomonas sp. B01: cytoplasmic enzyme, periplasmic enzyme, and cell membrane-bound enzyme. First, the cytoplasmic and periplasmic enzyme assay sample was obtained according to method “determination of enzyme activity,” and the nitrate reductase activity of this sample was measured. No nitrate reductase activity was detected. It is shown that the nitrate reductase of Halomonas sp. B01 does not exist in the cytoplasm and periplasm, and it is speculated to exist in the cell membrane debris of the centrifuged pellet. Second, in order to verify the above hypothesis, dodecyl-β-D-maltoside was used to dissolve the cytoplasmic membrane and release the membrane-bound protein according to method “determination of enzyme activity,” and the membrane protein was present in the supernatant after centrifugation. The nitrate reductase activity was then detected in the supernatant (no nitrate reductase activity was detected in the resuspended pellet). It was shown that the nitrate reductase of Halomonas sp. B01 is a membrane-bound enzyme (NAR). The following experiments used the extracted membrane-bound enzyme preparation solution.
Sodium azide inhibits the nitrate reductase activity of Halomonas sp. B01
A final concentration of 40 μM of sodium azide was added to determination system of nitrate reductase activity according to the literature (Bell et al. 1990), and the inhibitory effect of sodium azide on Halomonas sp. B01 nitrate reductase was investigated. The results showed that the nitrate reductase activity without sodium azide was 15.0 U/mg, the nitrate reductase activity adding sodium azide was 6.1 U/mg, and the enzyme activity was decreased by 8.9 U/mg. A low concentration of sodium azide inhibited the nitrate reductase activity of Halomonas sp. B01.
Reduced chlorate by nitrate reductase of Halomonas sp. B01
Potassium nitrate was replaced by a 1 mM concentration of potassium chlorate in the determination system of nitrate reductase activity (containing 0.2 mg/mL NADH as the electron donor). The activity of nitrate reductase reduced chlorate in Halomonas sp. B01 was investigated. The results showed that the nitrate reductase of this strain reduced the initial 83.6 mg/L of chlorate in the enzyme reaction system to 25.3 mg/L chlorite, and the conversion rate was 30.3%. Nitrate reductase of Halomonas sp. B01 can use NADH as an electron donor to reduce chlorate.
Combined with the above results, it was determined that the nitrate reductase of Halomonas sp. B01 is membrane-bound NAR.
Changes of NAR mRNA expression abundance during the induction process
The author of this article cloned an encoding gene narH of a subunit NarH of Halomonas sp. B01 NAR (membrane-bound enzyme) in previous studies (GenBank No. KT792958). To further verify that the nitrate reductase of the strain was membrane-bound NAR, the change of mRNA expression abundance of Halomonas sp. B01 narH with induced culture time was investigated by the RT-PCR method. The results are shown in Fig. 1. In the range of 0–36 h, the mRNA expression abundance of strain narH increased with prolonged induced culture time, which was synchronized with the effect of culture time on Halomonas sp. B01 NAR activity (Fig. 2b). The RT-PCR results further indicated that Halomonas sp. B01 nitrate reductase was a membrane-bound NAR and an inducible enzyme.
Effect of different induction culture times on mRNA expression abundance of Halomonas sp. B01 narH. a Agarose gel electrophoresis pattern of total RNA extracted from the strain samples cultured at different induced times (h). b RT-PCR amplification curves of narH from strain samples cultured at different induced times. c Dissolution curves of narH amplification products from strain samples cultured at different induced times. d mRNA expression abundance of narH from strain samples cultured at different induced times. Induction conditions of NAR: conducted using inducing medium, the induced substrate was 50 mg/L NO3−-N
Effect of the concentration of NO3−-N and induced time on Halomonas sp. B01 NAR activity. a Effect of the concentration of NO3−-N. Induction conditions of NAR: conducted using inducing medium, the induced time was 36 h. b Effect of induced time. Induction conditions of NAR: conducted using inducing medium, the induced substrate was 50 mg/L NO3−-N
It is generally believed that NAR is sensitive to oxygen molecules by nitrate respiration under anaerobic and hypoxic conditions and plays a major role in the biological N removal of anaerobic denitrification (Zumft 1997; Stolz and Basu 2002). While NAP is not sensitive to oxygen molecules, its physiological function may be related to aerobic denitrification. In this study, Halomonas sp. B01 NAP activity was not detected. In addition, the primers were designed by DNA similarity and PCR was carried out, and no encoding gene or DNA fragment of NAP was obtained. Both this study and the previous study (Wang et al. 2017) showed that Halomonas sp. B01 was able to perform N removal by SND (condition of aerobic and heterotrophic). The above indicated the NAR of the strain-tolerant oxygen molecule. This tolerance is a newly discovered trait of Halomonas sp. B01 NAR. This topic needs additional study.
NAR enzyme properties of Halomonas sp. B01
Effect of NO3−-N concentration and induced time on the NAR activity of Halomonas sp. B01 in induced culture
Halomonas sp. B01 was cultured in the growth medium for 36 h. The cells were collected by centrifugation (at 4 °C and 14,000×g for 15 min) and transferred to the inducing medium (the concentration of NO3−-N was set at 0, 25, 50, 75, and 100 mg/L). The cells were then cultured at 30 °C and 120 rpm in a rotary shaker, and the activity of NAR was measured. Figure 2 a shows that when the substrate concentration of NO3−-N was in the range of 0–100 mg/L, the activity of NAR increased with the increase in NO3−-N concentration. When the concentration of NO3−-N ≥ 50 mg/L, the activity of NAR did not increase significantly. Thus, a suitable concentration of NO3−-N was 50 mg/L. The activity of NAR was measured at different induced times (0, 12, 24, 36, and 48 h). Figure 2 b shows that the highest enzyme activity (15.0 U/mg) was obtained at 36 h after induction. The above results indicated that the suitable induced substrate concentration was 50 mg/L, and the induced time was 36 h for Halomonas sp. B01 NAR.
Effect of pH of enzyme reaction system on Halomonas sp. B01 NAR activity
The effect of the pH of the enzyme reaction system on the activity of Halomonas sp. B01 NAR was investigated. The pH of the NAR enzyme reaction system was set to 6, 7, 8, and 9 with a 0.1 M phosphate buffer solution and 0.1 M boric acid-potassium chloride-sodium hydroxide buffer, and the activity of the Halomonas sp. B01 NAR was determined. The results are shown in Fig. 3. The optimal pH of the Halomonas sp. B01 NAR enzyme reaction was 8 (18.1 U/mg). Halomonas sp. B01 NAR had a higher enzyme activity when the pH of the enzyme reaction was in the range of 6–8. When the pH of the enzyme reaction increased to 9, the NAR activity decreased significantly, which was 48.9% of the highest enzyme activity.
Effect of temperature of enzyme reaction system on the activity of Halomonas sp. B01 NAR
The effect of enzyme reaction temperature on the activity of Halomonas sp. B01 NAR was investigated. The enzyme reaction temperature was set to 20, 30, 40, 50, 60, 70, and 80 °C, and the activity of Halomonas sp. B01 NAR was measured. The results are shown in Fig. 4. The optimum temperature for the Halomonas sp. B01 NAR enzyme reaction was 30 °C (18.1 U/mg). The Halomonas sp. B01 NAR activity decreased with increasing temperature when the enzyme reaction temperature was in the range of 30–80 °C. When the enzyme reaction temperature was increased to 80 °C, the NAR activity decreased significantly, which accounted for only 28.8% of the highest enzyme activity.
Salt tolerance of Halomonas sp. B01 NAR
Expression abundance of Halomonas sp. B01 narH in inducing mediums at different NaCl concentrations
The effect of medium NaCl concentration on the mRNA expression abundance of the Halomonas sp. B01 narH was investigated by the RT-PCR method. The results are shown in Fig. 5. When the concentration of NaCl was in the range of 30–120 g/L, the mRNA expression abundance of narH was the highest (1.2) at a concentration of 60 g/L NaCl, and the mRNA expression abundance of narH was significantly decreased (≤ 0.04) when the concentration was ≥ 90 g/L NaCl. The NAR of this strain showed significant high salt adaptation at the gene expression level.
Effect of different NaCl concentrations on the mRNA expression abundance of Halomonas sp. B01 narH. a Agarose gel electrophoresis pattern of total RNA extracted from the strain samples cultured at different NaCl concentrations (g/L). b RT-PCR amplification curves of narH from strain samples cultured at different NaCl concentrations. c Dissolution curves of narH amplification products from strain samples cultured at different NaCl concentrations. d mRNA expression abundance of narH from strain samples cultured at different NaCl concentrations. Induction conditions of NAR: conducted using inducing medium, the induced substrate was 50 mg/L NO3−-N, and induced time was 36 h
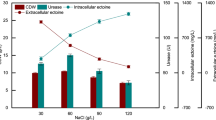
Halomonas sp. B01 NAR activity induced at different NaCl concentrations
The effect of inducing medium NaCl concentration on the activity of Halomonas sp. B01 NAR was investigated. The amount of cell growth (cell growth was defined as cell dry weight per liter culture broth, CDW, g/L) and the NAR activity of Halomonas sp. B01 in the inducing medium of 30, 60, 90, 120 g/L NaCl was determined. The results are shown in Fig. 6. The results show that the optimum NaCl concentration for Halomonas sp. B01 growth and the NAR inducing culture were both 60 g/L.
Effect of inducing medium NaCl concentration on Halomonas sp. B01 NAR activity. Induction conditions of NAR: conducting using inducing medium, the induced substrate was 50 mg/L NO3−-N, and induced time was 36 h. The enzyme reaction system substrate was 50 mg/L NO3−-N, pH was 8, reaction temperature was 30 °C, and reaction time was 1 h
In vitro enzymatic activity of Halomonas sp. B01 NAR at different NaCl concentrations
The in vitro enzymatic activity of Halomonas sp. B01 NAR at different NaCl concentrations was investigated. Two enzyme reaction systems were set up: no ectoine was added to the enzyme reaction system; the ectoine with a final concentration of 7 mM was added to the enzyme reaction system. The NaCl concentrations of the two reaction systems were set to 0, 30, 60, 90, and 120 g/L, respectively. The results are shown in Fig. 7. The NAR activity without ectoine decreased significantly as the concentration of NaCl increased. The NAR activity adding ectoine was very different from the case of above. In the range of 0–60 g/L NaCl concentration, the NAR activity was basically maintained at the same level (17.7 ± 0.3 U/mg), and when the NaCl concentration was ≥90 g/L, the NAR activity was reduced. The above results indicated that the ectoine added in the enzyme reaction system could maintain the NAR activity at a high concentration of NaCl. The enzyme reaction system adding ectoine was also a simulation of the intracellular environment of the strain.
Denitrification process of Halomonas sp. B01 under high salt
The denitrification progress of Halomonas sp. B01 at a concentration of 60 g/L NaCl was investigated. The results are shown in Fig. 8. The initial NO3−-N was 100 mg/L; after denitrification for 24 h, the residual NO3−-N concentration was 1.7 mg/L (the maximum NO2−-N concentration in the denitrification process was 0.9 mg/L), the denitrification rate was 97.1%, and the denitrification efficiency was 4.1 mg/L/h.
Most Halomonas bacteria can synthesize compatible solute ectoine under NaCl stress, and ectoine provides stress protection to the cell itself or to biological macromolecular substances (such as enzymes) in the cell’s internal and external environment, which allows it to resist the adverse effects of environmental osmotic pressure by synthesizing ectoine and also allows cell growth and metabolism to be maintained in a high osmotic pressure environment (including extreme temperature and extreme pH, desiccation, and radiation) (Pastor et al. 2010). Halomonas sp. B01 is also capable of synthesizing ectoine under NaCl stress (Wang et al. 2017), i.e., there is a certain concentration of ectoine in the cells in a high salt environment. In this paper, the in vitro enzymatic reaction of NAR with and without ectoine was performed for the first time. The results indicated that the salt tolerance of Halomonas sp. B01 NAR and the high efficiency of nitrate denitrification of this strain under high salt conditions were mainly attributed to the protective effect of ectoine synthesized by the cell itself under high salt. In addition, Halomonas sp. B01 is an ectoine-excreting strain, in which the ectoine synthesized by the cell itself is partially secreted outside the cell. In a specific microbial population, the secreted ectoine is absorbed by other microorganisms to obtain stress resistance. It is predicted that strains such as Halomonas sp. B01 will exert their unique advantages of salt tolerance assistance in the N removal of combined flora.
Conclusions
Halomonas sp. B01 showed nitrate reductase activity in nitrate-induced culture. The bacterial expression of dissimilatory nitrate reductase has two types: NAR and NAP. In this paper, the results of enzyme species identification showed that the nitrate reductase activity was only observed in the cell membrane-precipitated sample of the enzyme preparation. In addition, its enzyme activity was inhibited by a low concentration sodium azide, the enzyme could reduce chlorate to chlorite, and the mRNA expression abundance of narH varied with induced time. In summary, the enzyme was judged to be a membrane-bound nitrate reductase NAR. This study on the properties of Halomonas sp. B01 NAR showed that the enzyme was an induced enzyme, the suitable induced substrate concentration of NO3−-N was 50 mg/L, and the induced time was 36 h. Furthermore, the optimum pH of the enzyme reaction system is 8, and the optimum reaction temperature is 30 °C. Halomonas sp. B01 NAR had significant salt tolerance, and the mRNA expression abundance of narH was highest in the 60 g/L NaCl inducing matrix. The NaCl concentration of the optimum growth and optimum induction of NAR were both 60 g/L.
The ectoine added to the NAR in vitro enzyme reaction system could maintain the NAR activity under high concentration of NaCl. In the range of 0–60 g/L NaCl concentration, the NAR activity was stable at 17.7 (± 0.3) U/mg. The denitrification process of Halomonas sp. B01 at 60 g/L NaCl showed that, the initial NO3−-N was 100 mg/L, after denitrification for 24 h, the residual NO3−-N concentration was 1.7 mg/L, the denitrification rate was 97.1%, and the denitrification efficiency was 4.1 mg/L/h. This study provides a theoretical and technical basis for revealing the mechanism of Halomonas strain high salt tolerance to N removal.
References
APHA (1999) Standard methods for the examination of water and wastewater, 20th edn. American Public Health Association, Washington
Argandoña M, Martınez-Checa F, Llamas I, Arco Y, Quesada E, Moral AD (2006) A membrane-bound nitrate reductase encoded by the narGHJI operon is responsible for anaerobic respiration in Halomonas maura. Extremophiles 10:411–419. https://doi.org/10.1007/s00792-006-0515-2
Arnoux P, Sabaty M, Alric J, Frangioni B, Guigliarelli B, Adriano JM, Pignol D (2003) Structural and redox plasticity in the heterodimeric periplasmic nitrate reductase. Nat Struct Biol 10:928–934. https://doi.org/10.1038/nsb994
Barber MJ, Desai SK, Marohnic CC, Hernandez HH, Pollock VV (2002) Synthesis and bacterial expression of a gene encoding the heme domain of assimilatory nitrate reductase. Arch Biochem Biophys 402:38–50. https://doi.org/10.1016/S0003-9861(02)00035-8
Bell LC, Richardson DJ, Ferguson SJ (1990) Periplasmic and membrane-bound respiratory nitrate reductases in Thiosphaera pantotropha. FEBS 265:85–87. https://doi.org/10.1016/0014-5793(90)80889-Q
Berendes F, Gottschalk G, Heine-Dobbernack E, Moore ERB, Tindall BJ (1996) Halomonas desiderata sp. nov., a new alkaliphilic, halotolerant and denitrifying bacterium isolated from a municipal sewage works. Syst Appl Microbiol 19:158–167. https://doi.org/10.1016/S0723-2020(96)80041-5
Carter JP, Hsiao YH, Spiro S, Richardson DJ (1995) Soil and sediment bacteria capable of aerobic nitrate respiration. Appl Environ Microbiol 61:2852–2858. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC167561/. Accessed March 2018
de la Haba RR, Arahal DR, Sanchez-Porro C, Ventosa A (2014) The prokaryotes: Gammaproteobacteria: 17 the family Halomonadaceae. Springer, Heidelberg. https://doi.org/10.1007/978-3-642-38922-1_235
Guo Y, Zhou XM, Li YG, Li K, Wang CX, Liu JF, Yan DJ, Liu YL, Yang DH, Xing JM (2013) Heterotrophic nitrification and aerobic denitrification by a novel Halomonas campisalis. Biotechnol Lett 35:2045–2049. https://doi.org/10.1007/s10529-013-1294-3
Jetten MSM, Logemann S, Muyzer G, Robertson LA, Vries SD, Loosdrecht MCMV, Kuenen JG (1997) Novel principles in the microbial conversion of nitrogen compounds. Antonie Van Leeuwenhoek 71:75–93. https://doi.org/10.1023/A:1000150219937
Llamas I, Moral AD, Martı’nez-Checa F, Arco Y, Arias S, Quesada E (2006) Halomonas maura is a physiologically versatile bacterium of both ecological and biotechnological interest. Antonie Van Leeuwenhoek 89:395–403. https://doi.org/10.1007/s10482-005-9043-9
Marino JH, Cook P, Miller KS (2003) Accurate and statistically verified quantification of relative mRNA abundances using SYBR green I and real-time RT-PCR. J Immunol Methods 283:291–306. https://doi.org/10.1016/S0022-1759(03)00103-0
Mormile MR, Romine MF, Garcia MT, Ventosa A, Bailey TJ, Peyton BM (1999) Halomonas campisalis sp. nov., a denitrifying, moderately haloalkaliphilic bacterium. Syst Appl Microbiol 22:551–558. https://doi.org/10.1016/S0723-2020(99)80008-3
Morozkina EV, Zvyagilskaya RA (2007) Nitrate reductases: structure, functions, and effect of stress factors. Biochem Mosc 72:1151–1160. https://doi.org/10.1134/S0006297907100124
Pastor JM, Salvador M, Argandoña M, Bernal V, Reina-Bueno M, Csonka LN, Iborra JL, Vargas C, Nieto JJ, Cánovas M (2010) Ectoines in cell stress protection: uses and biotechnological production. Biotechnol Adv 28:782–801. https://doi.org/10.1016/j.biotechadv.2010.06.005
Stolz JF, Basu P (2002) Evolution of nitrate reductase: molecular and structural variations on a common function. Chembiochem 3:198–206. https://doi.org/10.1002/1439-7633(20020301)3:2/3<198::AID-CBIC198>3.0.CO;2-C
Sun HF, Wang HW, Yuan CY (2013) Optimization of zinc-cadmium reduction method for determination of nitrate in seawater. Adv Mater Res 864–867:1004–1007. https://doi.org/10.4028/www.scientific.net/AMR.864-867.1004
Wang T, Li J, Zhang LH, Yu Y, Zhu YM (2017) Simultaneous heterotrophic nitrification and aerobic denitrification at high concentrations of NaCl and ammonia nitrogen by Halomonas bacteria. Water Sci Technol 76:386–395. https://doi.org/10.2166/wst.2017.214
Zhang LH, Lang YJ, Nagata S (2009) Efficient production of ectoine using ectoine-excreting strain. Extremophiles 13:717–724. https://doi.org/10.1007/s00792-009-0262-2
Zhang SM, Li WG, Zhang DY, Huang XF, Qin W, Gu J (2015) Purification and characterization of a low-temperature ammonia monooxygenase from heterotrophic nitrifier Acinetobacter sp. Y16. Desalin Water Treat 53:257–262. https://doi.org/10.1080/19443994.2013.837002
Zumft WG (1997) Cell biology and molecular basis of denitrification. Microbiol Mol Biol Res 61:533–616. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC232623/. Accessed March 2018
Funding
This research was funded by the Fundamental Research Funds for the Central Universities and Collaborative Innovation Center for Vessel Pollution Monitoring and Control Seed Fund Project, Dalian Maritime University (20110216001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, T., Li, Y., Zhang, L. et al. Salt tolerance of nitrate reductase in Halomonas sp. B01. Folia Microbiol 65, 909–916 (2020). https://doi.org/10.1007/s12223-020-00801-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12223-020-00801-9