Abstract
Systemic sclerosis (SSc), an autoimmune disease of connective tissue, is characterized by inflammation, fibrosis, and vessel endothelial damage. Products of Integrin subunit beta 2 (ITGB2) and selectin L (SELL) genes participate in several functional pathways of immune system. The aim of this investigation was to survey the transcript level of ITGB2 and SELL genes as well as methylation status of CpG sites in promoter region of differently expressed gene in PBMCs of SSc patients. PBMCs were isolated from whole blood of 50 SSc patients and 30 healthy controls. Total RNA and DNA contents of PBMCs were extracted. Gene expression was analyzed by real-time PCR using the SYBR Green PCR Master Mix. To investigate the methylation status of CpG sites, DNA samples were treated by bisulfite, amplified through nested PCR, and sequenced through Sanger difficult sequencing method. ITGB2 gene in PBMCs of SSc patients was overexpressed significantly in comparison to healthy controls. However, no altered SELL expression was observed. Three CpG sites of 12, 13 and 14 were significantly hypomethylated in patients group, despite overall methylation status of ITGB2 gene promoter revealed no significant difference between study groups. There was no statistically significant correlation between methylation status of ITGB2 promoter and the gene expression in patients. Regarding to lack of correlation of increased expression of ITGB2 with its promoter hypomethylation in SSc patients, our study suggests that upregulation of ITGB2 in PBMCs from SSc patients is probably due to another mechanism other than methylation alteration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Systemic sclerosis (SSc) or scleroderma, a systemic and autoimmune disease of connective tissue, is characterized by vasculopathy, fibrosis, and the production of autoantibodies to nuclear proteins [1]. SSc can lead to death by involving the internal organs such as heart, lungs, kidneys, and intestine [2]. SSc is a rare rheumatic disease, which affects approximately 250 per million persons [3]. This disease associates with early vascular changes and cellular infiltration to target tissues. Although SSc pathogenesis is not completely understood, leukocyte infiltration and immigration to involved tissues and inflammation have been observed. Alternately, the infiltrated leukocytes release cytokines, growth factors, and chemokines, resulting in fibroblast activation, excess collagen deposition, fibrosis, and inflammation. These process may need to cell–cell and cell-extra cellular matrix (ECM) interactions, which is mediated by different cell surface adhesion molecules [4]. The main clinical subsets of SSc are limited cutaneous SSc (lSSc) and diffuse cutaneous SSc (dSSc), which are distinguished by fibrosis severity and autoantibodies features. The lSSc form is identified by slight vascular damages with slight fibrosis of skin and internal organs, and is accompanied with low progression rate. In contrast, the dSSc patients have progressive fibrosis of skin and internal organs, leading to high mortality rate [5,6,7,8,9,10,11]. However, it should be noted that despite lSSc form displays mild complications and the mortality and morbidity rates are slightly lower in them in comparison to dSSc form, the mortality and morbidity rate in lSSc form are still remarkable [12].
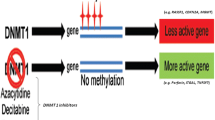
Both genetic and epigenetic factors play important roles in SSc etiopathogenesis [13,14,15]. Many studies demonstrated that the epigenetic mechanisms, including CpG DNA methylation, histones modifications, and miRNAs associate with vascular disorders, immune system activation, and accumulation of excessive ECM in SSc disease [16]. DNA Methylation occurs on the C5 position on a cytosine in the DNA sequence called CpG islands. This reaction is catalyzed by DNA methyltransferase (DNMT) enzymes, which transfer methyl (CH3) group of S-adenosylmethionine to the 5th carbon of cytosine molecule to form the 5-methylcytosine (5mC). About 70% of genes have CpG island(s) in their promoter region [17,18,19]. Universally, DNA hypermethylation leads to downregulation and its hypomethylation results in the upregulation of gene expression [20].
Peripheral blood cells (PBCs) of patients with SSc display increased expression of several selectin and integrin genes such as ITGB2, encoding integrin β2 protein, and SELL, encoding selectin L protein, which mediate cell adhesion to vascular endothelium and have important roles in vasculopathy, inflammation, and fibrosis in SSc disease [21]. ITGB2 gene is on chromosome 21 (21q22.3) and encodes integrin β2 protein (CD18) [22]. Integrin β2 family plays important roles in leukocyte adhesion, immune and inflammatory reactions, immigration through endothelial and chemotaxis [23]. Furthermore, selectin proteins family is expressed on most leukocytes such as neutrophils, monocytes, eosinophils, basophils, and many lymphocytes. SELL gene is located on chromosome 1 (1q23–q25) [24] and encodes l-selectin protein (CD62L), which interacts with inducible ligands on endothelium of non-lymphoid vessels, mediating first step of leucocyte immigration that called ‘rolling’ [4].
Due to the role of SELL and ITGB2 genes in inflammation and vasculopathy, and considering the important participation of DNA methylation in the regulation of gene expression, herein we intended to look deep inside the molecular mechanisms of inflammation and vascular disorders in patients with SSc with respect to analysis of mRNA expression and promoter methylation of the mentioned genes in PBMCs from SSc patients. Furthermore, the correlation of an important clinical manifestation of SSc patients, the Rodnan score, with RNA expression and promoter methylation of ITGB2 gene was investigated.
Materials and methods
Study participants
Fifty SSc patients (25 lSSc and 25 dSSc) were selected randomly from Rheumatology Research Center (RRC) clinic, Shariati Hospital. For all SSc patients, diagnosis of the disease were determined by American College of Rheumatology (ACR) criteria [25]. Moreover, 30 healthy controls without history of rheumatic and autoimmune diseases were selected that were matched base on gender, race and age with the patients. None of the healthy controls had rheumatic and autoimmune diseases in themselves and their close family members. The disease form was determined and Rodnan score of patients and erythrocyte sedimentation rate (ESR) for study subjects were measured. Baseline and demographic characteristics of dSSc and lSSc patients and healthy controls are listed in Table 1 with more details. The Human Research Ethics Committee of Tehran University of Medical Sciences confirmed this study. Written informed testimonial forms were obtained from all the participants. About 10 ml of blood samples was taken from SSc patients during clinical diagnosis; also, samples were taken from healthy controls.
PBMC isolation, RNA extraction and cDNA synthesis
The Ficoll-Hypaque density gradient centrifugation approach was applied to isolate PBMCs from peripheral blood of the cases. Total RNA of peripheral blood leukocytes of 80 samples were extracted by miRNeasy Mini Kit (Cat No. 217004, Qiagen, USA) according to the manufacturer’s instructions. Synthesis of cDNA was also performed by miScript II RT Kit (Cat No. 218161, Qiagen, USA) according to the manufacturer’s instructions. Yield and purity of extracted RNAs and synthesized cDNAs were assessed using a NanoDrop spectrophotometer at 260 and 280 nm (NanoDrop ND-2000C Spectrophotometer, Thermo Fisher Scientific, USA).
Gene expression by real-time quantitative PCR
Primers (ITGB2, SELL and β2M as a housekeeping gene) were designed using Primer Express 3.0 software (Applied Biosystems, Foster City, CA, USA). The Basic Local Alignment Search Tool on the US National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to determine accuracy and specificity of primers. Quantitative analysis of the genes expression in PBMCs was investigated by ABI StepOnePlus real-time PCR System and SYBR Green master mix. Each mixture of reaction (20 μl total volume) comprised of 10 μl of master mix, 2 μl of cDNA, 0.8 μl of forward primer, 0.8 μl of reverse primer, and 6.4 μl of H2O. The quantitative real-time PCR was fulfilled under these conditions: 10 min at 95 °C, then 40 cycles for 15 s at 95 °C, and 60 s at 60 °C. To analyze relative gene expression, a common approach, the comparative CT method was used as previously described by Schmittgen and Livak [26]. Relative mRNA expression level of target genes in samples was calculated and normalized to the corresponding β2M expression level as a housekeeping gene. After that, relative expression of each gene in the samples was calculated using this equation: relative mRNA expression = (2−ΔCt) × 103.
DNA extraction
DNA of 80 samples were extracted by phenol–chloroform method [27]. Yield and purity of extracted DNAs were determined using a NanoDrop spectrophotometer at 260 and 280 nm (NanoDrop ND-2000C Spectrophotometer, Thermo Fisher Scientific, USA).
DNA treatment via bisulfite conversion
The extracted DNA samples were used for bisulfite treatment according to a manual described by Kate Patterson et al. [28]. This method changes unmethylated C bases to U bases and has no effect on methylated C bases and was previously implemented at our lab [29,30,31].
Nested PCR, gel electrophoresis and sequencing
Our real-time quantitative PCR results led us to investigate methylation status of ITGB2 promoter region, due to its significantly different transcript profile between patients and controls.
The amplification target DNA was 251 bp on chromosome 21q22.3, located in a CpG island with 576 bp length, from promoter to exon 1. There was 22 CpG sites in the target DNA (Fig. 1) but 16 CpG sites were detectable in our sequencing and took the analysis criteria.
Schematic picture of the target CpG sites of selected CpG island in the promoter of ITGB2 gene. The amplification target DNA was 251 bps on chromosome 21q22.3, which was covered within a CpG island with 576 bps length from promoter to exon 1. Twenty-two CpG sites were found in the chosen region of ITGB2 promoter. Sixteen CpG sites methylation status were surveyed
Because the amplification of bisulfite treated DNA was difficult, nested PCR was designed for the amplification of treated DNA. To amplify the sequence containing CpG islands in promoter of ITGB2 (21q22.3), in the first stage, an outer forward primer (F1 sequence: AAGGGTGATGTCGTTAGGGTT) and an outer reverse primer (R1 sequence: CGCGCACCGACTAAACAAC) with 919 bp product length was designed. After that, this PCR product was used as templet for second run. At this run, an inner forward primer (F2 sequence: GGATAGTGAGGTTGGGAGTTTAT) and an inner reverse primer (R2 sequence: TCCAAAACACCTAACTAAAACCTAC) with 251 bps product size was designed. Both pairs of primers were designed by Methprimer website (http://www.urogene.org/cgibin/methprimer/methprimer.cgi).
Each tube for PCR comprised of 10 μl of PCR master mix (including PCR Taq polymerase and buffer), 0.75 μl of forward primer, 0.75 μl of reverse primer, 4 μl of bisulfite-treated DNA, and H2O to a total volume of 25 μl. It is important to note that at second run 1.5 μl of each forward and reverse primers were used. PCR was fulfilled under these conditions: 35 cycles for DNA denaturation at 94 °C for 30 s, primer annealing at an optimized temperature of 58.4 °C (outer primer) and 61.5 °C (inner primer) for 45 s, and an extension stage at 72 °C for 45 s. Finally, the extension at 72 °C for 10 min was fulfilled.
Gel electrophoresis of final PCR product was done to validate amplification (Fig. 2) and, then, sequenced via difficult Sanger sequencing method (Macrogen, Seoul, Korea). The results of DNA sequencing were analyzed using CodonCode Aligner version 2 software.
Statistical analysis
Analysis of data was performed using SPSS software version 22 (SPSS, Chicago, IL, USA). The Kolmogorov–Smirnov test was used to normality evaluation of Scale variables. Group comparisons of non-parametric variables were performed via the Mann–Whitney U test. To determine the relationship between parametric and nonparametric scale variables, Pearson’s and Spearman’s correlations were applied, respectively. The GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, California, USA; http://www.graphpad.com) was used for plotting the graphs. The study results were represented as Median and Interquartile Range (IQR), with statistical significance set at 0.05.
Results
mRNA expression level of ITGB2 and SELL in PBMCs
It was observed that mRNA expression level of ITGB2 gene was significantly upregulated in PBMCs of SSc patients compared to that of healthy controls (fold change = 1.44; P = 0.047; Fig. 3A). ITGB2 mRNA expression was nearly increased significantly twofold in PBMCs from dSSc patients compared to healthy controls (fold change = 1.81; P = 0.038; Fig. 3B). However, nearly similar level of ITGB2 mRNA was found in lSSc patients compared to healthy controls; but the expression level difference was insignificant (fold change = 1.07; P = 0.195; Fig. 3C). On the other side, expression of SELL in SSc patients was a little lower than in healthy subjects; but this difference was not significant (fold change = 0.86; P = 0.901; Fig. 3D). As well, SELL mRNA expression in PBMCs of dSSc patients was nearly identical to that of healthy controls, and the difference of expression was not significant (fold change = 0.97; P = 0.372; Fig. 3E). Despite insignificantly, lSSc patients expressed lower SELL mRNA level compared to healthy controls (fold change = 0.74; P = 0.480; Fig. 3F).
Relative mRNA expression level of ITGB2 and SELL in PBMCs of SSc patients vs. healthy control (A, D), dSSc patients vs. healthy control (B, E) and lSSc patients vs. healthy control (C, F). The Kolmogorov–Smirnov test revealed non-normal distribution of data and, therefore, the Mann–Whitney U test applied to calculating represented P values (SSc systemic sclerosis, dSSc diffuse SSc, lSSc limited SSc, FC fold change)
Methylation status of ITGB2 promoter in PBMCs
The methylation status of ITGB2 promoter CpG sites in SSc and control groups has been shown in Table 2. Sixteen CpG sites in ITGB2 promoter region were surveyed. CpG 12, 13 and 14 were hypomethylated significantly in patients compared to the control groups (P = 0.046, 0.018 and 0.043, respectively). In other words, the methylation status of the other 13 CpG sites were not significantly different. CpG12 showed decreased methylation level in patients compared with controls and its methylation level was associated significantly with the decreased risk of SSc (P = 0.046, OR 0.33, 95% CI 0.11–0.98). Moreover, methylation level of CpG13 in patients compared with controls was decreased and methylation of this CpG site demonstrated association with lower SSc risk (P = 0.018, OR 0.14, 95% CI 0.03–0.71). As well, a negative association was observed between decreased methylation level of CpG14 and SSc risk (P = 0.043, OR 0.36, 95% CI 0.14–0.97).
Taking all the CpG sites together, analysis of methylation status of CpG sites of ITGB2 promoter region revealed that there was no significant difference between PBMCs from patients and control groups (P = 0.46, Fig. 4A). Furthermore, comparison of ITGB2 promoter methylation level between dSSc PBMCs and healthy subjects showed no significant difference (P = 0.24, Fig. 4B). As well, there was no statistically significant difference of ITGB2 promoter methylation status between PBMCs from lSSc and healthy subjects (P = 0.36, Fig. 4C).
Correlation of ITGB2 expression with its promoter methylation
In SSc patients, methylation percentage of ITGB2 gene promoter in PBMCs did not display correlation with mRNA expression level of this gene (P = 0.76, ρ = 0.044; Supplementary Fig. 1A). Moreover, the methylation level of ITGB2 promoter in PBMCs did not correlate significantly with mRNA expression level of the gene in dSSc patients (P = 0.253, ρ = 0.237; Supplementary Fig. 1B). Although methylation level of ITGB2 gene promoter correlated negatively with mRNA expression of this gene in PBMCs of lSSc patients, the correlation was not significant (P = 0.47, ρ = − 0.153; Supplementary Fig. 1C).
ITGB2 mRNA expression level in PBMCs from SSc patients in both methylated and unmethylated CpG sites is shown in Table 3. In some CpG sites, including CpG7, 9, 10, 11, 14, 15, and 16 expression level of ITGB2 in methylated state was lower than in unmethylated state. Although, in other CpG sites, including CpG2, 5, 8, 12, and 13 ITGB2 mRNA level was overexpressed in methylated state compared with unmethylated state. It was also observed that the expression level of ITGB2 was significantly increased in hypermethylated status of CpG12 and 13 compared with unmethylated status (Table 3).
Correlation of ITGB2 expression and methylation with Rodnan score of SSc patients
The Rodnan score was used to determining severity of skin involvement in SSc patients. There was no significant correlation between ITGB2 expression level in PBMCs and Rodnan Score of SSc, dSSc, and lSSc patients, respectively (Supplementary Fig. 2A, C and E). Moreover, the methylation status of ITGB2 promoter in PBMCs did not demonstrate statistically significant correlation with Rodnan score in SSc, dSSc, and, lSSc patients, respectively (Supplementary Fig. 2B, D and F).
Discussion
Nowadays, there is no doubt that epigenetic dysregulations play a paramount role in the etiopathogenesis of autoimmune disorders. Epigenetic modulations alter gene expression without alteration in the gene sequence [32]. Epigenetic researches have concentrated preliminarily on the role of 5-methylcytosine (5-mC) in CpG islands in regulation of gene transcription in autoimmune disorders [33,34,35,36,37]. In this investigation, the possible implication of epigenetic dysmodulation in promoter of ITGIB2 gene was evaluated in PBMCs of SSc patients compared with that of healthy subjects. The results of the experiments supported somewhat the role of CpG DNA methylation dysregulation in pathogenesis of SSc disease.
SSc pathogenesis includes an intricate interaction between vascular disorder, inflammation and autoimmunity, and severe tissue fibrosis [38]. Overexpression of cellular adhesion molecules, for example integrin beta 2 and Selectin L, provides the direct cellular binding to vascular endothelial cells and fibroblasts, which facilitates the infiltration of inflammatory cells to the sites of involvement. This interplay is believed to lead to vascular endothelial cells and fibroblasts activation, which manifests as vascular dysfunction and tissue fibrosis in SSc patients, respectively [39]. Moreover, DNA hypomethylation of dermal fibroblasts from dSSc and lSSc patients is related to the disease pathogenesis [40].
In the present study, it was shown that ITGB2 mRNA level in patients with SSc was significantly increased compared to healthy individuals. This observation is in agreement with the study of Tan et al. [41], who reported several differently expressed genes in SSc PBC such as ITGB2 and SELL. In contrast, we did not find significant difference in SELL mRNA level between patients and controls. In dSSc patients, ITGB2 mRNA expression was upregulated significantly compared to healthy controls. But lSSc patients did not express ITGB2 mRNA differently compared to healthy subjects.
Considerably, CpG islands are placed upstream of the transcriptional start site in promoter of gene [42]. DNA methyltransferase (DNMT) enzymes transfer methyl (CH3) group to the C5 position on a cytosine molecule to form the 5-methylcytosine (5mC). In this reaction, S-adenosyl-methionine (SAM) plays as the methyl (CH3) group donor. Evidence shows that increased methylation level of CpGs in the promoter of genes leads to the suppression of gene transcription [18].
Furthermore, it has been found that DNA methyltransferase inhibitors are able to reverse epigenetic alterations and have protective effect against pulmonary fibrosis in mice [43]. Accordingly, epigenetic alterations appear to be reversible, suggesting they could emerge as new therapeutic options as anti-fibrotic therapy [44]. On the other side, given to important contribution of integrin β2 family to inflammation [23], it seems that targeting ITGB2 gene with elements modulating the methylation status might be promising in controlling the unwanted immune response and, therefore, disease treatment.
We observed that methylation status of ITGB2 promoter had no significant difference between the patients and healthy individuals. Our study demonstrated that three CpGs 12, 13 and 14 were significantly hypomethylated in patients group. The hypomethylation of these three CpG sites was associated significantly with decreased risk of SSc. If this finding approve by other investigations, the methylation evaluation of these three CpG sites in ITGB2 promoter can be beneficial measurement to predict the risk of SSc.
Methylation of DNA can repress gene expression in two ways: directly by preventing the binding of transcription factors and indirectly by employing the histone deacetylases (HDACs) via DNMT proteins and binding of proteins to methylated CpG [45]. Histone modifications, on the other hand, have been postulated to regulate gene expression [46]. It has also demonstrated that methylation and histone modifications are dependent events [47]. As well, DNMTs upregulation has been shown as a probable mechanism of gene hypermethylation [48]. In addition, Methyl-CpG-binding proteins (MECPs) were proposed as another mechanism of transcription inhibition through hypermethylation and following inhibition of transcription factors from binding to gene promoters [49]. Many studies have shown that CpGs methylation in promoter region of gene is only one of many possible regulatory mechanisms [50,51,52]. Other than these two, microRNAs might also potentially regulate the expression of ITGB2 gene.
In our study, no statistically significant correlation was seen between methylation status of ITGB2 promoter and mRNA expression of the gene in SSc patients compared to healthy group. Methylation events, as an important epigenetic mechanism, have wide effects during the evolution of immune system. By repression the immune system-related genes, epigenetic dysmodulation can deeply break self-tolerance and lead to autoimmunity disorder. This epigenetic dysmodulation has especially been observed in the genes involved in thymus formation, T cells development, and differentiation and effector function of these cells [53].However, no significant correlation was observed between ITGB2 mRNA expression level as well as the methylation status of ITGB2 promoter with Rodnan score of SSc patients in the present survey. Regarding to the overexpression of ITGB2 was not correlated with ITGB2 promoter hypomethylation in SSc patients, our study offers that upregulation of ITGB2 expression in PBMCs from SSc patients probably is due to another regulatory mechanism other than the alteration of methylation.
In consideration of all, this study was performed to evaluate the methylation status of ITGB2 gene and its possible effect on the mRNA expression of this gene in PBMCs from SSc patients. Here, we realized that methylation of ITGB2 promoter did not impress the mRNA expression regulation of ITGB2 in PBMCs from SSc patients. The mRNA expression of ITGB2 was upregulated in PBMCs from SSc patients, which may participate in contribution of immune cells to migrate to the involved tissues. However, this contribution might not be mediated through CpG DNA methylation. Alternately, the methylation and expression levels of ITGB2 in PBMCs from SSc patients did not correlate with disease severity with respect to Rodnan score. Epigenetic dysmodulation plays a role in SSc pathogenesis, requiring further evaluations to shed deep light on this puzzle.
References
Perosa F, Prete M, Di Lernia G, Ostuni C, Favoino E, Valentini G (2016) Anti-centromere protein A antibodies in systemic sclerosis: significance and origin. Autoimmun Rev 15:102–109. https://doi.org/10.1016/j.autrev.2015.10.001
Denton C (2015) Systemic sclerosis: from pathogenesis to targeted therapy. Clin Exp Rheumatol 33:S3-S7
Hutterer E, Asslaber D, Caldana C, Krenn PW, Zucchetto A, Gattei V, Greil R, Hartmann TN (2015) CD18 (ITGB2) expression in chronic lymphocytic leukaemia is regulated by DNA methylation-dependent and-independent mechanisms. Br J Haematol 169:286–289. https://doi.org/10.1111/bjh.13188
Shimada Y, Hasegawa M, Takehara K, Sato S (2001) Elevated serum l-selectin levels and decreased l-selectin expression on CD8+ lymphocytes in systemic sclerosis. Clin Exp Immunol 124:474–479. https://doi.org/10.1046/j.1365-2249.2001.01514.x
Miao C-g, Xiong Y-y, Yu H, Zhang X-l, Qin M-s, Song T-w, Du C-l (2015) Critical roles of microRNAs in the pathogenesis of systemic sclerosis: new advances, challenges and potential directions. Int Immunopharmacol 28:626–633. https://doi.org/10.1016/j.intimp.2015.07.042
Karimizadeh E, Gharibdoost F, Motamed N, Jafarinejad-Farsangi S, Jamshidi A, Mahmoudi M (2015) c-Abl silencing reduced the inhibitory effects of TGF-β1 on apoptosis in systemic sclerosis dermal fibroblasts. Mol Cell Biochem 405:169–176. https://doi.org/10.1007/s11010-015-2408-0
Karimizadeh E, Motamed N, Mahmoudi M, Jafarinejad-Farsangi S, Jamshidi A, Faridani H, Gharibdoost F (2015) Attenuation of fibrosis with selective inhibition of c-Abl by siRNA in systemic sclerosis dermal fibroblasts. Arch Dermatol Res 307:135–142. https://doi.org/10.1007/s00403-014-1532-0
Jafarinejad-Farsangi S, Farazmand A, Gharibdoost F, Karimizadeh E, Noorbakhsh F, Faridani H, Mahmoudi M, Jamshidi AR (2016) Inhibition of MicroRNA-21 induces apoptosis in dermal fibroblasts of patients with systemic sclerosis. Int J Dermatol 55:1259–1267. https://doi.org/10.1111/ijd.13308
Yousefi B, Mahmoudi M, Sarafnejad A, Karimizadeh E, Farhadi E, Jamshidi AR, Kavosi H, Aslani S, Gharibdoost F (2017) Downregulation of Aquaporin3 in systemic sclerosis dermal fibroblasts. Iran J Allergy Asthma Immunol 16:228
Almasi S, Aslani S, Poormoghim H, Jamshidi A, Poursani S, Mahmoudi M (2016) Gene expression profiling of toll-like receptor 4 and 5 in peripheral blood mononuclear cells in rheumatic disorders: ankylosing spondylitis and rheumatoid arthritis. Iran J Allergy Asthma Immunol 15:87
Jafarinejad-Farsangi S, Farazmand A, Mahmoudi M, Gharibdoost F, Karimizadeh E, Noorbakhsh F, Faridani H, Jamshidi AR (2015) MicroRNA-29a induces apoptosis via increasing the Bax: Bcl-2 ratio in dermal fibroblasts of patients with systemic sclerosis. Autoimmunity 48:369–378. https://doi.org/10.3109/08916934.2015.1030616
Ferri C, Sebastiani M, Monaco AL, Iudici M, Giuggioli D, Furini F, Manfredi A, Cuomo G, Spinella A, Colaci M (2014) Systemic sclerosis evolution of disease pathomorphosis and survival. Our experience on Italian patients’ population and review of the literature. Autoimmun Rev 13:1026–1034. https://doi.org/10.1016/j.autrev.2014.08.029
Li Y, Huang J, Guo M, Zuo X (2015) MicroRNAs regulating signaling pathways: potential biomarkers in systemic sclerosis. Genom Proteom Bioinform 13:234–241. https://doi.org/10.1016/j.gpb.2015.07.001
Mahmoudi M, Fallahian F, Sobhani S, Ghoroghi S, Jamshidi A, Poursani S, Dolati M, Hosseinpour Z, Gharibdoost F (2017) Analysis of killer cell immunoglobulin-like receptors (KIRs) and their HLA ligand genes polymorphisms in Iranian patients with systemic sclerosis. Clin Rheumatol 36:853–862. https://doi.org/10.1007/s10067-016-3526-0
Abtahi S, Farazmand A, Mahmoudi M, Ashraf-Ganjouei A, Javinani A, Nazari B, Kavosi H, Amirzargar A, Jamshidi A, Gharibdoost F (2015) IL-1A rs1800587, IL-1B rs1143634 and IL-1R1 rs2234650 polymorphisms in Iranian patients with systemic sclerosis. Int J Immunogenet 42:423–427. https://doi.org/10.1111/iji.12212
Luo Y, Wang Y, Shu Y, Lu Q, Xiao R (2015) Epigenetic mechanisms: an emerging role in pathogenesis and its therapeutic potential in systemic sclerosis. Int J Biochem Cell Biol 67:92–100. https://doi.org/10.1016/j.biocel.2015.05.023
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492. https://doi.org/10.1038/nrg3230
Quintero-Ronderos P, Montoya-Ortiz G (2012) Epigenetics and autoimmune diseases. Autoimmun Dis. https://doi.org/10.1155/2012/593720
Estécio MR, Issa J-PJ (2011) Dissecting DNA hypermethylation in cancer. FEBS Lett 585:2078–2086. https://doi.org/10.1016/j.febslet.2010.12.001
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324:930–935. https://doi.org/10.1126/science.1170116
Tan F, Zhou X, Mayes M, Gourh P, Guo X, Marcum C, Jin L, Arnett F (2006) Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology 45:694–702. https://doi.org/10.1093/rheumatology/kei244
Yassaee VR, Hashemi-Gorji F, Boosaliki S, Parvaneh N (2016) Mutation spectra of the ITGB2 gene in Iranian families with leukocyte adhesion deficiency type 1. Hum Immunol 77:191–195. https://doi.org/10.1016/j.humimm.2015.11.019
Nasiri Kalmarzi M (2015) Investigation of ITGB2 gene in 12 new cases of leukocyte adhesion deficiency-type I Revealed four novel mutations from Iran. Arch Iran Med 18:760
Stavarachi M, Apostol P, CIMPONERIU D, Toma M, Butoianu N, and GAVRILĂ L (2009) Possible association between l-selectin gene P213S polymorphism and respiratory complications of childhood spinal muscular atrophy patients. Rom Biotechnol Lett 14:4119–4122
Hoogen F, Khanna D, Fransen J, Johnson SR, Baron M, Tyndall A, Matucci-Cerinic M, Naden RP, Medsger TA, Carreira PE (2013) 2013 classification criteria for systemic sclerosis: an American College of Rheumatology/European League against Rheumatism collaborative initiative. Arthritis Rheum 65:2737–2747. https://doi.org/10.1002/art.38098
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat protoc 3:1101. https://doi.org/10.1038/nprot.2008.73
Sambrook J, Russell DW (2006) Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harb Protoc 2006:pdb. https://doi.org/10.1101/pdb.prot4455
Patterson K, Molloy L, Qu W, Clark S (2011) DNA methylation: bisulphite modification and analysis. J Vis Exp. https://doi.org/10.3791/3170
Aslani S, Mahmoudi M, Garshasbi M, Jamshidi AR, Karami J, Nicknam MH (2016) Evaluation of DNMT1 gene expression profile and methylation of its promoter region in patients with ankylosing spondylitis. Clin Rheumatol 35:2723–2731. https://doi.org/10.1007/s10067-016-3403-x
Karami J, Mahmoudi M, Amirzargar A, Gharshasbi M, Jamshidi A, Aslani S, Nicknam M (2017) Promoter hypermethylation of BCL11B gene correlates with downregulation of gene transcription in ankylosing spondylitis patients. Genes Immun 18:170–175. https://doi.org/10.1038/gene.2017.17
Rezaei R, Mahmoudi M, Gharibdoost F, Kavosi H, Dashti N, Imeni V, Jamshidi A, Aslani S, Mostafaei S, Vodjgani M (2017) IRF7 gene expression profile and methylation of its promoter region in patients with systemic sclerosis. Int J Rheum Dis 20:1551–1561. https://doi.org/10.1111/1756-185X.13175
Feinberg AP, Ohlsson R, Henikoff S (2006) The epigenetic progenitor origin of human cancer. Nat Rev Gen 7:21–33. https://doi.org/10.1038/nrg1748
Aslani S, Mahmoudi M, Karami J, Jamshidi AR, Malekshahi Z, Nicknam MH (2016) Epigenetic alterations underlying autoimmune diseases. Autoimmunity 49:69–83. https://doi.org/10.3109/08916934.2015.1134511
Aslani S, Jafari N, Javan MR, Karami J, Ahmadi M, Jafarnejad M (2017) Epigenetic modifications and therapy in multiple sclerosis. NeuroMol Med 19:11–23. https://doi.org/10.1007/s12017-016-8422-x
Mahmoudi M, Aslani S, Nicknam MH, Karami J, Jamshidi AR (2017) New insights toward the pathogenesis of ankylosing spondylitis; genetic variations and epigenetic modifications. Mod Rheumatol 27:198–209. https://doi.org/10.1080/14397595.2016.1206174
Ahmadi M, Gharibi T, Dolati S, Rostamzadeh D, Aslani S, Baradaran B, Younesi V, Yousefi M (2017) Epigenetic modifications and epigenetic based medication implementations of autoimmune diseases. Biomed Pharmacother 87:596–608. https://doi.org/10.1016/j.biopha.2016.12.072
Foma AM, Aslani S, Karami J, Jamshidi A, Mahmoudi M (2017) Epigenetic involvement in etiopathogenesis and implications in treatment of systemic lupus erythematous. Inflamm Res. https://doi.org/10.1007/s00011-017-1082-y
Varga J, Abraham D (2007) Systemic sclerosis: a prototypic multisystem fibrotic disorder. J Clin Invest 117:557–567. https://doi.org/10.1172/JCI31139
Altorok N, Almeshal N, Wang Y, Kahaleh B (2014) Epigenetics, the holy grail in the pathogenesis of systemic sclerosis. Rheumatology 54:1759–1770. https://doi.org/10.1093/rheumatology/keu155
Altorok N, Tsou P-S, Coit P, Khanna D, Sawalha AH (2014) Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis 74:1612–1620. https://doi.org/10.1136/annrheumdis-2014-205303
Tan F, Zhou X, Mayes M, Gourh P, Guo X, Marcum C, Jin L, Arnett F Jr. (2006) Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatology 45:694–702. https://doi.org/10.1093/rheumatology/kei244
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484. https://doi.org/10.1038/nrg3230
Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS (2008) Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol 39:610–618. https://doi.org/10.1165/rcmb.2007-0322OC
Wynn TA, Ramalingam TR (2012) Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med 18:1028–1040. https://doi.org/10.1038/nm.2807
Bird AP, Wolffe AP (1999) Methylation-induced repression—belts, braces, and chromatin. Cell 99:451–454. https://doi.org/10.1016/S0092-8674(00)81532-9
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45. https://doi.org/10.1038/47412
Stancheva I (2005) Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochem Cell Biol 83:385–395. https://doi.org/10.1139/o05-043
Mizuno S-i, Chijiwa T, Okamura T, Akashi K, Fukumaki Y, Niho Y, Sasaki H (2001) Expression of DNA methyltransferases DNMT1, 3A, and 3B in normal hematopoiesis and in acute and chronic myelogenous leukemia. Blood 97:1172–1179. https://doi.org/10.1182/blood.V97.5.1172
Fujita N, Takebayashi S-i, Okumura K, Kudo S, Chiba T, Saya H, Nakao M (1999) Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol 19:6415–6426. https://doi.org/10.1128/MCB.19.9.6415
Gregory RI, Randall TE, Johnson CA, Khosla S, Hatada I, O’Neill LP, Turner BM, Feil R (2001) DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpnand U2af1-rs1. Mol Cell Biol 21:5426–5436. https://doi.org/10.1128/MCB.21.16.5426-5436.2001
Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze S-H, Chenoweth J, Schwartz P, Pevzner PA, Glass C, Mandel G (2002) Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 298:1747–1752. https://doi.org/10.1126/science.1076469
Lande-Diner L, Zhang J, Ben-Porath I, Amariglio N, Keshet I, Hecht M, Azuara V, Fisher AG, Rechavi G, Cedar H (2007) Role of DNA methylation in stable gene repression. J Biol Chem 282:12194–12200. https://doi.org/10.1074/jbc.M607838200
Lawson BR, Eleftheriadis T, Tardif V, Gonzalez-Quintial R, Baccala R, Kono DH, Theofilopoulos AN (2012) Transmethylation in immunity and autoimmunity. Clin Immunol 143:8–21. https://doi.org/10.1016/j.clim.2011.10.007
Acknowledgements
Authors are deeply grateful of the individuals who contributed to the accomplishment of this study.
Funding
This study was funded by the Deputy of Research, Tehran University of Medical Sciences (Grant no. 95-01-30-31356).
Author information
Authors and Affiliations
Contributions
ND: email: naviddashtigoolahgoolah@yahoo.com. Performed the experiments and participated in manuscript drafting. MM: email: mahmoudim@tums.ac.ir. Developed the main idea and red the manuscript critically. FG: email: gharibdoost@sina.tums.ac.ir. Provided the financial support of the project. HK: email: h-kavosi@sina.tums.ac.ir. Examined the SSc patients. RR: email: ramin.rezaei25@gmail.com. Participated in performing the experiments. VI: email: vahide.amini@yahoo.com. Participated in questionnaire filling of the patients and conducting experiments. AJ: email: jamshidia@tums.ac.ir. Provided the financial support of the project. SA: email: s-aslani@razi.tums.ac.ir. Participated in manuscript writing. SM: email: mostafa.shayan@modares.ac.ir. Performed the statistical analysis. MV: email: vojganim@sina.tums.ac.ir. Provided the financial support of the project.
Corresponding authors
Ethics declarations
Ethical approval
All participants in the study were treated in agreement with the ethical standards of the Ethics Committee at Tehran University of Medical Sciences and with the revised Helsinki Declaration in 2000.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
296_2017_3915_MOESM1_ESM.tif
Correlation of mRNA expression level of ITGB2 with methylation percentage in (A) SSc patients, (B) dSSc patients, and (C) lSSc patients (rho; Spearman’s correlation coefficient) (TIF 51 KB)
296_2017_3915_MOESM2_ESM.tif
Scatter plots demonstrating the correlation of Rodnan scores with mRNA expression of ITGB2 (A) and methylation level of ITGB2 promoter in SSc patients. Rodnan score correlation with mRNA expression and promoter methylation of ITGB2 in dSSc patients is shown in plots C and D, respectively. Rodnan score correlation with mRNA expression and promoter methylation of ITGB2 in lSSc patients is shown in plots E and F, respectively (r; Pearson’s correlation coefficient) (TIF 186 KB)
Rights and permissions
About this article
Cite this article
Dashti, N., Mahmoudi, M., Gharibdoost, F. et al. Evaluation of ITGB2 (CD18) and SELL (CD62L) genes expression and methylation of ITGB2 promoter region in patients with systemic sclerosis. Rheumatol Int 38, 489–498 (2018). https://doi.org/10.1007/s00296-017-3915-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00296-017-3915-y