Abstract
Background
Genetic and epigenetic changes have strong role in the development of gastric cancer. The mutation of the MIR129-2 gene is one of the major causes in many cancers, especially gastric cancer. The aim of this study was to investigate the methylation changes of the MIR129-2 gene in tumor and normal tissue of patients with gastric cancer.
Method
In this study, 50 gastric cancer patients with Iranian Azari ethnic origin without any familial relations were included. Genomic DNAs was extracted from the tumoral and normal tissues. Then the promotor regions of the MIR129-2 gene were analyzed by methylation-specific PCR (MSP) to evaluate the presence or absence of methylated CpG sites.
Results
There was a statistically significant difference in methylation level of MIR129-2 gene between tumoral and normal tissues. It was observed that 84 out of 100 CpG cites were methylated in tumoral tissues in compression to 13 out of 100 CpG cites in normal tissues.
Conclusion
MIR129-2 gene was hypermethylated in tumoral tissues, suggesting that methylation is involved in the development of gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is an asymmetric division of the cells of the body, in which the cells have no longer normal mechanisms of cell growth and division. The exact reason of this phenomenon is uncertain, but genetic factors or cases that interfere with the activity of the cells are likely to play a role in this occurrence [1, 2]. Gastric cancer, is a malignancy caused by the proliferation and spread of gastrointestinal cells [3]. Gastric cancer is responsible for 7% of the total cancers and is ranked fifth among the most common cancers whereas it is responsible for 9% of cancer deaths worldwide [4]. Gastrointestinal cancer predisposing factors are categorized into two genetic and environmental factors; environmental factors include infection with Helicobacter pylori or Epstein–Barr virus, smoking, and diet with greasy foods and high amount of salt [2, 5]. After environmental factors, the second important factor involved in the development of cancer is genetics. About 10% of all people with gastric cancer present the disease as heredity, which suggests that, like other cancers, genetics has an outstanding role in the creation and development of this malignancy [6, 7]. The most important and famous gene involved in gastric cancer is the Cadherin 1 (CDH1) gene, also known as Hereditary Detune Gastric Cancer (HDGC), which encodes the cadherin E protein and is located on chromosome 6. Although mutation in CDH1 has not been the only genetic cause of gastric cancer, yet many mutations in several genes, such as P53, BRCA2, and epigenetic processes, have been responsible for causing this disease [8, 9].
MicroRNAs (miRNAs) are regulatory, small, and noncoding RNAs and are known as gene expression modulators that act at the posttranscriptional level [10], controlling the expression of most of the human protein-coding genes. Recently, it has been observed that different miRNAs are controlling cell signaling pathways in normal and tumor tissues [11, 12]. Some of the roles of miRNAs include degradation or translational inhibition of the target mRNAs by base-pairing with their 3′ untranslated region (3′UTR) [13]. Another role include regulation of thousands of genes, playing an essential role in cell development, proliferation, differentiation, chromatin structure, apoptosis, metabolism, and morphogenesis [14]. The role of epigenetics with processes such as methylation and alkylation and acetylation has been shown in the regulation of expression profiles of miRNAs in gastric cancer [15,16,17]. DNA methylation is a process by which methyl groups are added to the cytosine nucleotides of DNA molecules, resulting in suppression of gene expression. MIR129-2 is an important miRNA involved in different cancers [18]. Generally, MIR129 has been reported as a tumor suppressor gene in most cancers, and overall downregulated expression of this molecule has been reported in cancers and also, it should be considered that there are different mechanisms for reducing the expression of a miRNA in cancers, and the most important of which are deletion, point mutations, regulatory effects of other genes, and ultimately epigenetic changes, such as methylation, which seems to be the last option in the regulation of MIR129-2 gene and is playing the most important role [19,20,21]. The purpose of the present study was to investigate the methylation changes of the MIR129-2 gene in tumoral and normal tissue of patients with gastric cancer.
Material and Methods
Population and Sampling
In this study, 50 patients were diagnosed with gastric cancer. The disease was identified by a gastroenterologist and patients were referred to the surgery. Participants in this disease were all from the Azari population living in the northwest of Iran. Tumoral and healthy marginal tissues around the tumor were obtained from each study subject. Inclusion criteria of this study were gastric cancer disease in stage 2, having Azerbaijani ethnicity and habitation in the northwest of Iran. Exclusion criteria included existence of family relationship with other patients in the study, metastasis, unwillingness to continue cooperation in research, simultaneous diagnosis of another malignant disease other than gastric cancer, history of chemotherapy, and radiation therapy. Consent forms were obtained from each study subject, and the local ethical committee of Tabriz University of Medical Sciences approved the protocol of the study.
DNA Extraction and Bisulfite Treatment
DNA extraction was performed according to the protocol of phenol-chloroform approach. The quality and quantity of the DNAs were evaluated by nanodrop spectrophotometer. Before performing methylation-specific PCR (MSP), extracted DNA was treated with sodium bisulfite according to the protocol of ZYMO RESEARCH kit. During this process, all non-methyl cytosine is converted to uracil, but methyl-cytosine remains intact.
Methylation-Specific PCR
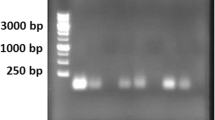
This technique requires use of two primers for the methylated DNA (M primer) and unmethylated DNA (U primer). In fact, for each sample, two PCR reactions were performed separately with each of the primers. The amplification with M primer represents methylation in CpG islands and amplification with U primer represents the lack of methylation in the examined region. Amplification with both primers represents partial methylation in the target area. The product of the M primer was 189 bp length and the length of the U primer was 188 bp. After performing the PCR reaction, the product of all samples (300 reactions) was electrophoresed on the agarose gel to ensure the reaction of the products. Examples of agarose gel electrophoresis related to the met primer with sequences of 189 nucleotides and unmet primer with sequences of 189 nucleotides with 100 bp ladder are shown in Fig. 1.
The Primer 3 software was used for designing the U and M primers (Table 1). The primers were then examined by NCBI Blast for specificity and proper melting temperature was determined for each primer with a temperature gradient.
PCR conditions were determined by using various variables including DNA and MgCl2 concentrations and temperature profiles for PCR reaction conditions is presented below. The amount of consumables in the PCR reaction for U primer is Master Mix Red 10 μl, DNA Template 2 μl, Primer 1 μl, Depc Water 7 μl, and for M primer is Master Mix Red 10 μl, DNA template 2 μl, Primer 1 μl, Depc Water 7 μl, and Mgcl2 (25 Mm).
Statistical Analysis
Statistical package for the Social Sciences (SPSS) software v.22 used to analyze data and plotting was performed using GraphPad Prism software v.6. The Pearson’s chi-square and Fisher’s exact test were used to check if there is a significant difference between the two groups in terms of methylation. The significant level was considered as P < 0.05/100 (Fig. 2).
Results
Totally, in tumor samples, the prevalence of methylation of the gene was 84 out of 100 CpG sites and in healthy samples, this number was 12. This indicates an increase in a significant difference between the two groups with respect to methylation of MIR129-2 gene. Regarding the pattern of the methylation of tumors and marginal samples, there were no significant differences in the prevalence of methylation with respect to sex, age, history of cancer, smoking, and alcohol consumption.
Discussion
In this study, 50 samples were examined for the methylation level of the CpG sites in the promoter of MIR129-2 gene. There was a significant difference in the distribution of methylated sites between tumoral and marginal tissues from gastric cancer subjects.
For the first time, Katada and his colleagues in a project published in 2009 revealed miR-129 as a distinct risk factor for gastric cancer [22]. Another study by Bandres et al. evaluated the methylation of MIR129-2 gene in colorectal cancer and it was introduced as a potential biomarker for colorectal cancer. One of the differences between this study and our research was the type of involved tissue and another was the method of investigation. A similar study evaluated the expression profile of miR-129 in colorectal cancer and introduced it as a biomarker for this disease [23].
A study by Liu and colleagues in hepatocellular cancer also confirmed the role of miR-129 in this cancer [24]. Although the study by Chen et al. on esophagus cancer with respect to the DNA methylation of MIR129-2 gene did not report a relationship between methylation level and development of this cancer [25]. A study by Kang and colleagues on esophageal cancer [26], as confirmed by the results of the Chen study, is in contradiction with our studies, which contradicts the role of different gene and molecular pathways in various cancers. The study by Chen is one of the few studies performed on methylation of gastric cancer [25], the results of which generally confirm the results of our review.
Although the P value and odds ratio varies with the results of our study, this difference can be due to a variety of reasons, including the difference in the population studied and the method used in the two studies. In all of the above studies, the sample population is different from our society. Generally, these studies are a statistical community with a higher frequency of our study and at least 100 patients. However, in this study, only 50 patients were studied and the difference in the volume of the studied population can be considered as one of the most important factors justifying the differences. The second leading cause of this difference can be the difference in the type of study, as in this study, we examined the level of methylation of the desired gene, while some previous studies only evaluated the expression. The third major reason for using methyl specific (MS)-PCR in this study is a qualitative method, while some previous studies have used quantitative methods to investigate the amount of methylation. The fourth factor that can somehow explain the differences is race. This raises the issue of racial differentiation as one of the factors contributing to the development of the disease. The fifth factor that can be the most important factor in making the difference in the data of this gene in our study compared with other similar studies is the limitation in collecting clinical data from patients. Generally, due to limitations in collecting samples and access to patient’s information, we were unfortunately unable to perform appropriate statistical surveys.
In conclusion, the methylation of MIR129-2 gene in patients with gastric cancer in the northwestern community of Iran has a significant difference in tumor samples with healthy samples and these changes can be considered as a risk factor for gastric cancer. The drugs that reverse the methylation of CpG sites can be further investigated for possible treatment of patients with gastric cancer.
References
Sim, F. and M. McKee, Issues in public health. 2011: McGraw-Hill Education (UK).
Shomali N, Mansoori B, Mohammadi A, Shirafkan N, Ghasabi M, Baradaran B. MiR-146a functions as a small silent player in gastric cancer. Biomed Pharmacother. 2017;96:238–45.
Ruddon RW. Cancer biology: Oxford University Press; 2007.
Park JY, von Karsa L, Herrero R. Prevention strategies for gastric cancer: a global perspective. Clinical endoscopy. 2014;47(6):478.
Tomasetti C, Li L, Vogelstein B. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017;355(6331):1330–4.
Devi, L.I., L. Ralte, and M.A. Ali, Serum biochemical profile of gastric cancer patients.
Ghavifekr Fakhr M, Rezaie Kahkhaie K, Shanehbandi D, Farshdousti Hagh M, Zarredar H, Safarzadeh E, et al. Scrophularia atropatana extract reverses tp53 gene promoter hypermethylation and decreases survivin antiapoptotic gene expression in breast cancer cells. Asian Pac J Cancer Prev. 2018;19(9):2599–605.
Pharoah PD, Guilford P, Caldas C. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology. 2001;121(6):1348–53.
Hansford S, Kaurah P, Li-Chang H, Woo M, Senz J, Pinheiro H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1(1):23–32.
Asadi M, Shanehbandi D, Mohammadpour H, Hashemzadeh S, Sepehri B. Expression level of miR-34a in tumor tissue from patients with esophageal squamous cell carcinoma. J Gastrointest Cancer. 2018.
Mayank, Jaitak V. Drug target strategies in breast cancer treatment: recent developments. Anti Cancer Agents Med Chem. 2014;14(10):1414–27.
Park HS, Jang MH, Kim EJ, Kim HJ, Lee HJ, Kim YJ, et al. High EGFR gene copy number predicts poor outcome in triple-negative breast cancer. Mod Pathol. 2014;27(9):1212–22.
Shirafkan N, Mansoori B, Mohammadi A, Shomali N, Ghasbi M, Baradaran B. MicroRNAs as novel biomarkers for colorectal cancer: new outlooks. Biomed Pharmacother. 2018;97:1319–30.
Bagaria SP, Ray PS, Sim MS, Ye X, Shamonki JM, Cui X, et al. Personalizing breast cancer staging by the inclusion of ER, PR, and HER2. JAMA Surg. 2014;149(2):125–9.
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101(9):2999–3004.
Farazi TA, et al. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20.
Shrestha S, Hsu SD, Huang WY, Huang HY, Chen WL, Weng SL, et al. A systematic review of microRNA expression profiling studies in human gastric cancer. Cancer Med. 2014;3(4):878–88.
Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17(8):991–1008.
Shen R, Pan S, Qi S, Lin X, Cheng S. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 in gastric cancer. Biochem Biophys Res Commun. 2010;394(4):1047–52.
Lee YY, Derakhshan MH. Environmental and lifestyle risk factors of gastric cancer. Arch Iran Med. 2013;16(6):358–65.
Tsai KW, Wu CW, Hu LY, Li SC, Liao YL, Lai CH, et al. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer. 2011;129(11):2600–10.
Katada T, Ishiguro H, Kuwabara Y, Kimura M, Mitui A, Mori Y, et al. microRNA expression profile in undifferentiated gastric cancer. Int J Oncol. 2009;34(2):537–42.
Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125(11):2737–43.
Liu Y, Hei Y, Shu Q, Dong J, Gao Y, Fu H, et al. VCP/p97, down-regulated by microRNA-129-5p, could regulate the progression of hepatocellular carcinoma. PLoS One. 2012;7(4):e35800.
Chen X, Zhang L, Zhang T, Hao M, Zhang X, Zhang J, et al. Methylation-mediated repression of microRNA 129-2 enhances oncogenic SOX4 expression in HCC. Liver Int. 2013;33(3):476–86.
Kang M, et al. miR-129-2 suppresses proliferation and migration of esophageal carcinoma cells through downregulation of SOX4 expression. Int J Mol Med. 2013;32(1):51–8.
Funding
This project was supported by a grant from research committee of Tabriz Medical University, Tabriz, Iran.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Informed consent was obtained from each patient and the ethical approaches were approved by the ethic committee of Tabriz University of Medical Sciences.
Conflict of Interest
The authors declare that they have no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Alizadeh, N., Asadi, M., Shanehbandi, D. et al. Evaluation of the Methylation of MIR129-2 Gene in Gastric Cancer. J Gastrointest Canc 51, 267–270 (2020). https://doi.org/10.1007/s12029-019-00239-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-019-00239-4