Abstract
Nitrogen removal by microorganisms has attracted increasing attention in wastewater treatment. In the present study, a heterotrophic nitrification bacterium was isolated from tannery wastewater and identified as Klebsiella sp. TN-10 based on phenotypic and phylogenetic characteristics. The optimal conditions for cell growth and nitrogen removal were investigated, and the results showed that the greatest ammonium removal rate and maximum biomass were achieved by using sodium pyruvate (7 g/L) as carbon source, C/N 12, pH 7, and temperature 30 °C. Under optimal conditions, the removal rate of ammonia nitrogen reached 96%. Besides, the growth characteristic and the ability of utilizing nitrate and nitrite were investigated. The results demonstrated that strain TN-10 exhibited excellent characteristics to remove both nitrate and nitrite, with the removal rate of 95.44% and 99.87%, respectively. In addition, the nitrite reductase (NiR) and nitrate reductase (NR) involved in denitrification were both active, with the activities of 0.0815 and 0.0283 U/mg proteins, respectively. Furthermore, the aggregation ability, auto-aggregation kinetics, and the relationship between zeta potentials and flocculating efficiency were determined. These results indicated that the strain Klebsiella sp. TN-10, with efficient heterotrophic nitrification-aerobic denitrification ability, has potential application in wastewater treatment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Along with industrialization development and urbanization, the concentration of nitrogen discharged from municipal sewage and industrial sewage has soared [1]. Furthermore, uncontrolled discharge of nitrogenous wastewater has caused serious deterioration of ecological health and brought about some environmental problems such as eutrophication in surface water and contamination in groundwater [2]. In addition, eutrophication problem, which caused algal bloom and the massive death of fishes, has become one of the main environmental problems in water bodies. Therefore, the removal of nitrogenous substances has received increasing attention owing to the damage they cause to receiving waters [3].
Conventional methods dealing with wastewater containing unacceptable concentrations of nitrogen include chemical, physical, biological, biochemical, and physicochemical processes [3]. In recent years, biological means have been widely adopted for nitrogen removal based on the convenience of operation, effective performance, and low cost. Even though nitrogen recovery by chemical precipitation and ammonium removal using ion exchange resin are attractive alternatives [4, 5], wastewater with elevated content of nitrogen is mainly treated through nitrification process by autotrophic nitrifiers and denitrification process by heterotrophic denitrifiers [6, 7]. However, autotrophic nitrification bacteria with slow specific growth rates and biomass yields are sensitive to environment conditions such as pH, heavy metals, temperature, and chemical toxicants, which make nitrification become the rate-limiting step during biological nitrogen removal [8, 9].
Compared with autotrophic nitrifying bacteria, heterotrophic bacteria have obvious advantages in organic substrate utilization and oxygen tolerance properties [10, 11]. In addition, during the process of simultaneous nitrification and denitrification (SND) in single reactor, there is no need to add additional organic substances like methanol or acetate [12]. Alkalinity caused by denitrification and acidification generated by nitrification could help maintain pH in the SND, which achieves less additional cost and easy operation [13].
Recently, many microorganisms with the capacity of heterotrophic nitrifying and aerobic denitrifying have been screened and applied to biological nitrogen removal systems and exhibited greater nitrogen removal performances [14], such as Rhodococcus sp. CPZ24 [15], Pseudomonas mendocina [16], and Acinetobacter junii YB [17]. A previous report demonstrated that the strain Pseudomonas stutzeri YZN-001 was isolated from swine manure effluent and that the bacterium could utilize 95% of ammonium in 18 h [3]. As a result of undetectable nitrate reductase and nitrite reductase activity, it was speculated that the possible nitrogen removal pathway was via hydroxylamine intermediate rather than nitrite. However, Chune et al. [18] isolated the nitrifying bacterium Pseudomonas stutzeri YG-24 from sediment of the eutrophic Taihu Lake, and the possible nitrogen removal pathway was via nitrite rather than hydroxylamine. Obviously, the biochemical processes of nitrification-denitrification are still unclear because of undetectable intermediate products and limited enzyme activities. It is necessary to investigate the possible process for different nitrifying bacteria in detail. Moreover, the auto-aggregation of microorganisms would influence biofilm formation and sludge-water separation, which are crucial for biological wastewater treatment [19]. To the best of our knowledge, only Klebsiella pneumoniae CF-S9 [1], Acinetobacter junii YB [17], and Enterobacter sp. strain FL [20] have been demonstrated to possess the ability to produce cell flocs, but change in zeta potentials and auto-aggregation in various nitrification-denitrification media have yet to be measured in combination.
Although many nitrification-denitrification microorganisms have been isolated, bacteria with the nitrification ability to process effectively ammonia nitrogen produced by leather factories are still few in number. Furthermore, it is necessary to elucidate in greater detail the nitrogen removal mechanisms. In the present study, a nitrification-denitrification bacterium was isolated from tannery wastewater and identified based on phenotypic and phylogenetic characteristics. The nitrification and denitrification performances of the strain were explored. Moreover, the activities of key enzymes proposed in the process of nitrogen removal were measured. In addition, zeta potentials, flocculating efficiency, and auto-aggregation kinetics were investigated as well. Results presented in the present study may provide an alternate microbial resource and a potential candidate to treat nitrogen-containing wastewater.
Materials and Methods
Medium
The selective medium (SM) used for isolation of heterotrophic nitrifying bacteria consisted of the following components (per liter): 5 g sucrose, 0.5 g (NH4)2SO4, 0.4 g FeSO4·7H2O, 0.5 g MgSO4, 1 g K2HPO4, 0.8 g EDTA, and pH 7.0. The basic medium (BM) used for studying the capacity of amino-nitrogen removal contained the following components (per liter): 7 g sodium pyruvate, 0.5 g (NH4)2SO4, 0.4 g FeSO4·7H2O, 0.5 g MgSO4, 1 g K2HPO4, 0.8 g EDTA, and pH 7.0. The denitrification medium 1 (DM1) used for nitrate reduction studies contained the substances as follows (per liter): 7 g sodium pyruvate, 0.5 g NaNO3, 0.4 g FeSO4·7H2O, 0.5 g MgSO4, 1 g K2HPO4, 0.8 g EDTA, and pH 7.0. The denitrification medium 2 (DM2) used for nitrite reduction studies contained the following components (per liter): 7 g sodium pyruvate, 0.5 g NaNO2, 0.4 g FeSO4·7H2O, 0.5 g MgSO4, 1 g K2HPO4, 0.8 g EDTA, and pH 7.0.
Isolation and Identification of Nitrification Bacteria
To isolate the nitrification bacteria, a sludge sample was collected from the sequencing batch reactor in Ruixing Leather Industry Co. Ltd., Zhejiang, China. Ten milliliters of sludge sample was transferred into 90 mL of sterile selective medium (SM) in 250-mL Erlenmeyer flasks. After cultivation at 30 °C and 150 rpm for 24 h, 1 mL of bacterial suspension was inoculated into 100 mL of fresh SM supplementing with the 1 g/L of (NH4)2SO4. After cultivation for 24 h, 1 mL of bacterial suspension was again transferred into 100 mL of fresh SM with the concentration of (NH4)2SO4 2.5 g/L. The fermentation broth was gradient dilutions, and the bacterial suspensions were spread onto agar SM plates. Then, the plates were incubated at 30 °C for 48 h. Isolated colonies were removed from the plates and individually tested for their ability to remove nitrogen. The strain with the greatest nitrogen removal ability was placed in 30% glycerol solution and stored at − 80 °C.
To identify the isolated strain, the genomic DNA of the isolate was extracted using a DNA extraction kit (Sangon, Shanghai, China). The 16S rDNA gene was amplified by PCR using the primers 5′-AGAGTTTGATCCTGGCTAG and 5′-TACGGTTACCTTGTTACGACTT [21] and sequenced by Sangon Biotech Co., Ltd. The sequence of 16S rDNA was compared with that of other bacteria in the Genbank using BLAST (https://www.ncbi.nlm.nih.gov/). A phylogenetic tree was constructed in MEGA7 using the neighbor-joining method.
Optimization of the Conditions for Removal of Ammonia-Nitrogen
To investigate the effects of different culture conditions on the heterotrophic nitrification ability of the isolate, single-factor experiments were conducted, including carbon source, C/N ratio, pH, and temperature. As for the carbon source experiments, different carbon sources (sodium succinate, sodium citrate, sodium pyruvate, and sucrose) were added, respectively, to the basic medium (BM) as the sole carbon source. The cells were cultivated at 30 °C for 24 h, and the nitrifying capacities were monitored to determine the optimal carbon source. In the C/N ratio experiments, different C/N ratios (2, 4, 8, 12, and 16) were obtained by adjusting the content of sodium pyruvate. To investigate the effects of pH and temperature on nitrification efficiency and cell growth, the initial pH was adjusted to either 4, 5, 6, 7, 8, or 9, and the temperature was set at either 10, 20, 30, or 37 °C in the BM. All of the above experiments were conducted in triplicate in 100 mL of sterile medium with 1% (v/v) inoculation size. After incubation for 24 h, samples were harvested to determine the OD600 and the concentration of ammonium nitrogen (NH4+-N).
Utilization of Nitrite and Nitrate
To determine the denitrification ability and growth characteristic of the isolate in DM1 and DM2, 1 mL cell suspension (1% inoculum) from the seed fermentation broth was inoculated into DM1 and DM2. The cells were aerobically incubated at 30 °C and 150 rpm. Samples were taken from Erlenmeyer flasks periodically to measure the OD600, NH4+-N, NO2−-N, NO3−-N, and TN. Each treatment was performed in triplicate.
Enzyme Assay
The strain TN-10 was harvested by centrifugation (10,000×g, 4 °C) for 5 min, washed three times with physiological saline (0.9% NaCl), and then resuspended in the same solution. The bacterial suspensions were sonicated on ice (20 kHz, 15 min), followed by centrifugation (10,000×g, 4 °C) for 5 min. The crude enzyme gathered from BM was used to determine the ammonia monooxygenase (AMO), nitrite reductase (NiR), and nitrate reductase (NR). Formation of nitrite from ammonium, production of nitrite from nitrate, and reduction of nitrite were taken as measures for AMO [22], NR, and NiR, respectively [23]. Protein concentration from the cell extract was determined by Bradford Protein Assay Kit (Biyuntian, Haimen, China). Enzyme-specific activity was defined as the amount of enzyme that transformed 1 μmol substrate per minute per milligram protein.
Analysis of the Visual Aggregation Degree of the Isolate
The isolate was cultivated in BM, DM1, and DM2 for 56 h, respectively, and the cells were harvested from the Erlenmeyer flask at 12-h intervals to assess the flocculability. Cells were collected via centrifugation (10,000×g, 4 °C) for 5 min, washed three times by physiological saline solution, and resuspended in 0.9% saline solution to adjust the OD600 to 1.5. Two milliliter purified cell suspensions from different culture media were transferred to glass tubes and vortexed for 1 min and remained stationary for 10 min. The visual aggregation scoring criteria of the strain TN-10 were performed as previously described [17] with minor modifications. The detailed scoring criteria were “−,” no flocculation; “+,” small amount of uniform floc, turbid suspension; “++,” visible floc, turbid suspension; “+++,” apparent floc, clear supernatant after settling; “++++,” large floc and clear supernatant, instantaneous settling.
Zeta Potentials and Auto-Aggregation Assays
The zeta potentials of cell suspensions from BM, DM1, and DM2 after 24-h cultivation were measured by Zetasizer Nano ZS90 (Malvern Instruments, UK). Cells were harvested by centrifugation (10,000×g, 4 °C) for 5 min, washed for three times by physiological saline (0.9% NaCl), and then resuspended in deinozed water. The OD546 of cell suspensions was adjusted to 0.1, and the zeta potentials were measured using the electrophoretic mobility of bacteria [24, 25]. For the auto-aggregation ability, the cells were resuspended in 0.9% sodium chloride and initial OD600 of cell suspension was adjusted to 0.6. Subsequently, 5 mL suspensions were transferred into a test tube. The test tube was left standing and the OD600 of upper suspension was measured after 1, 2, 3, 4, and 5 h. All tests were performed in triplicate. The auto-aggregation index (At) of microbial cells was calculated by Eq. (1) [26].
The auto-aggregation kinetics of the cells were described by Eq. (2) [27].
where k1 = h−1, Ae is the equalizing auto-aggregation (%), and At is the aggregation index (%) (t = 1, 2, 3, 4, and 5 h).
Analytical Methods
Optical density of the culture broth was measured at 600 nm using a spectrophotometer (UV-1901, Puxi, Beijing). The ammonium concentrations were determined using Nessler’s reagent spectrophotometry [28]. The nitrite concentrations were measured by N-(1-naphthalene)-diaminoethane ultraviolet spectroscopy at 540 nm [29]. The nitrate and total nitrogen (TN) concentrations were determined by phenol disulfonic acid method and phenol disulfonic acid ultraviolet spectroscopy, respectively [28].
Results and Discussion
Isolation and Identification of Nitrifying Bacterium
In the present study, a total of 200 isolates were grown on selective agar medium and preliminary screening and second screening were carried out in SM to determine ammonium content removal rates. The strain, named TN-10, demonstrated the greatest ammonium removal ability in the SM. The morphological characteristics of the strain were investigated, and it was found to form a circular colony that displayed creaminess, smooth surface, surface embossment, and ropiness (data not shown). In addition, the strain TN-10 was gram-positive with no spore formation. The 16S rDNA sequence of strain TN-10 was determined and deposited in the GenBank database under the accession number of MF461048. Homology comparison of the sequence showed that strain TN-10 had the greatest similarity (100%) to Klebsiella oxytoca BCGB2. A neighbor-joining phylogenetic tree was constructed in the MEGA 7 based on the partial 16S rDNA sequence (Fig. 1). According to this tree, the strain TN-10 was identified as Klebsiella sp. TN-10. Previous research also reported that Klebsiella species isolated from domestic sewage water had the capability of heterotrophic nitrification and denitrification [30].
Effect of Carbon Source on Nitrification
The carbon source usually has been considered to be an important factor influencing nitrogen removal because it could provide the energy source for heterotrophic bacteria [17]. In the present study, effects of different carbon sources including sodium pyruvate, sodium citrate, sodium succinate, and sucrose on nitrification ability and cell growth were investigated (Fig. 2a, b). As expected, significant differences were observed by using different carbon sources. Among the tested carbon sources, when sodium citrate and sodium pyruvate were added as carbon sources, cells exhibited relatively greater nitrification efficiencies, with the NH4+-N removal rate of 95.89% and 96.45%, respectively. In addition, analysis of the biomass revealed that OD600 reached 2.51 and 2.39, respectively, when sodium citrate and sodium pyruvate were supplemented as sole carbon source. Generally, both sodium citrate and sodium pyruvate could be used as carbon source for most heterotrophic nitrifying bacteria [31]. Thus, in the present study, sodium pyruvate was chosen as the carbon source in subsequent experiments in terms of nitrification performance.
Effect of carbon source (a, b) and C/N ratio (c, d) on the growth and ammonium removal of strain TN-10. Cells were cultivated with different carbon sources and C/N ratios. Biomass and nitrogen removal rate were determined after cultivation for 24 h. Different uppercase letters are used to indicate the significant differences of OD600 and ammonium degradation at p < 0.05 (n = 3)
Effect of C/N Ratio on Nitrification
The influence of different C/N ratios on cell growth and nitrogen removal in the BM was investigated in shaking cultures by changing the concentration of sodium pyruvate (Fig. 2c, d). As for the cell growth, the biomass increased with the C/N and the greatest biomass was obtained at C/N 14:1. As for ammonium removal, significant differences were observed along the C/N ratio gradient from 2:1 to 18:1. The ammonium removal ratio increased with C/N ratios, and then leveled off with terminal ammonium removal greater than 96%. Little NH4+-N degradation occurred when the C/N was either 2:1 or 6:1, and this may result from the insufficient carbon source and the paid exhaustion of pyruvate quickly. Li et al. [18] observed that the optimum C/N ratio was 8:1 for the Pseudomonas stutzeri YG-24, and similar results were also obtained with Agrobacterium sp. LAD9 [32] and Bacillus subtilis A1 [31] with C/N 8:1 and 6:1, respectively. Taking nitrification efficiency and OD600 into consideration, C/N 14 was chosen in the subsequent experiments.
Effects of pH and Temperature on Nitrification
Cell growth and ammonium removal were determined at different pH (Fig. 3a). Strain TN-10 presented efficient nitrification characteristics at initial pH ranging from 5 to 9 with NH4+-N removal percentages greater than 90%. Obviously, Klebsiella sp. TN-10 could grow well and had efficient nitrogen removal ability over a relatively wide pH range. The greatest removal rates of 96.4% and OD600 3.32 both were observed at initial pH 7 after cultivation for 24 h. Strain TN-10 barely grew at pH 4 as a result of the harm caused by strong acidic condition. Surprisingly, the strain TN-10 could grow well with OD600 2.78 and kept high nitrification efficiency when the pH was adjusted to 9. The pH in the system affects the nitrification process because of its effect on the NH3/NH4+-N and HNO2/NO2− equilibrium [33]. It has been reported that slightly alkaline environment is beneficial for nitrification bacterium because more free ammonium is contained in medium, and ammonia monooxygenase would take advantage of NH3 as the actual substrate rather than having a priority for NH4+ [13]. The evident adaptability of Klebsiella sp. TN-10 over a wide pH range is helpful for practical applications.
Effects of pH (a) and temperature (b) on ammonium removal and the growth of strain TN-10. Cells were cultivated at different pH and temperatures, and the biomass and nitrogen removal rate were determined after cultivation for 24 h. Different letters indicate significant differences at p < 0.05 (n = 3)
The effect of temperature on biomass and nitrogen removal characteristics of Klebsiella sp. TN-10 was investigated (Fig. 3b); cultivation at 10 °C and 37 °C resulted in little growth and nitrogen removal. The optimal temperature for Klebsiella sp. TN-10 was 30 °C, with the greatest ammonium removal rate 96.41% and the maximum OD600 3.32. In addition, it is worth noting that the strain could maintain strong nitrification efficiency at 20 °C. Similar results were also reported for Alcaligenes faecalis No. 4, which exhibited great ammonium removal efficiency at 20 °C and 30 °C [34]. Up to now, most reported heterotrophic nitrification bacteria were mesophiles, and they showed little or only began to grow after a long lag period at 10 °C and nearly were unable to grow at all at 4 °C. However, it was interesting that some microorganism possessed excellent cold resistance, for example Acinetobacter sp. HA2 whose ammonium removal rate was 3.03 mg/L/h at 10 °C [14]. Zhang et al. [3] reported that Pseudomonas stutzeri YZN-001 could grow at temperature between 4 °C and 45 °C; however, 2 weeks were required for this strain to progress to complete ammonium removal and to advance in growth from the lag stage to the decline stage.
Nitrifying Characteristics of Strain TN-10 Under the Optimal Conditions
Under the optimal conditions, the time courses of OD600, NH4+-N, NO2−-N, NO3−-N, and TN were investigated during nitrification by Klebsiella sp. TN-10 (Fig. 4). As for cell growth, the biomass of strain TN-10 increased rapidly from 4 to 24 h, corresponding to the OD600 increased from 0.01 to 3.04. Meanwhile, the biomass of the strain TN-10 maintained nearly constant for 24 h. Ammonium concentration (77.93 mg/L initial NH4+-N) decreased sharply during the initial 12 h, and it was completely removed within approximately 24 h with the nitrification rate of NH4+-N about 3.02 mg/L/h. It has been reported that Bacillus sp. LY could remove NH4+-N at a rate of 0.43 mg/L/h in 96 h with the initial NH4+-N 42.5 mg/L [10]. Similar results were reported by Su et al. who isolated a strain designated Pseudomonas sp. AS-1 from piggery wastewater, and the NH4+-N removal rate of the strain was 1.15 mg/L/h [35]. It is easy to come to the conclusion that the strain TN-10 had better nitrification ability than Bacillus sp. LY and Pseudomonas sp. AS-1. As for total nitrogen (TN), the contents decreased from 176.07 to 83.54 mg/L during the initial 16 h, and decreased more gradually after 16 h. The relatively great terminal TN concentration may be ascribed to the existence of unavailable nitrogen in EDTA. Overall, these results showed that there was a direct relationship between ammonium removal and bacterial growth, which is consistent with previous research [3, 36].
Aerobic-Nitrate and Nitrite Removal Efficiency
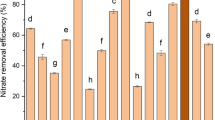
To clarify the denitrification characteristics of TN-10, nitrate and nitrite as the intermediates were used during the nitrification as the sole N-source in the medium under aerobic conditions, and denitrification efficiencies were determined (Fig. 5). In the sodium nitrate medium (DM1), cell growth reached a peak with the OD600 2.8 in 56 h, with biomass declining thereafter. As for the sole N-source NO3−-N, a significant decrease was observed at the first 32 h, which related to the rate of cell growth. The maximum removal rate of NO3−-N was 2.22 mg/L/h, which occurred between 8 and 24 h, and a similar removal rate was achieved by Klebsiella pneumoniae CF-S [30]. After cultivation for 40 h, the strain TN-10 could remove approximately 95.44% of NO3−-N, which was similar to Pseudomonas stutzeri SU2 with NO3−-N removal rate 98.01% in 44 h, but ammonium removal capability was undetermined for this strain [37]. Chen et al. [15] isolated a nitrification-denitrification strain Rhodococcus sp. CPZ24 which could completely transform NH4+-N in 16 h with initial NH4+-N 50 mg/L and remove 67% of NO3−-N after 36-h cultivation with the initial NO3−-N 50 mg/L. A detailed analysis showed that the initial NO2−-N concentration was 44.45 mg/L when NO3−-N was added, probably because partial nitrate was reduced to nitrite during the medium sterilization. The concentration of NO2−-N increased slightly in the beginning, which may result from the conversion of partial NO3−-N to NO2−-N. Subsequently, the concentration of NO2−-N had a sharp decline from 16 to 24 h, and was nearly undetectable after 32 h. The concentration of TN decreased with the decline of nitrite and nitrate and kept a relatively great termination concentration (85.16 mg/L) (Fig. 5a). This may be explained by unutilized nitrogen in the EDTA.
In the DM2 where NO2−-N was added as the sole N source, the strain TN-10 began to grow rapidly after 24-h cultivation, but it had a lesser growth rate in DM2 than that in DM1. After 56-h cultivation, the biomass of strain TN-10 reached maximum with OD600 2.43; then, the biomass began to decrease. As for the sole N source NO2−-N, the concentration increased continuously and accumulated from 87.67 to 98.34 mg/L in the first 16 h, which may be the conversion of NO3−-N to NO2−-N by strain TN-10. In addition, the maximum removal efficiency of nitrite, 10.83, occurred between 16 and 24 h, and the nitrite was nearly undetectable after 56 h. The removal rate of NO2−-N was 99.87% after cultivation for 56 h. Besides TN-10, many similar aerobic denitrification microorganisms have been investigated in the denitrification performance such as Pseudomonas stutzeri YZN-001 [3]. The strain P. stutzeri YZN-001 could remove all nitrates in 24 h with initial nitrate of 200 mg/L. A detailed analysis of the NO3−-N concentration showed that 15.43 mg/L NO3−-N was detected at the beginning of cultivation when NO2−-N was added, and this may be caused by oxidization during the medium sterilization. Nevertheless, NO3−-N was nearly undetectable after 8-h cultivation because of the utilization by strain TN-10. Besides, the concentration of TN declined from 191.33 to 91.33 mg/L in 16–48 h, and remained constant after 48 h, corresponding with the decline of NO2−-N and NO3−-N. From the above discussion, the results revealed that Klebsiella sp. TN-10 could not only utilize NO2−-N and NO3−-N but also exhibit excellent denitrifying performances. Different nitrifying bacteria may have distinct nitrification-denitrification characteristics (Table S1).
Enzyme Assay
In order to explore the possible pathway by which the nitrifying-denitrifying process occurs, the activities of ammonia monooxygenase (AMO), nitrite reductase (NiR), and nitrate reductase (NR) were measured (data not shown). AMO is the key enzyme in the metabolism of ammonium [38], but the effort to measure the AMO was unsuccessful. NiR and NR as two key enzymes involved in denitrification process were proposed to be responsible for converting nitrate to nitrite and nitrite to nitrogenous gas, respectively [23]. The specific enzyme activities of NiR and NR were 0.0815 and 0.0283 U/mg proteins, respectively, when the strain was cultivated in DM1 and DM2 for 24 h. The NR activity is much greater than that in Klebsiella pneumoniae CF-S9 [30], but less than Acinetobacter junii YB [17]. The NiR activity is greater than that for Klebsiella pneumoniae CF-S9 [30] and Acinetobacter junii YB [17]. The great activity of NiR may lead to little accumulation of nitrite during the ammonium removal process. Up to now, the nitrogen removal pathway under heterotrophic conditions is not fully understood, and further investigation remains required to elucidate it. Ren et al. [17] investigated the activities of three potential enzymes (hydroxylamine oxidase, nitrate reductase, nitrite reductase) involved in heterotrophic nitrification denitrification process, and the possible nitrogen removal pathway of Acinetobacter junii was determined (NH4+ → NH2OH → NO2− → NO3− → NO2− → N2O, N2). Padhi et al. [30] measured the specific activities of three enzymes (hydroxylamine oxidase, nitrate reductase, nitrite reductase) of Klebsiella pneumonia, which were found to be 0.011, 0.0074, and 0.018 U/mg proteins, respectively. The present work may provide additional evidence to improve the understanding of the heterotrophic nitrification and denitrification pathway.
The Visual Aggregation Degree and Auto-Aggregation Kinetic
The variations of aggregation ability in different media over 56 h were evaluated (Table 1). + was used to indicate the degree of aggregation. Obviously, the degree of aggregation increased during the cultivation, and aggregation appeared earlier for strain TN-10 in BM than in DM1 and DM2. Small aggregates (+) were readily observed in BM and DM1 after 12 h and 24 h, respectively. Easily visible aggregates (++) were observed in BM and DM1 and DM2 after 24, 36, and 48 h, respectively. It seems that the degrees of aggregation were relevant to cell growth in different media. The aggregation ability of strain TN-10 in the BM after 48 h was different from Acinetobacter junii YB [17]. It has been realized that bacterial aggregation has an essential effect for biofilm development [39]. High-efficiency flocculating ability and proper setting of flocs are important for activated sludge to generate good-quality effluent [39]. The flocculating efficiency and zeta potentials of strain TN-10 from nitrification-denitrification medium were determined (Fig. 6a). Obviously, the zeta potentials of cells grown in BM were more negative than that of cells grown in DM1 and DM2. Flocculating efficiencies occurred in the descending order of BM > DM1 > DM2, and the variation trend of zeta potentials was consistent with the flocculating efficiencies. The cells are required to overcome electrostatic repulsion to develop aggregation capability [40]. The auto-aggregation kinetics of TN-10 were evaluated in different media (Fig. 6b). The aggregation index of strain TN-10 in BM was greater than that in DM1 and DM2, which correlated with zeta potentials and flocculating efficiencies. It seems that the flocculation efficiencies of strain TN-10 were not strong and they may need more time to reach equilibrium. Some researchers have investigated the aggregation ability of EPS in activated sludge [25, 41, 42], but the studies of individual strains are still few in number. Strain TN-10 exhibited aggregation ability which could contribute to practical application in wastewater treatment. However, more attention to the cell binding ability as influenced by extracellular polymeric substances (EPS) is required. Thus, our research group will devote more energy to determination of the factors that influence cell binding ability in our future research.
Conclusion
In the present study, a bacterium Klebsiella sp. TN-10 with heterotrophic nitrifying and aerobic denitrifying ability was isolated and identified. The optimal conditions for ammonium removal and cell growth were investigated, and the maximum removal rate and biomass under optimized conditions were 93.93% and 3.06, respectively. Additionally, Klebsiella sp. TN-10 exhibited the ability to metabolize nitrate and nitrite efficiently. Moreover, the variation of aggregation ability when TN-10 was grown in different media was observed during the entire range of cultivation times. Nitrite reductase (NiR) and nitrate reductase (NR) were detected in the medium. Results of the present study indicate a promising future to apply this aeroboc heterotrophic nitrifying-denitrifying microorganism in wastewater.
References
Zhu, L., Ding, W., Feng, L. J., Kong, Y., Xu, J., & Xu, X. Y. (2012). Isolation of aerobic denitrifiers and characterization for their potential application in the bioremediation of oligotrophic ecosystem. Bioresource Technology, 108(4), 1–7.
Feng, C., Huang, L., Yu, H., Yi, X., & Wei, C. (2015). Simultaneous phenol removal, nitrification and denitrification using microbial fuel cell technology. Water Research, 76(2), 160–170.
Zhang, J., Wu, P., Hao, B., & Yu, Z. (2011). Heterotrophic nitrification and aerobic denitrification by the bacterium Pseudomonas stutzeri YZN-001. Bioresource Technology, 102(21), 9866–9869.
Sica, M., Duta, A., Teodosiu, C., & Draghici, C. (2014). Thermodynamic and kinetic study on ammonium removal from a synthetic water solution using ion exchange resin. Clean Technologies & Environmental Policy, 16(2), 351–359.
Khardenavis, A. A., Kapley, A., & Purohit, H. J. (2007). Simultaneous nitrification and denitrification by diverse Diaphorobacter sp. Applied Microbiology & Biotechnology, 77(2), 403–409.
Ji, Z., & Chen, Y. (2010). Using sludge fermentation liquid to improve wastewater short-cut nitrification-denitrification and denitrifying phosphorus removal via nitrite. Environmental Science & Technology, 44(23), 8957–8963.
Leta, S., Assefa, F., Gumaelius, L., & Dalhammar, G. (2004). Biological nitrogen and organic matter removal from tannery wastewater in pilot plant operations in Ethiopia. Applied Microbiology & Biotechnology, 66(3), 333–339.
Kim, Y. M., Park, D., Lee, D. S., & Park, J. M. (2008). Inhibitory effects of toxic compounds on nitrification process for cokes wastewater treatment. Journal of Hazardous Materials, 152(3), 915–921.
Kim, Y. M., Park, H., & Chandran, K. (2016). Nitrification inhibition by hexavalent chromium Cr (VI)—microbial ecology, gene expression and off-gas emissions. Water Research, 92, 254–261.
Zhao, B., He, Y. L., & Zhang, X. F. (2010). Nitrogen removal capability through simultaneous heterotrophic nitrification and aerobic denitrification by Bacillus sp. LY. Environmental Technology, 31(4), 409–416.
Third, K. A., Gibbs, B., Newland, M., & Cordruwisch, R. (2005). Long-term aeration management for improved N-removal via SND in a sequencing batch reactor. Water Research, 39(15), 3523–3530.
Liu, L. H., & Koenig, A. (2002). Use of limestone for pH control in autotrophic denitrification: batch experiments. Process Biochemistry, 37(8), 885–893.
Zhang, Q. L., Liu, Y., Ai, G. M., Miao, L. L., Zheng, H. Y., & Liu, Z. P. (2012). The characteristics of a novel heterotrophic nitrification-aerobic denitrification bacterium, Bacillus methylotrophicus strain L7. Bioresource Technology, 108(3), 35–44.
Yao, S., Ni, J., Ma, T., & Li, C. (2013). Heterotrophic nitrification and aerobic denitrification at low temperature by a newly isolated bacterium, Acinetobacter sp HA2. Bioresource Technology, 139(13), 80–86.
Chen, P., Li, J., Li, Q. X., Wang, Y., Li, S., Ren, T., & Wang, L. (2012). Simultaneous heterotrophic nitrification and aerobic denitrification by bacterium Rhodococcus sp. CPZ24. Bioresource Technology, 116(13), 266–270.
Zhu, L., Ding, W., Feng, L. J., Dai, X., & Xu, X. Y. (2012). Characteristics of an aerobic denitrifier that utilizes ammonium and nitrate simultaneously under the oligotrophic niche. Environmental Science & Pollution Research, 19(8), 3185–3191.
Ren, Y. X., Yang, L., & Liang, X. (2014). The characteristics of a novel heterotrophic nitrifying and aerobic denitrifying bacterium, Acinetobacter junii YB. Bioresource Technology, 171, 1–9.
Li, C., Yang, J., Wang, X., Wang, E., Li, B., He, R., & Yuan, H. (2015). Removal of nitrogen by heterotrophic nitrification-aerobic denitrification of a phosphate accumulating bacterium Pseudomonas stutzeri YG-24. Bioresource Technology, 182, 18–25.
Badireddy, A. R., Chellam, S., Gassman, P. L., Engelhard, M. H., Lea, A. S., & Rosso, K. M. (2010). Role of extracellular polymeric substances in bioflocculation of activated sludge microorganisms under glucose-controlled conditions. Water Research, 44(15), 4505–4516.
Wang, X., An, Q., Zhao, B., Guo, J. S., Huang, Y. S., & Tian, M. (2018). Auto-aggregation properties of a novel aerobic denitrifier Enterobacter sp. strain FL. Applied Microbiology & Biotechnology, 102(4), 2019–2030.
Heuer, H., Krsek, M., Baker, P., Smalla, K., & Wellington, E. M. (1997). Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Applied & Environmental Microbiology, 63(8), 3233–3241.
Juliette, L. Y., Hyman, M. R., & Arp, D. J. (1993). Inhibition of ammonia oxidation in Nitrosomonas europaea by sulfur compounds: thioethers are oxidized to sulfoxides by ammonia monooxygenase. Applied & Environmental Microbiology, 59(11), 3718–3727.
Richardson, D. J., & Watmough, N. J. (1999). Inorganic nitrogen metabolism in bacteria. Current Opinion in Chemical Biology, 3(2), 207–219.
Pembrey, R. S., Marshall, K. C., & Schneider, R. P. (1999). Cell surface analysis techniques: what do cell preparation protocols do to cell surface properties? Applied & Environmental Microbiology, 65(7), 2877–2894.
Zhang, P., Fang, F., Chen, Y. P., Shen, Y., Zhang, W., Yang, J. X., Li, C., Guo, J. S., Liu, S. Y., & Huang, Y. (2014). Composition of EPS fractions from suspended sludge and biofilm and their roles in microbial cell aggregation. Chemosphere, 117, 59–65.
Sheng, G. P., & Yu, H. Q. (2006). Chemical-equilibrium-based model for describing the strength of sludge: taking hydrogen-producing sludge as an example. Environmental Science & Technology, 40(4), 1280–1285.
Lin, L., Rosenberg, M., Taylor, K. G., & Doyle, R. J. (1995). Kinetic analysis of ammonium sulfate dependent aggregation of bacteria. Colloids & Surfaces B Biointerfaces, 5(3–4), 127–134.
APHA. (1998). Standard methods for the examination of water and waste water (20th ed.). Washington, DC: American Public Health Association.
Clesceri, L., Greenberg, A. S., & Eaton, D. L. (2005) Standard methods for the examinations of water and wastewater, ed., American Journal of Public Health & the Nations Health, Washington, DC, USA.
Padhi, S. K., Tripathy, S., Sen, R., Mahapatra, A. S., Mohanty, S., & Maiti, N. K. (2013). Characterisation of heterotrophic nitrifying and aerobic denitrifying Klebsiella pneumoniae CF-S9 strain for bioremediation of wastewater. International Biodeterioration & Biodegradation, 78(3), 67–73.
Yang, X.-P., Wang, S.-M., Zhang, D.-W., & Zhou, L.-X. (2011). Isolation and nitrogen removal characteristics of an aerobic heterotrophic nitrifying–denitrifying bacterium, Bacillus subtilis A1. Bioresource Technology, 102(2), 854–862.
Huang, X., Li, W., Zhang, D., & Wen, Q. (2013). Ammonium removal by a novel oligotrophic Acinetobacter sp. Y16 capable of heterotrophic nitrification–aerobic denitrification at low temperature. Bioresource Technology, 146(10), 44–50.
Mosquera-Corral, A., González, F., Campos, J. L., & Méndez, R. (2005). Partial nitrification in a SHARON reactor in the presence of salts and organic carbon compounds. Process Biochemistry, 40(9), 3109–3118.
Joo, H. S., Hirai, M., & Shoda, M. (2005). Characteristics of ammonium removal by heterotrophic nitrification-aerobic denitrification by Alcaligenes faecalis no. 4. Journal of Bioscience & Bioengineering, 100(2), 184–191.
Su, J. J., Yeh, K. S., & Tseng, P. W. (2006). A strain of Pseudomonas sp. isolated from piggery wastewater treatment systems with heterotrophic nitrification capability in Taiwan. Current Microbiology, 53(1), 77–81.
Chen, Q., & Ni, J. (2012). Ammonium removal by Agrobacterium sp. LAD9 capable of heterotrophic nitrification-aerobic denitrification. Journal of Bioscience & Bioengineering, 113(5), 619–623.
Su, J. J., Liu, B. Y., & Liu, C. Y. (2001). Comparison of aerobic denitrification under high oxygen atmosphere by Thiosphaera pantotropha ATCC 35512 and Pseudomonas stutzeri SU2 newly isolated from the activated sludge of a piggery wastewater treatment system. Journal of Applied Microbiology, 90(3), 457–462.
Bédard, C., & Knowles, R. (1989). Physiology, biochemistry, and specific inhibitors of CH4, NH4 +, and CO oxidation by methanotrophs and nitrifiers. Microbiological Reviews, 53(1), 68–84.
Malik, A., Sakamoto, M., Hanazaki, S., Osawa, M., Suzuki, T., Tochigi, M., & Kakii, K. (2003). Coaggregation among nonflocculating bacteria isolated from activated sludge. Applied & Environmental Microbiology, 69(10), 6056–6063.
Renner, L. D., & Weibel, D. B. (2011). Physicochemical regulation of biofilm formation. MRS Bulletin, 36(5), 347–355.
Sheng, G. P., Yu, H. Q., & Li, X. Y. (2010). Extracellular polymeric substances (EPS) of microbial aggregates in biological wastewater treatment systems: a review. Biotechnology Advances, 28(6), 882–894.
Gao, J. F., Zhang, Q., Wang, J. H., Wu, X. L., Wang, S. Y., & Peng, Y. Z. (2011). Contributions of functional groups and extracellular polymeric substances on the biosorption of dyes by aerobic granules. Bioresource Technology, 102(2), 805–813.
Funding
This study was funded by the Special Foundation for State Key Research and Development Program of China (2017YFB0308500), the National Natural Science Foundation of China (31671849), and Talent Training platform construction of Sichuan University (SCUKG017).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOCX 25 kb)
Rights and permissions
About this article
Cite this article
Li, D., Liang, X., Jin, Y. et al. Isolation and Nitrogen Removal Characteristics of an Aerobic Heterotrophic Nitrifying-Denitrifying Bacterium, Klebsiella sp. TN-10. Appl Biochem Biotechnol 188, 540–554 (2019). https://doi.org/10.1007/s12010-018-02932-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-018-02932-9