Abstract
Low-grade gliomas is the malignant nervous tumor with distinct biological and clinical characteristics. Despite advances in diagnostic and therapeutic methods, how to significantly elongate the survival of low-grade gliomas is still challengeable. Complement 3, as the critical component in the innate immune system, plays an essential role in local immune response and participating into regulation of the epithelial–mesenchymal transition and tumor microenvironment. In this study, we systematically determined the expression levels and immunological roles of C3 in low-grade gliomas using various public databases. Then, we further identified the impact of C3 expression on immune cell infiltration compared to normal tissue, indicating the effect of cellular microenvironment on overall survival of LGG patients. We obtained clinical characteristics, transcriptome, and survival of C3 in LGG from the TCGA, GEPIA2.0, and cBioportal databases. Two differentially expressed genes (DEGs) were obtained, DEGs compared to normal tissue (DEG_G1) and DEGs between C3 high expression and C3 low expression in LGG patients (DEG_G2). By performing the GO analysis and protein–protein interaction (PPI) network of DEG_G1, we have identified the top-ranked 10 hub genes, which are highly associated with regulation of cell cycle. The gene set enrichment analysis demonstrated that overexpression of C3 in LGG patient is positively correlated with regulation of cell cycle. The relative PPI analysis and GSEA of DEG_G2 were performed and analysis results indicated that higher expression of C3 in the LGG can activate immune-related pathways. Finally, immune cell infiltration analysis of C3 in the LGG patients was employed and clearly indicated that higher neutrophil infiltration can worsen the survival of the LGG patients with higher expression of C3. These results were confirmed by the Human Protein Atlas database, in which expression level of C3 protein in gliomas patients always higher. This investigation implied that C3 can be as diagnostic biomarker and potential targets of precise therapy for the LGG patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Low-grade gliomas (LGG) was classified as the Grade I and II by World Health Organization, accounting for approximately 17% of all primary nervous tumors [1, 2]. Owing to the diverse pathology, median survival for LGG patients is only ranged from 5.6 to 13.3 years [3]. To further elongate survival of the LGG patients, many emerging diagnostic and therapeutic methods have been well developed in the past decade year, for example, precise therapy depended on specific molecular characteristics and histological classification [4,5,6] and chemotherapy combined with radiation therapy [7, 8]. Recently, the LGG patients can benefit from molecular diagnosis, for example, mutations of IDH1 and IDH2 [9, 10], and patients can further receive precise therapeutic interventions. To determine whether more genes can be the therapeutic targets or diagnostic biomarkers, more bioinformatics investigation to explore the role of differentially expressed genes in the LGG patients will provide the emerging molecular characteristics with precise treatment.
Tumor environment plays an important role in tumorigenesis, metastasis, and affecting clinical therapy [11]. In the past decade, emerging interventions to target tumor microenvironment have significantly improved the median survival of patients [12, 13], for example, apatinib for lung cancer immunotherapy [14], immune checkpoint blockers to improve the tumor microenvironment [15], and chimeric antigen receptor T cells in refractory B-Cell non-Hodgkin’s lymphoma [16]. However, owing to presentation of the compact blood–brain barrier, it is hard to deliver most of therapeutic cargos into brain parenchyma [17, 18]. To identify indirect molecular targets (e.g., vasculature molecular targets), designation of therapeutic cargo, including siRNA or antibodies [19,20,21], will be useful for the LGG therapy. These results indicated that targeting tumor microenvironment in the nervous system can be an emerging therapeutic strategies.
C3, so-called complement 3 protein, is one of critical components in the complement system, which contains two subtypes of proteins (C3a and C3b) and was further modified by C3 convertase complex [22]. The subtypes of C3 proteins (C3a and C3b) can strongly bind with many cell surface receptors to further activate downstream pathways, for example, C3b binding with complement receptor 1 (CR1 or CD35) to blockade the immune adherence [23] and binding with CD21 to promote the generation of B memory cell [24]. Activation of C3/C3a are the potent pro-inflammatory molecules and can induce the cascade response on affecting tumor microenvironment, for example, recruiting neutrophils and monocytes [25]. In the tumor microenvironment, recent reports clearly showed that cancer cell also can generate complement proteins to further modulate relative molecular pathways [26,27,28]. Overexpression of complement proteins suggested that C3a/C3aR participated into the regulation of epithelial–mesenchymal transition [29]. This literature demonstrated that complement can be as the potential therapeutic targets for cancer therapy.
In this study, we performed the comprehensive investigation to determine the correlation between C3 expression and LGG progression. By the Kaplan–Meier analysis, overall survival and progression-free survival of higher expression of C3 in the LGG patients can be identified. To explore role of overexpression of C3 in the LGG patients, we identified two DEGs groups, DEGs compared to normal tissue (DEG_G1) and DEGs between C3 high expression and C3 low expression in LGG patients (DEG_G2). After identifying the differentially expressed genes (DEGs), the PPI analysis provides the hub genes and relative roles of C3 in tumorigenesis. The gene set enrichment analysis (GSEA) of DEG_G1 and DEG_G2 was performed to explore how overexpression of the C3 gene affects the cellular networks. Finally, we study the immune cell infiltration between C3 expression and immune cells using TIMER database and confirmed using different algorithm methods. Our investigation provided that C3 may be as diagnostic biomarker and potential therapeutic target for the precise LGG therapy.
Methods
Database and clinical information about low-grade gliomas (LGG) patients
In this investigation, the clinical information about LGG patients (n = 515) was obtained from TCGA database (https://portal.gdc.cancer.gov/; Data Release 29.0-March 31, 2021) and cBioportal database (http://www.cbioportal.org/). The mRNA expression matrix was analyzed using R software package (Version 4.1.2, https://www.r-project.org/) and the mutation information was obtained from the cBioportal website. The information of C3 expression level was obtained from TIMER2.0 database.
Overall survival and progression-free survival analysis
The OS and PFS were obtained from GEPIA2.0 database (http://gepia2.cancer-pku.cn/#index), with threshold value as 50 to 50%. The clinical information was obtained from TCGA database.
The human protein atlas analysis
The protein expression level of relative biomarkers was analyzed using The Human Protein Atlas (https://www.proteinatlas.org/). The IHC staining of normal tissue was selected as cerebral cortex and the IHC staining of tumor tissue was selected as gliomas. All the IHC images were directly downloaded from HPA database without any further modification.
Identification of differentially expressed genes (DEGs)
The DEGs of C3 gene compared to normal tissues was obtained from GEPIA2.0 database, in which the threshold value considered as significant difference is |log2FoldChange|> 1.0 and p value < 0.01. The mRNA expression level of C3 and other critical genes compared from normal tissue (GTEx database) was also obtained from GEPIA 2.0 database.
These LGG patients (n = 515) can be divided into two individual groups, low expression group (n = 253) and high expression group (n = 252), by the median value in C3 expression matrix. To identify the DEGs between high expression group and low expression group, gene expression matrix was firstly obtained from TCGA database using R software TCGAbiolinks and further obtained using several R software packages (limma and edgeR). The threshold value of DEGs was set as p-value as 0.01 & |log2FoldChange|> = 1.0.
Protein–protein interaction (PPI) network analysis
The PPI analysis of DEGs obtained from GEPIA2.0 database (|log2FoldChange|> 1.5 and p value < 0.01) was analyzed using STRING website (https://www.string-db.org/), in which minimum required interaction score was set as 0.9 and cluster analysis parameter, number of k-mean, is set as 5. The interaction analysis was further performed using Cytoscape version 3.6.0.
Moreover, DEGs obtained between C3 high expression group and C3 low expression group were set as (|log2FoldChange|> 1.60 & p value < 0.01). PPI analysis of these DEGs was performed using STRING website, and images of network were generated using Cytoscape.
Gene set enrichment analysis (GSEA)
The overlaps of DEGs with MigDB gene set was performed using GSEA website (http://www.gsea-msigdb.org/gsea/msigdb/annotate.jsp), selecting hallmark gene sets, KEGG gene sets, reactome gene sets, and WikiPathways gene sets (FDR q-value < 0.05 considered as significant difference). GSEA scoring profile was analyzed using GSEA software (GSEA v4.1.0) using Molecular Signatures Database (MSigDB, http://www.gsea-msigdb.org/gsea/msigdb/index.jsp). The parameters of GSEA was set as default, except especially mentioned. Number of permutations was set as 2000 and collapse to gene symbols was set as No_collapse. Normalized enrichment score (NES) more than 1.0 was considered as up-regulation and NES less than -1.0 was considered as down-regulation. The threshold value with NOM p-value < 0.05 & FDR q-value < 0.25 is considered as significant difference.
Immune cell infiltration analysis
The immune cell infiltration analysis was performed using TIMER 2.0 database (http://timer.cistrome.org/). Correlation between gene expression and immune infiltration was performed using Gene blocks with purity adjustment, which contains 21 types of immune cells. The outcome of immune cell infiltration with clinical and gene expression was performed using TIMER 2.0 outcome block: Z-score > 0 & p < 0.05 was considered as increased risk and Z-score < 0 & p < 0.05 was considered as decreased risk. Moreover, immune cell infiltration by different algorithm methods, i.e., EPIC, XCELL, and CIBERSORT, was employed to validate the TIMER results.
Statistical analysis
All values in this investigation were presented as mean ± standard deviation (SD).The chi-square testing was employed to analyze the relationship of C3 expression between LGG group and normal tissue. The OS (overall survival) and PFS (progression-free survival) were defined as clinical endpoints. The GSEA analysis and images were generated using GSEA software using log-rank test and FDR q-value < 0.25 was considered as significance. Survival curve were curve using Kaplan–Meier method using log-rank test to evaluate whether can be consider as significance. p-value < 0.05 is the cutoff value to identify significance. Other statistical analysis was presented in relative section. All the images were generated using relative software described in aforementioned sections.
Results
Clinical information of LGG
In the World Health Organization (WHO) classification system, gliomas can be categorized from grade I to grade IV based on histopathological features. The WHO defined grade I–II tumors as LGG, which is different from the TCGA classification system (grade I–III) [30]. To identify the difference between WHO LGG and TCGA LGG, we firstly analyzed the clinical information of LGG samples in TCGA database, as shown in Table 1. Five types of gliomas were collected for further genomic investigation, i.e., anaplastic astrocytoma, astrocytoma NOS, mixed glioma, anaplastic oligodendroglioma, and oligodendroglioma NOS. Except astrocytoma NOS subtype, all the LGG samples in the TCGA database can be attributed as grade III. These results suggested that our investigation focused on effect of C3 expression on grade II–III gliomas.
Expression level and genetic status of C3 in the LGG patients
C3 is the critical component in complement system and plays an important role in immune response. We firstly examine copy number of C3 genes in the LGG patients using cBioportal website (Fig. 1A). There are four different types of genetic status identified in the LGG patients, containing deep deletion, shallow deletion, diploid, gain, and amplification. Among these genetic statuses in the LGG patients, diploid and gain are the major status, implying the overexpression levels of C3. By analyzing the mutation frequency of C3 depended on subtypes of the LGG patients, mutation and amplification are major status, i.e., oligoastrocytoma, oligodendrogliomas, and astrocytoma, which confirmed this genetic status is ordinary in the LGG patients. By performing the analysis of mRNA expression levels, we found that the C3 expression level in the LGG patients is significantly higher than normal brain tissue, about 2 times, implying that overexpression of C3 may play an important role in the LGG tumorigenesis.
A Copy number alterations of C3 genes in brain low-grade gliomas. B Mutation frequency of C3 genes dependence on subtypes of LGG. C mRNA expression levels of C3 genes compared to normal tissue. D Mutation sites of C3 genes in the LGG patients. E mRNA expression levels of C3 genes depended on various cancer types. *p < 0.05; **p < 0.01; ***p < 0.001
Gene mutation always plays the critical role in modulation of genetic network. For C3 protein, it always contains six sub-domains, i.e., A2M_N, A2M_N_2, A2M, A2M_comp, A2M_re, and NTR. Among these domains, there are 13 sites identified as mutation, especially for S770R. This mutation in C3 can affect the FGFR2 IIIb C3-transforming activity, causing aberrant receptor recycling and persistent FRS2-dependent signaling [31]. To further identify the role of C3 in various cancers, we found that C3 always displays higher expression level and may highly related with patient prognosis.
Effect of higher C3 expression on OS and PFS
Although expression level of C3 in the LGG patients is significantly higher in normal tissue, the protein level is still unknown. Here, we utilized the Human Protein Atlas to study the protein level in LGG patient tissues. As shown in Fig. 2A, we can observe that the expression level of C3 protein in the LGG patients is significantly higher than normal nervous tissue, consistent with mRNA level. To further investigate the impact of overexpression C3 in the LGG patients, we performed the Kaplan–Meier analysis utilizing GEPIA2.0 database. As shown in Fig. 2B-C, the medium survival time (OS and PFS) of LGG patients with low expression of C3 is significantly better than higher expression level of C3 (log-rank p values are 0.0031 and 0.0055, respectively). These results implied that overexpression of C3 protein worsens the survival time of the LGG patients. If C3 protein levels can be inhibited by specific therapeutic methods, e.g., antibody or siRNA silencing, the overall survival of LGG patients may benefit from these interventions.
Hub genes of C3 overexpression in LGG
To investigate the molecular mechanism of C3 overexpression in LGG, we utilized the protein–protein interaction network to identify the hub genes. The differentially expressed genes (DEGs_G1, |log2FoldChange|> 1.5 & FDR q-value < 0.01) were obtained from GEPIA2.0 website, compared to normal tissue (GTEx data). These DEGs were analyzed using STRING website and the interaction network was regenerated using CytoScape software, as shown in Fig. 3. The top 10 hub genes are C3AR1, CDK1, ITGAM, UBE2C, CCNB1, THBS1, CXCL12, POMC, CCNB2, and ADCY4, respectively. The biological function of these hub gens are listed in Table 2, which are highly associated with regulation of cell cycle. To further analyze expression levels of these hub genes, we utilized the GEPIA2.0 database to obtain the expression level compared to normal tissue. As shown in Fig. 3, we can find that C3AR1, CDK1, ITGAM, UBE2C, CCNB1, THBS1, CXCL12, and CCNB2 are significantly higher than normal tissue, while ADCY4 and POMC are lower than normal tissue.
Correlation between critical biomarkers and C3
By analyzing the transcriptome levels of GBM, Cameron W Brennan et al. [32] have identified several critical pathways to affect the tumorigenesis of glioblastoma, i.e., RTK pathway, PI3K pathway, MAPK pathway, p53 pathway, RB1 pathway, and ChrMod regulation, respectively. Here, we utilized cBioportal database to analyze the correlation between critical genes of these pathways and C3 expression level. As shown in Fig. 4, these results only displayed the correlation with significant difference. Among these genes, MET, FGFR3, RB1, IDH1, CDK6, and CDKN2C display the positive correlation with C3 expression level in the LGG patients (Spearman co-efficiency > 0 & p-value < 0.05), meanwhile PI3KR1, PDFFRA, PTEN, ND1, BRAF, and ATRX display the negative correlation with C3 (Spearman co-efficiency < 0 & p-value < 0.05) (Table 3).
Biological influence of C3 higher expression in LGG
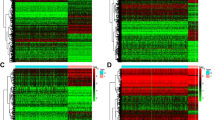
Compared to normal tissue, higher expression of C3 may promote the tumorigenesis of the LGG patients by several critical pathways as aforementioned, for example, MAPK pathway, p53 pathway, and RB1 pathway. However, how higher expression of C3 worsened the survival of LGG patients is still unclear. Here, we firstly obtained the DEGs between high expression group and low expression group, in which the threshold value to consider as significance is p-value < 0.01 & |log2FoldChange|> 1.6. Volcano plot of identified gene is shown in Fig. 5A, and there are 577 genes identified as DEGs (including 439 up-regulated genes and 138 down-regulated genes). To determine the interaction networks of these DEGs, we performed the PPI analysis in STRING website. As shown in Fig. 5B, we can find that several genes play a critical role in this regulation network, i.e., IL10, ITGB2, ITGAM, and CSF1R. Reported by previous literature [33,34,35], these genes are highly associated with tumorigenesis and tumor microenvironment, implying that higher expression C3 in the LGG patients may influence the tumor microenvironment. The impact of C3 on immune cell infiltration was further investigated as follows.
A Volcano plot of all identified genes in the LGG patients with low expression of C3 compared to high expression of C3. Down-regulated DEGs (n = 138) are labeled by green dots and up-regulated DEGs (n = 439) are labeled by orange dots. Threshold value to identify the DEGs are |log2FoldChange|> 1.60 & p-value < 0.01. B Protein–Protein interaction of differentially expressed genes. IL10, ITGB2, ITGAM, and CSF1R are identified as key hub genes, labeled by red circles
Gene set enrichment analysis of DEGs
To explore potential molecular mechanism for the LGG tumorigenesis, we firstly analyzed transcription factors of these DEGs_G1 using the TRRUST database (https://www.grnpedia.org/trrust/). As shown in Fig. 6A and B, we can find that the major transcription factor of up-regulated DEGs are TP53 and E2F1, related with the proliferation pathway and implied that overexpression of C3 may promote the tumor growth. Meantime, the major transcription factors of down-regulated genes are NFKB1, RELA, STAT3, and SP3. NFKB1, RELA, and STAT3 are attributed as the NFκB pathway-related factors and SP3 also can strongly interact with NR1 NFκB site [36]. The major transcription factors of down-regulated genes can be attributed as NFκB-related pathway, implying down-regulation of these genes may be owing to inhibition of NFκB pathway.
Then, we also analyzed the impact of DEGs on pathway levels using GSEA database. As shown in Fig. 6C, we can find that up-regulated DEGs are highly associated with Reactome_signaling_by_receptor_tyrosine_kinase, Hallmark_hypoxia, Reactome_neutrophil_degranulation, and Hallmark_TNFA_signaling_via_NFKB pathways. Moreover, we also observed that the down-regulated DEGs are highly related with Hallmark_E2F_targets, Reactome_cell_cycle, Reactome_cell_cycle_mototic, and Reactome_cell_cycle_checkpoints pathways, as shown in Fig. 6D.
To further explore the impact of these DEGs, we analyzed the GSEA profile of DEGs using GSEA software with Reactome gene sets, which can identify the impact of DEGs in LGG, as shown in Fig. 7. The top 5 up-regulated pathways are Reactome_cell_cycle, Reactome_cell_ccle_mitotic, Reactome_cell_cycle_checkpoints, Reactome_mitotic_metaphase_and_anaphase, and Reactome_M_phase. These pathways are highly related with tumor growth and cell cycle regulation. Moreover, the top 5 down-regulated pathways are Reactome_atimicrobial_peptides, Reactome_phase_I_functionalization_of_compounds, Reactome_disease_of_glycosylation, Reactome_diseases_of_metabolism, and Reactome_disoders_of_transmembrane_transporters. These inhibited pathways are highly associated with metabolism, indicating overexpression of C3 may affect the energy metabolism of cancer cells. We also observed the inhibition of _TNFA_signaling_via_NFKB (NES = − 1.555 with FDR q-value = 0.216), which is consistent with the results of transcription factor analysis (Fig. 6B). Among these pathways, regulation of cell cycle pathway plays the central role in LGG tumorigenesis.
GSEA scoring profiles of top 5 up-regulated and top 5 down-regulated pathways. Differentially expressed genes of the LGG samples compared to normal tissue were obtained from GEPIA 2.0 database. The threshold value of GSEA results to be considered as significance is |NES|> 1.0 & FDR value < 0.25 & p-value < 0.05
Moreover, we also performed the GSEA analysis using expression matrix, which is obtained by analyzing between C3 high expression group and C3 low expression group. As shown in Fig. 8, we found that higher expression of C3 in LGG patient may activate several pathways, i.e., KEGG_systemic_lupus_erythematosus, KEGG_cytokine_cytokine_receptor_interaction, KEGG_allograft_rejection, KEGG_leishamania_infection, KEGG_complement_and_coagulation_cascades, KEGG_asthma, and KEGG_graft_versus_host_disease. These annotations are highly associated with immunological response, implying that higher expression levels of C3 in LGG may affect the tumorigenesis of LGG by modulating tumor-related immune response. However, we also observed several inhibited pathways, i.e., KEGG_cardiac_muscle_contraction, KEGG_terpenoid_backbone_biosynthesis, and KEGG_neuroactive_ligand_receptor_interaction. These pathways are associated with biosynthesis or interaction of signaling molecules and these results indicated that higher expression of C3 in LGG may participate into dysfunction of neurological system. These results suggested that it may be helpful to improve the quality of patients’ life by inactivating bioactivity of C3.
GSEA scoring profiles of top 7 up-regulated and top 3 down-regulated pathways. Differentially expressed genes of LGG samples with high expression of C3 compared to the LGG samples with low expression of C3 were obtained from TCGA database. The threshold value of GSEA results to be considered as significant when |NES|> 1.0 & FDR value < 0.25 & p-value < 0.05
Immune cell infiltration analysis
Tumor microenvironment always plays critical roles in tumorigenesis and affects the prognosis of LGG patients [11]. To explore how overexpression of C3 gene affects the immune cell filtration in the LGG patients, we further analyzed the correlation between immune cells and C3 expression level. We analyzed the correlation between C3 expression level and 20 types of immune cells using TIMER2.0 database. As shown in Fig. 9, we can find that major types of immune cells are highly associated with C3 expression in the LGG patients. We found that immune cell infiltration levels of neutrophil is positive correlation with C3 expression (Cor = 0.808 & p value = 1.26e-111), indicating that higher infiltration level of immune worsens the survival of the LGG patients (Fig. 9G-H). To further validate whether immune cell plays a critical role in modulation of tumor microenvironment, immune cell infiltration levels of these six types of immune cells were performed using four different methods, as shown in Table 4. We can find that immune cell infiltration levels of macrophage and neutrophil cells can be observed in three algorithm methods, indicating that macrophage and neutrophil may play an important role in modulation of tumor microenvironment.
To our knowledge, the biomarkers of neutrophil cells are MPO, CD11b, CD66b, and CD16, respectively. To further analyze protein expression of neutrophil biomarkers in the LGG patients, we utilized HPA database to analyze the expression levels of these biomarkers. As shown in Fig. 10, IHC staining results clearly showed that the expression levels of these biomarkers are highly expressed in gliomas tissues, confirming the immune cell infiltration results.
Discussion
Complement system is the major immune response system in blood circulation system, involving in host innate immune response [37]. In recently investigation, the innate immune system may play an important role in tumorigenesis and proliferation, indirectly affecting the survival of cancer patients [38,39,40]. In complement system, C3 is the central role in activation of complement system by cleaving C3 molecules to C3a through C3 convertase complex [22]. Binding of C3 molecules to targeted cell surface can further recruit the immunological cells to infiltrate tumor tissue, including neutrophils [41]. How C3 protein affects the progression of the LGG patients should be determined.
In this investigation, we firstly explored the C3 expression levels in various tumor tissues (Fig. 1E), clearly demonstrated that the C3 expression level in the LGG patients is significantly higher than normal tissue, and also displayed the significantly up-regulated status in most of cancers. Then, we analyzed the C3 copy number, frequency, and mutation sites in the LGG patients, which confirmed the overexpression of C3 in the LGG patients.
To explore the impact of C3 on survival of the LGG patients, the OS and PFS curves (Fig. 2B) corroborated that the higher expression of C3 in the LGG patients worsens the survival time, confirming the negative correlation between C3 expression and survival. Then, we examined the protein expression of C3 protein in normal tissue and tumor tissue. In the normal tissue, the C3 proteins mainly expressed around the vasculature. While, the C3 expression in gliomas tissue are highly expressed, not only endothelial cells (Fig. 2A). These results confirmed that higher expression of C3 protein in tumor tissue is positively associated with tumorigenesis.
To explore the intrinsic molecular mechanism of C3 overexpression to promote tumor proliferation, we obtained the DEGs in the LGG patients compared to normal tissues. The PPI analysis (Fig. 3) clearly showed the top 10 hub genes, i.e., C3AR1, CDK1, ITGAM, UBE2C, CCNB1, THBS1, CXCL12, POMC, CCNB2, and ADCY4. These genes are mainly related with cell cycle, implying that regulation of cell cycle may play the critical role in tumorigenesis. By analyzing the correlation between C3 gene and critical biomarkers (Fig. 4), these results implied the inhibition of PI3K and MAPK pathways, while the RTK, p53, and RB1 pathways are over-activated in the LGG patients. Moreover, we also analyzed the expression profile of identified genes between C3 high expression group and C3 low expression group (Fig. 5A). The PPI network clearly showed that the critical genes modulating the molecular network are IL10, ITGB2, ITGAM, and CSF1R. These genes are highly associated with immunological response, implying that higher expression of C3 in the LGG patients may activate immune-related pathways.
To explore molecular mechanisms in the LGG tumorigenesis, we firstly analyzed the transcription factors of up-regulated and down-regulated genes. As shown in Fig. 6A and B, the transcription factors of up-regulated genes are TP53 and E2F series, demonstrating the activation of p53 pathway and regulation of cell cycle. Moreover, the transcription factors of down-regulated genes is NFKB1, RELA, STAT3, and SP series. NFKB1, RELA, and STAT3 are highly associated with TNFα/NFκB pathway, implying the inhibition of TNFα/NFκB pathway. Then, the GSEA overlaps (Fig. 6C-D) and profile analysis (Fig. 7) clearly showed that cell cycle-related pathways are activated in LGG and confirmed the inhibition of TNFα/NFκB pathway. Moreover, GSEA analysis (Fig. 8) between C3 high expression group and C3 low expression group implied that higher expression levels of C3 in the LGG patients may activate the immune-related pathways and inhibit biosynthesis of several critical signaling molecules. These results suggested that higher expression of C3 in the LGG patients may affect the survival via modulating the tumor microenvironment.
As abovementioned, most of affected pathways are associated with immune response. For the tumor microenvironment, it always participated into the modulation of several critical cells, for example, cancer-associated fibroblast, cancer stem cell, endothelial cell, pericyte, immune inflammatory cells, and invasive cancer [11]. The immune cell infiltration in tumor tissue is highly associated with tumor proliferation, drug resistance, and epithelial–mesenchymal transition (EMT) [11]. Consequently, we analyzed the immune cell infiltration in the LGG tissue. By analyzing the 21 types of immune cells in the LGG tissue, we find that the neutrophil infiltration is highly associated with C3 expression (Fig. 9). Higher neutrophil infiltration can worsen survival of the LGG patients and inhibition of C3 expression may improve survival of the LGG patients. To validate the neutrophil infiltration in gliomas tissue, we analyzed the neutrophil biomarkers using HPA database (Fig. 10). The IHC results clearly demonstrated that the biomarkers of neutrophil in the LGG patients is highly expressed, consistent with neutrophil infiltration.
Conclusion
In summary, our study showed that the overexpression of C3 in the LGG patients can worsen survival of patients. The overexpression of C3 in the LGG patients can lead the over-activation of cell cycle-related pathways, highly associated with tumorigenesis. The LGG patients may benefit from the improvement of neutrophil infiltration. These results indicated that the LGG patients may benefit from the inhibition of C3 in the LGG patients and the results displayed C3 can be the excellent therapeutic target for the LGG therapy.
Data Availability
All data and materials are available at TCGA, TIMER, and GEPIA2.0 databases.
Abbreviations
- LGG:
-
Low-grade gliomas
- GSEA:
-
Gene set enrichment analysis
- DEGs:
-
Differentially expressed genes
- IHC:
-
Immunohistochemistry
- HPA:
-
Human protein atlas
References
Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64(6):479–89.
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–20.
Buckner JC, Shaw EG, Pugh SL, Chakravarti A, Gilbert MR, Barger GR, Coons S, Ricci P, Bullard D, Brown PD, Stelzer K, Brachman D, Suh JH, Schultz CJ, Bahary JP, Fisher BJ, Kim H, Murtha AD, Bell EH, Won M, Mehta MP, Curran WJ. Radiation plus Procarbazine, CCNU, and Vincristine in Low-Grade Glioma. N Engl J Med. 2016;374(14):1344–55.
Tan Y, Zhang S, Xiao Q, Wang J, Zhao K, Liu W, Huang K, Tian W, Niu H, Lei T, Shu K. Prognostic significance of ARL9 and its methylation in low-grade glioma. Genomics. 2020;112(6):4808–16.
Li X, Meng Y. Survival analysis of immune-related lncRNA in low-grade glioma. BMC Cancer. 2019;19(1):813.
Liu J, Chen C, Wei T, Gayet O, Loncle C, Borge L, Dusetti N, Ma X, Marson D, Laurini E. In Dendrimeric nanosystem consistently circumvents heterogeneous drug response and resistance in pancreatic cancer. Exploration, Wiley Online Library. 2021;1:21–34.
Shaw EG, Wang M, Coons SW, Brachman DG, Buckner JC, Stelzer KJ, Barger GR, Brown PD, Gilbert MR, Mehta MP. Randomized trial of radiation therapy plus procarbazine, lomustine, and vincristine chemotherapy for supratentorial adult low-grade glioma: initial results of RTOG 9802. J Clin Oncol. 2012;30(25):3065–70.
Guo, S.; Li, K.; Hu, B.; Li, C.; Zhang, M.; Hussain, A.; Wang, X.; Cheng, Q.; Yang, F.; Ge, K. In Membrane‐destabilizing ionizable lipid empowered imaging‐guided siRNA delivery and cancer treatment, Exploration, Wiley Online Library: 2021; pp 35–49.
Jones DTW, Kieran MW, Bouffet E, Alexandrescu S, Bandopadhayay P, Bornhorst M, Ellison D, Fangusaro J, Fisher MJ, Foreman N, Fouladi M, Hargrave D, Hawkins C, Jabado N, Massimino M, Mueller S, Perilongo G, Schouten van Meeteren AYN, Tabori U, Warren K, Waanders AJ, Walker D, Weiss W, Witt O, Wright K, Zhu Y, Bowers DC, Pfister SM, Packer RJ. Pediatric low-grade gliomas: next biologically driven steps. Neuro Oncol. 2018;20(2):160–73.
van den Bent MJ, Looijenga LH, Langenberg K, Dinjens W, Graveland W, Uytdewilligen L, Sillevis Smitt PA, Jenkins RB, Kros JM. Chromosomal anomalies in oligodendroglial tumors are correlated with clinical features. Cancer. 2003;97(5):1276–84.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Terrén I, Orrantia A, Vitallé J, Zenarruzabeitia O, Borrego F. NK cell metabolism and tumor microenvironment. Front Immunol. 2019;10:2278.
Meurette O, Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34(4):536–48.
Zhao S, Ren S, Jiang T, Zhu B, Li X, Zhao C, Jia Y, Shi J, Zhang L, Liu X, Qiao M, Chen X, Su C, Yu H, Zhou C, Zhang J, Camidge DR, Hirsch FR. Low-dose apatinib optimizes tumor microenvironment and potentiates antitumor effect of PD-1/PD-L1 blockade in lung cancer. Cancer Immunol Res. 2019;7(4):630–43.
Datta M, Coussens LM, Nishikawa H, Hodi FS, Jain RK. Reprogramming the tumor microenvironment to improve immunotherapy: emerging strategies and combination therapies. Am Soc Clin Oncol. 2019;39:165–74.
Yan ZX, Li L, Wang W, OuYang BS, Cheng S, Wang L, Wu W, Xu PP, Muftuoglu M, Hao M, Yang S, Zhang MC, Zheng Z, Li J, Zhao WL. Clinical efficacy and tumor microenvironment influence in a dose-escalation study of anti-CD19 chimeric antigen receptor T cells in refractory b-cell non-Hodgkin’s Lymphoma. Clinical Cancer Res. 2019;25(23):6995–7003.
Arvanitis CD, Ferraro GB, Jain RK. The blood–brain barrier and blood–tumour barrier in brain tumours and metastases. Nat Rev Cancer. 2020;20(1):26–41.
Zhao, Y.; Zhang, Z.; Pan, Z.; Liu, Y. In Advanced bioactive nanomaterials for biomedical applications, Exploration, Wiley Online Library: 2021: 0089.
Qiao, N.; Du, G.; Zhong, X.; Sun, X. In Recombinant lactic acid bacteria as promising vectors for mucosal vaccination, Exploration, Wiley Online Library: 2021; p 0026.
Tang, G.; He, J.; Liu, J.; Yan, X.; Fan, K. In Nanozyme for tumor therapy: Surface modification matters, Exploration, Wiley Online Library: 2021; pp 75–89.
Tu, L.; Liao, Z.; Luo, Z.; Wu, Y. L.; Herrmann, A.; Huo, S. In Ultrasound‐controlled drug release and drug activation for cancer therapy, Exploration, Wiley Online Library: 2021; 0023.
Afshar-Kharghan V. The role of the complement system in cancer. J Clin Investig. 2017;127(3):780–9.
Erdei A, Sándor N, Mácsik-Valent B, Lukácsi S, Kremlitzka M, Bajtay Z. The versatile functions of complement C3-derived ligands. Immunol Rev. 2016;274(1):127–40.
Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, Lemere CA, Selkoe DJ, Stevens B. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352(6286):712–6.
Kleczko EK, Kwak JW, Schenk EL, Nemenoff RA. Targeting the complement pathway as a therapeutic strategy in lung cancer. Front Immunol. 2019;10:954.
Cho MS, Vasquez HG, Rupaimoole R, Pradeep S, Wu S, Zand B, Han HD, Rodriguez-Aguayo C, Bottsford-Miller J, Huang J, Miyake T, Choi HJ, Dalton HJ, Ivan C, Baggerly K, Lopez-Berestein G, Sood AK, Afshar-Kharghan V. Autocrine effects of tumor-derived complement. Cell Rep. 2014;6(6):1085–95.
Thurman JM, Le Quintrec M. Targeting the complement cascade: novel treatments coming down the pike. Kidney Int. 2016;90(4):746–52.
Zhou, J.; Zhang, Z.; Joseph, J.; Zhang, X.; Ferdows, B. E.; Patel, D. N.; Chen, W.; Banfi, G.; Molinaro, R.; Cosco, D. In Biomaterials and nanomedicine for bone regeneration: Progress and future prospects, Exploration, Wiley Online Library: 2021; p 0011.
Cho MS, Rupaimoole R, Choi HJ, Noh K, Chen J, Hu Q, Sood AK, Afshar-Kharghan V. Complement component 3 is regulated by TWIST1 and mediates epithelial-mesenchymal transition. J Immunol. 2016;196(3):1412–8.
Forst DA, Nahed BV, Loeffler JS, Batchelor TT. Low-grade gliomas. Oncologist. 2014;19(4):403–13.
Cha JY, Maddileti S, Mitin N, Harden TK, Der CJ. Aberrant receptor internalization and enhanced FRS2-dependent signaling contribute to the transforming activity of the fibroblast growth factor receptor 2 IIIb C3 isoform. J Biol Chem. 2009;284(10):6227–40.
Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, Beroukhim R, Bernard B, Wu CJ, Genovese G, Shmulevich I, Barnholtz-Sloan J, Zou L, Vegesna R, Shukla SA, Ciriello G, Yung WK, Zhang W, Sougnez C, Mikkelsen T, Aldape K, Bigner DD, Van Meir EG, Prados M, Sloan A, Black KL, Eschbacher J, Finocchiaro G, Friedman W, Andrews DW, Guha A, Iacocca M, O’Neill BP, Foltz G, Myers J, Weisenberger DJ, Penny R, Kucherlapati R, Perou CM, Hayes DN, Gibbs R, Marra M, Mills GB, Lander E, Spellman P, Wilson R, Sander C, Weinstein J, Meyerson M, Gabriel S, Laird PW, Haussler D, Getz G, Chin L. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–77.
Dashti N, Mahmoudi M, Gharibdoost F, Kavosi H, Rezaei R, Imeni V, Jamshidi A, Aslani S, Mostafaei S, Vodjgani M. Evaluation of ITGB2 (CD18) and SELL (CD62L) genes expression and methylation of ITGB2 promoter region in patients with systemic sclerosis. Rheumatol Int. 2018;38(3):489–98.
Ouyang W, O’Garra A. IL-10 Family Cytokines IL-10 and IL-22: from Basic Science to Clinical Translation. Immunity. 2019;50(4):871–91.
Rojo R, Raper A, Ozdemir DD, Lefevre L, Grabert K, Wollscheid-Lengeling E, Bradford B, Caruso M, Gazova I, Sánchez A, Lisowski ZM, Alves J, Molina-Gonzalez I, Davtyan H, Lodge RJ, Glover JD, Wallace R, Munro DAD, David E, Amit I, Miron VE, Priller J, Jenkins SJ, Hardingham GE, Blurton-Jones M, Mabbott NA, Summers KM, Hohenstein P, Hume DA, Pridans C. Deletion of a Csf1r enhancer selectively impacts CSF1R expression and development of tissue macrophage populations. Nat Commun. 2019;10(1):3215.
Liu A, Hoffman PW, Lu W, Bai G. NF-kappaB site interacts with Sp factors and up-regulates the NR1 promoter during neuronal differentiation. J Biol Chem. 2004;279(17):17449–58.
Gialeli C, Gungor B, Blom AM. Novel potential inhibitors of complement system and their roles in complement regulation and beyond. Mol Immunol. 2018;102:73–83.
Corrales L, Matson V, Flood B, Spranger S, Gajewski TF. Innate immune signaling and regulation in cancer immunotherapy. Cell Res. 2017;27(1):96–108.
Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14(10):1014–22.
Zhang, Y.; Yang, H.; Wei, D.; Zhang, X.; Wang, J.; Wu, X.; Chang, J. In Mitochondria‐targeted nanoparticles in treatment of neurodegenerative diseases, Exploration, Wiley Online Library: 2021; p 0115.
Matthews KW, Mueller-Ortiz SL, Wetsel RA. Carboxypeptidase N: a pleiotropic regulator of inflammation. Mol Immunol. 2004;40(11):785–93.
Funding
Funding was provided by Henan University for Young Teachers.
Author information
Authors and Affiliations
Contributions
JY together performed the analysis and generated all the figures in this manuscript; KM, LW, and YM participated in the preparation of manuscript and revised the manuscript; HW designed and supervised this assay, writing this manuscript, and further revision.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, S., Miao, K., Wang, L. et al. Bioinformatics analysis of C3 in brain low-grade gliomas as potential therapeutic target and promoting immune cell infiltration. Med Oncol 39, 27 (2022). https://doi.org/10.1007/s12032-022-01647-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12032-022-01647-6