Abstract
The thermostable phytase gene was isolated from Bacillus subtilis ARRMK33 (BsPhyARRMK33). The gene has an ORF of 1152 bp and that encodes a protein of 383 amino acids. Sequence analysis showed high homology with Bacillus sp. phytase proteins, but no similarity was found with other phytases. SDS-PAGE analysis exhibited a predicted molecular mass of 42 kDa. Homology modeling of BsPhyARRMK33 protein based on Bacillus amyloliquefaciens crystal structure disclosed its β-propeller structure. BsPhyARRMK33 recombinant plasmid in pET-28a(+) was expressed in Rosetta gami B DE3 cells and the maximum phytase activity 15.3 U mg−1 obtained. The enzyme exhibits high thermostability at various temperatures and broad pH ranges. The recombinant protein retained 74 % of its original activity after incubation at 95 °C for 10 min. In the presence of Ca2+, the recombinant phytase activity was maximal where as it was inhibited by EDTA. The optimal pH and temperature for the recombinant phytase activity is achieved at 7.0 and 55 °C, respectively. Thermostable nature and wide range of pH are promising features of recombinant BsPhyARRMK33 protein that may be employed as an efficient alternative to commercially known phytases and thereby alleviate environmental eutrophication.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phytic acid is the major storage form of phosphorus in plant tissues, especially grains or seeds and bran. Plant seeds have 80 % of the phosphorus in the form of phytic acid. The chemical description for phytic acid is myo-inositol (1, 2, 3, 4, 5, and 6) hexakisdihydrogen phosphate [1]. This chemical attribute makes phytic acid a strong chelator of several crucial dietary minerals such as calcium, magnesium, iron, and zinc [2], rendering them inaccessible for absorption in intestines of monogastric animals, such as human, poultry, fish, and swine [3]. Phytate has been considered the precipitation of metal-binding enzymes and also in the reduction of the digestibility of proteins, starch, and lipids [4]. Monogastric animals have limited ability to digest the phytates in their diet [5]. Hence, unabsorbed phytate, which has abundant phosphorus content, gets excreted through feces, leading to environmental eutrophication [6].
Addition of inorganic phosphorus to animal feed as a crucial dietary requirement not only causes environmental pollution but also expensive and non-sustainable [7, 8]. Introduction of phytases into animal feed is anticipated to resolve phosphorus pollution [9]. For the past few years, phytases have been studied extensively due to the potential in using these enzymes for diluting phosphorus supplements in animal feed and food items. Phytases not only fulfil the high phosphorus requirements but also help in efficient assimilation of other required dietary elements [10].
Bioengineering of microbial phytase genes for heterologous expression in edible parts of food crops, such as sweet potato, rice, and microalgae, improves bioavailability of mineral nutrients [9, 11, 12]. Scope for nutritional enrichment of animal feed and reduction in phosphorus pollution opened up vivid aspects for research on phytase. Phytases are ubiquitously present in kingdom of life such as microorganisms, plants, and animals. First-generation fungal phytases have been commonly employed for use on a commercial scale in feed industry, due to their activity in acidic conditions. However, researchers are now focusing on the use of bacterial phytases, as Bacillus phytase exhibits that strict substrate (phytate) specific activity, which would not disturb other metabolic pathways. These phytases have been studied intensively due to their unique features and feasible mass production for market and applicability in animal feed [13–16].
Fungal phytase expressions in bacterial systems cause glycosylation, which may alter the enzyme activity [17]. Hence, using Bacillus phytase in E. coli would not alter the protein function, few studies targeted on economically competitive expression and secretion systems for phytase were accomplished [18]. The present study deals with cloning and molecular, biochemical characterization of phytase derived from Bacillus subtilis sp. (BsPhyARRMK33). We performed the molecular characterization of BsPhyARRMK33 and its recombinant protein. Homology modeling was carried out to reveal its structural role. The recombinant phytase may serve as an economical ingredient of feed given to monogastric animal.
Materials and Methods
Isolation and Cloning of BsPhyARRMK33 Gene
Genomic DNA was purified from the Bacillus sp., isolated at the School of Life Sciences, Hyderabad, India, according to the method of Ausubel [19]. The BsPhyARRMK33 was amplified with gene-specific primers, i.e., forward 5′-ATACTACATATGAATCATTCAA-3′ and reverse 5′-TAATGCGGC CGCTTATTTTCC-3′ flanked by restriction sites NdeI and NotI, respectively. The primers were designed based on conserved sequences and used for amplification of the phytase coding sequence. PCR amplifications of BsPhyARRMK33 was carried out using 50 ng of genomic DNA as template with 1.5 units of Taq DNA polymerase, 200 mol l−1 dNTP and 1.5 mmol l−1 MgCl2 in Biorad Thermal Cycler. The conditions of PCR were as follows: 94 °C for 45 s, 56 °C for 45 s, and 72 °C for 70 s for 35 cycles. The PCR-purified product was cloned into the TOPO TA2.1 vector linearized with NdeI. The gene was subcloned into the bacterial expression vector pET-28a(+) downstream to the T7 promoter within NdeI and NotI sites. The recombinant plasmid harboring the full-length B. subtilis phytase was designated as BsPhyARRMK33 (Fig. 1) and subjected to sequence analysis. The deduced amino acid sequence was compared with the NCBI protein database using BLAST search. The pET recombinant plasmid construct was transformed into the Rosetta gami® B DE3 cells for protein expression analysis.
Homology Modeling of BsPhyARRMK33
BsPhyARRMK33 molecular model was generated using the homology modeling server SWISS-MODEL [20] utilizing the Bacillus amyloliquefaciencs 13213 protein crystal structure as a template (PDB ID 1POO). Pymol and discovery studio (http://pymol.sourceforge.net/) program was employed to depict the Bsphy ARRMK33 molecular model.
Phytase Expression and Purification of Recombinant BsPhyARRMK333 Phytase
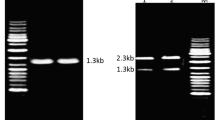
The recombinant Rosetta gami B DE3 cells containing pET-28a(+)-BsPhyARRMK33 were grown on Luria-Bertani (LB) agar medium containing kanamycin (50 μg ml−1) at 30 °C. A single colony was picked for culture in 10-ml LB broth containing kanamycin (50 μg ml−1) and grown for 3 h at 37 °C and 220 rpm. The secondary culture was set up for 18 °C overnight from the primary culture in 1-l broth. Once the culture reached an optical density of 0.6 (log phase), the cells were induced with 1 mmol l−1 isopropyl β-d-1-thiogalactopyranoside (IPTG). Induced culture was harvested after 4 h, and cells were lysed by sonication. Clarified bacterial lysates were fractionated on native as well as SDS polyacrylamide gel. Further, SDS polyacrylamide gel was stained with Coommassie Blue to visualize protein bands (Fig. 2). Recombinant phytase protein was purified to near homogeneity on a Ni–NTA column chromatography following the manufacturer’s instructions (Qiagen, Germany).
SDS-PAGE (12 %) analysis BsPhyARRMK33 protein expression in E. coli BL21 (Commassie blue staining). Lane M, protein marker (kDa); L1, whole lysate of uniduced BL21 E. coli cells containing the plasmid pET-28a(+)-BsPhyARRMK33; L2, whole cell lysate of the same cells obtained after 3-h induction with 1 mM IPTG; L3, purified recombinant BsPhyARRMK33 protein
Biochemical Characterization of BsphyARRMK33 Recombinant Protein
Phytase Assay
Protein concentration was measured by Bradford method using bovine serum albumin as a standard [21]. The recorded values were an average of measurements of triplicate samples. Phytase activity was estimated by incubating 750 μl of recombinant purified phytase enzyme with 600 μl of 2.0 mmol l−1 sodium phytate in 100 mmol−l Tris–Cl buffer (pH 7.0), supplemented with and without 2.0 mmol l−1 CaCl2·2H2O. The reaction was carried out at 37 °C for 30 min, and then, 750 μl of 5 % trichloroacetic acid was added to stop the reaction. The free phosphate was measured at 700 nm by following the production of phosphomolybdate with 1.5 ml of color reagent (freshly prepared by mixing four volumes of 1.5 % ammonium molybdate solution in 5.5 % sulfuric acid and one volume of 2.7 % ferrous sulfate solution). The enzyme kinetics of recombinant protein was determined using various sodium phytate substrate concentrations (0.25, 0.5, 1.0, 2.0, 3.0, and 4.0 mmol l−1).
One unit of phytase activity was defined as the quantity of enzyme required to liberate 1 μmol of phosphate per minute under the assay conditions [22]. Three independent experiments were performed for enzyme activity to check the various parameters influencing recombinant enzyme activity.
pH Optima
To assess the pH optima of phytase, enzyme-specific buffers were utilized in enzyme activity assays to examine the pH optima: 100 mmol l−1 Tris–Cl (pH 7.0, 8.0, 9.0), 100 mmol l−1 sodium acetate (pH 5.0, 6.0), and 100 mmol l−1 glycine (pH 3.0, 4.0).
Thermostability
To check the thermal stability of recombinant purified phytase, enzyme were examined in various temperatures ranging from 75 to 95 °C for 10 min in 100 mmol l−1 Tris–HCl (pH 7.0) along with 5 mmol l−1 Ca2+, followed by cooling to 28 °C for 1 h before enzyme assay was carried out at 37 °C.
Proteolytic Resistance
In order to determine the susceptibility of the BsPhyARRMK33 phytase to digestive proteases, purified enzyme was pre incubated with 0.1 mg ml−1 trypsin and pepsin at 37 °C and activity was measured after 1 h.
Results
Isolation and Sequence Analysis of Full-Length BsPhyARRMK33
The phytase gene (BsPhyARRMK33) was isolated from B. subtilis with an ORF of 1152 bp which encodes for protein 383 amino acids possessing a putative signal peptide of 27 amino acids with a calculated molecular mass of 42 kDa. In silico analysis of the deduced amino acid sequence revealed only four amino acid differences between BsPhyARRMK33 from existing source B. amyloliquefacins, i.e., instead of alanine, valine (81), histidine, proline (94), asparagine, aspartic acid (148), leucine, and phenylalanine (331). The nucleotide sequence analysis BsPhyARRMK33 revealed 99 % identity with phytase gene (phy) of B. subtilis, IDCC1102 strain, and B. amyloliquefaciens strain CC178. At the amino acid level, it showed 99 % identity with B. subtilis of, AAK97047.1, ABC75080.1, WP_007407926.1, AEN75156.1, 92 % with Bacillus amyloliquefaciens (AAL59320.1). The complete BsPhyARRMK33 cDNA sequence was deposited in GenBank with accession number EF092835.
Sequence Homology and Molecular Modeling of BsPhyARRMK33
Homology model for BsPhyARRMK33 protein was built on the basis of existing structural information from a closely related heterologous source. The crystal structure of B. amyloliquefacins protein (PDB ID 2POO) [23] was chosen as a template for BsPhyARRMK33 model building using the homology modeling server SWISS-MODEL [20]. The BsPhyARRMK33 from B. subtilis and B. amyloliquefacins shared 98 % similarity at their primary amino acid sequence level with only four amino acids mismatch. Structure of the BsPhyARRMK33 monomer revealed six Ca2+ binding sites (Fig. 3a) and six ß-stranded sheets in the active site architecture (Fig. 3b)
The predicted 3D structure of BsPhyARRMK33. a BsPhyARRMK33 protein model showing six beta-propeller sheets; each blade is shown in different colors. b Six Ca2+ binding sites within the active site showing Ca1, Ca2, and Ca3 ions responsible for high thermal stability, whereas Ca4, Ca5, are Ca6 are for catalytic activity
Expression and Purification of Recombinant BsPhyARRMK33 Protein
Rosetta gami B DE3 cells containing recombinant phytase plasmid (pET-28a(+)-ARRMK33) were grown at 37 °C and induced expression with IPTG for 3 h. The major induced portion of protein was observed in inclusion bodies. A temperature-course study was executed to analyze the protein releasing into cytosol at different temperatures (18, 25, 30, and 37 °C). Soluble fraction of recombinant protein was noticed at 18 °C. The recombinant protein was purified to near homogeneity from clarified E. coli lysate by passing through Ni–NTA agarose beads as the N-termini of the recombinant proteins possessed a 6× His tag. Molecular weight of the BsPhyARRMK33 protein was estimated by SDS-PAGE, as approximately 42 kDa (Fig. 2). The recombinant purified phytase protein was used for all the enzyme assays, which showed a specific activity of 13.5 U mg−1. The recombinant strain activity was found to be 43 U ml−1 in shake flask conditions at 220 rpm. The predicted pI and molecular weight of the enzyme was estimated by Expassy Compute PI/Mw tool as 4.97 and 41.8 kDa, respectively.
Substrate Specificity and Enzyme Kinetics
Observations with purified phytase protein revealed a maximum enzyme activity of (13.5 U mg−1) at pH 7. Recombinant phytase enzyme activity was tested using various concentrations of sodium phytate as the substrate. The K m and V max values were 0.95 mM and 15.3 μmol l−1 (Fig. 4). Enzyme was more active in the presence of Ca2+ and showed hydrolysis of the phytate, where as it failed to hydrolyse the p-nitrophenylphosphate. It has not been detected activity with other substrates like (2 mmol l−1) ATP, ADP, and glucose-6-phosphate. Addition of (2 mmol l−1) EDTA to the reaction mix resulted in complete inhibition of the enzyme activity.
Kinetic analysis of the recombinant BsPhyARRMK33 protein. X-axis: various substrate concentrations of sodium phytate effect on BsPhyARRMK33 activity. Y-axis: represents liberated phosphate in micromoles per liter. The K m and V max values were 0.95 mM and 15.3 μmol l−1. The optimum pH and temperature for the phytase activity was 7 and 55 °C, respectively. Data represent mean ± SD (n = 9)
Effect of pH and Temperature
The recombinant enzyme was found active at 37 °C between pH 5.0 and 8.0 with maximum in activity at pH 7.0. Although the enzyme exhibited activity at various temperatures ranging from 25 to 70 °C, an optimum activity has been observed at 55 °C and pH 7.0. The enzyme showed a recovery of 30 % activity when it was preincubated at pH 3.0 and 37 °C for 6 h prior to phytase assay. The enzyme upon denaturation for 10 min at 75, 85, and 95 °C, followed by 1-h renaturation at the room temperature (28 °C, in the presence of 5 mmol l−1 Ca2+), restored the phytase activity by 74, 52, and 28 % respectively.
Proteolysis Resistance
The recombinant purified phytase from BsPhyARRMK33 showed resistance to proteases, and the activity retained 80 % after 1-h incubation in the presence of pepsin and trypsin.
Discussion
The global phytase market value as estimated by the first international summit in 2010 is $350 million annually sharing 60 % of the total feed enzyme market. Currently, 70 % of the swine and poultry feed includes phytase enzyme [24]. Such a tremendous application of phytase has tempted researchers to attempt economical strategies to synthesize this enzyme in the bacterial system. Although several phytases from fungi, yeast, and bacteria have been characterized, commercial production of phytase from Aspergillus niger was started in 1990 and extending to several microbial phytases. However, these enzymes are not thermostable at elevated temperatures, as well as they have broad substrate specificity which could alter the other metabolic pathways [25, 26]. Incidentally, BsPhyARRMK33 phytase is an ideal substitute that has a great potential to replace other commercial phytases due to its higher thermostability, strict substrate specificity, and resistance to proteolysis [27].
In silico analysis of the phytase structure analysis revealed, β-propeller confirmation with six stranded blades with six Ca2+ binding sites; Ca1, Ca2, are Ca3 are responsible for thermostability, whereas Ca4, Ca5, and Ca6 are responsible for catalytic activity (Fig. 3). The purified enzyme exhibited a wide range of pH (5–8), with optimal pH 7, and is active in the small intestine where the micronutrient’s absorption takes place. In the neutral pH conditions, phytic acid easily combines with divalent cations especially with Ca2+ and makes insoluble Ca-phytate, which is a strict substrate of B. subtilis phytase [28]. This enzyme is particularly beneficial for layers whose diet includes high calcium concentration due to its strict substrate-specific activity for Ca-phytate [29]. Further, it would be beneficial for aquatic animals due to alkaline pH. We found that the activity of the purified recombinant phytase is highest at 55 °C, and the enzyme was stable up to 95 °C for 10 min. The enzyme showed maximum activity at 55 °C in the presence of Ca2+ probably due to increase in enzyme stability in the presence of Ca2+. Heterologous expression of the BsPhyARRMK33 phytase in the E. coli showed a maximal activity of 13.5 U mg−1. However, in the presence of calcium, the recombinant purified phytase showed maximum activity of 15.3 U mg−1.
Michaelis–Menten kinetics principle reveals phytase activity and phosphorus liberation is dependent on the concentration of substrate. BsPhyARRMK33 enzyme showed metal dependency for its activity. While on addition of EDTA, enzyme inhibition occurred due to metal depletion. Phytase activity inhibition of metal-depleted enzyme proves the metal dependency for its stability and integrity [26].
The phytase activity almost doubled at 55 °C in comparison to the activity at 37 °C. However, when it was exposed for elevated temperatures like 75–95 °C, it exhibited a declined activity. Our study reveals that BsPhyARRMK33 is Ca2+-dependent at higher temperatures, and this is in conformation in the reports [30]. However, the animal feed undergoes a feed pelleting process and the ideal phytase added should be stable at up to 80–85 °C. Among all phytases, B. subtilis phytase would be suitable due to its virtue of thermal stability even at 95 °C for 10 min. The phytase BsPhyARRMK33 belongs to alkaline phytase, which has a great resistance to papain and pancreatin, thus compatible with the intestinal pH.
Hence, BsPhyARRMK33 is an ideal ingredient for animal feed due to higher thermal stability, strict substrate specificity, and neutral pH, which are the conditions in the small intestine, where phosphate absorption takes place. The phytase BsPhyARRMK33 did not show activity with p-nitrophenylphosphate, thus indicating no phosphatase activity. Therefore, BsPhyARRMK33 has been considered the best candidate for transgenic expression in plants whereas fungal phytase expression in plants may disturb other metabolic pathways due to their additional phosphatase activity and broad substrate specificity. In addition, this phytase has an ability to degrade 3-inositol ring from 6-inositol ring which would help for signal transduction [8, 26]. In the present investigation, reproducible protocols for cloning and expression of Bacillus phytase enzyme from the recombinant E. coli have been successfully developed.
Conclusion
Phytases are crucial ingredients of diets given to monogastric animals that include poultry, fish, and swine. Animal feed preparation involves feed pelleting that requires high temperatures (80–85 °C). We propose that our BsPhyARRMK33 would be a suitable candidate for animal feed as it is thermostable at extreme temperatures required in the pelleting process. Moreover, being strictly substrate-specific for phytate and alkaline pH is particularly beneficial for poultry, transgenic plant expression, and aquaculture, respectively. Expression of BsPhyARRMK33 in plants would enable a cost-effective and propitious strategy for livestock industry. Thus, all the above characteristic making it an ideal feed supplement.
References
Yao, M. Z., Wang, X., Wang, W., Fu, Y. J., & Liang, A. H. (2013). Improving the thermostability of Escherichia coli phytase, appA, by enhancement of glycosylation. Biotechnology Letters, 35(10), 1669–1676.
Kumar, V., Sinha, A. K., Makkar, H. P., & Becker, K. (2010). Dietary roles of phytate and phytase in human nutrition: a review. Food Chemistry, 120, 945–959.
Hmida-Sayari, A., Elgharbi, F., Farhat, A., Rekik, H., Blondeau, K., & Bejar, S. (2014). Overexpression and biochemical characterization of a thermostable phytase from bacillus subtilis US417 in Pichia pastoris. Molecular Biotechnology, 1–10.
Suhairin, A., Manap, A., Yazid, M., Hussin, M., Shobirin, A., & Mustafa, S. (2010). Phytase: application in food industry. International Food Research Journal, 17, 13–21.
Ali, N., Paul, S., Gayen, D., Sarkar, S. N., Datta, K., & Datta, S. K. (2013). Development of low phytate rice by RNAi mediated seed-specific silencing of inositol 1, 3, 4, 5, 6-pentakisphosphate 2-kinase gene (IPK1). PLoS One, 8(7), e68161.
Singh, B., Singh, D., & Sharma, K. K. (2013). Microbial phytases in skirmishing and management of environmental phosphorus pollution. In Biotechnology for environmental management and resource recovery (pp. 239–260). India: Springer.
Brinch-Pedersen, H., Sorensen, L. D., & Holm, P. B. (2002). Engineering crop plants: getting a handle on phosphate. Trends in Plant Science, 7, 118–125.
Chinreddy, S. Reddy., Vani, K., Pandey, S., Gupta, G., Lakshmi, M. V., Reddy, P. C. O., Kaul, T., (2013). Manipulating microbial phytases for heterologous expression in crops for sustainable nutrition. Annals of Plant Sciences Review ISSN: 2287-688X
Yoon, S. M., Kim, S. Y., Li, K. F., Yoon, B. H., Choe, S., & Kuo, M. M. (2011). Transgenic microalgae expressing Escherichia coli AppA phytase as feed additive to reduce phytate excretion in the manure of young broiler chicks. Applied Microbiology and Biotechnology, 91, 553–563.
Yao, M. Z., Zhang, Y. H., Lu, W. L., Hu, M. Q., Wang, W., & Liang, A. H. (2012). Phytases: crystal structures, protein engineering and potential biotechnological applications. Journal of Applied Microbiology, 112(1), 1–14.
Lucca, P., Hurrell, R., & Potrykus, I. (2001). Genetic engineering approaches to improve the bioavailability and the level of iron in rice grains. Theoretical and Applied Genetics, 102, 392–397.
Hong, Y. F., Liu, C. Y., Cheng, K. J., Hour, A. L., Chan, M. T., Tseng, T. H., & Yu, S. M. (2008). The sweet potato sporamin promoter confers high-level phytase expression and improves organic phosphorus acquisition and tuber yield of transgenic potato. Plant Molecular Biology, 67(4), 347–361.
Tran, T. T., Mamo, G., Mattiasson, B., & Hatti-Kaul, R. (2010). A thermostable phytase from Bacillus sp. MD2: cloning, expression and high-level production in Escherichia coli. Journal of Industrial Microbiology & Biotechnology, 37, 279–287.
Hong, S. W., Chu, I. H., & Chung, K. S. (2011). Purification and biochemical characterization of thermostable phytase from newly isolated Bacillus subtilis CF92. Journal of the Korean Society for Applied Biological Chemistry, 54(1), 89–94.
Kammoun, R., Farhat, A., Chouayekh, H., Bouchaala, K., & Bejar, S. (2012). Phytase production by Bacillus subtilis US417 in submerged and solid state fermentations. Annals of Microbiology, 62(1), 155–164.
Farhat-Khemakhem, A., Farhat, M. B., Boukhris, I., Bejar, W., Bouchaala, K., Kammoun, R., & Chouayekh, H. (2012). Heterologous expression and optimization using experimental designs allowed highly efficient production of the PHY US417 phytase in Bacillus subtilis 168. AMB Express, 2(1), 1–11.
Rao, D. E. C. S., Rao, K. V., Reddy, T. P., & Reddy, V. D. (2009). Molecular characterization, physicochemical properties, known and potential applications of phytases: an overview. Critical Reviews in Biotechnology, 29(2), 182–198.
Miksch, G., Kleist, S., Friehs, K., & Flaschel, E. (2002). Overexpression of the phytase from Escherichia coli and itsextracellular production in bioreactors. Applied Microbiology and Biotechnology, 59, 85–694.
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., & Struhl, K. (1993). Current protocols in molecular biology, Vol: 1. USA: Geene Publishing Associates and John Wiley & Sons.
Arnold, K., Bordoli, L., Kopp, J., & Schwede, T. (2006). The SWISS-MODEL workspace: a web based environment for protein structure homology modeling. Bioinformatics, 22, 195–201.
Bradford, M. M. (1976). A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry, 72, 248–254.
Choi, Y. M., Suh, H. J., & Kim, J. M. (2001). Purification and properties of extracellular phytase from Bacillus sp. KHU-10. Journal of Protein Chemistry, 20, 287–292.
Ha, N. C., Oh, B. C., Shin, S., Kim, H. J., Oh, T. K., Kim, Y. O., & Oh, B. H. (2000). Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states. Nature Structural & Molecular Biology, 7, 147–153.
Lei, X. G., Weaver, J. D., Mullaney, E., Ullah, A. H., & Azain, M. J. (2013). Phytase, a new life for an “old” enzyme. Annual Review of Animal Biosciences, 1, 283–309.
Pen, J., Venvoerd, T. C., van Paridon, P. A., Beudeker, R. F., VandenElzen, P. J. M., Geerse, K., van der Klis, J. D., Versteegh, A. J., van Ooyen, A. J. J., & Hoekema, A. (1993). Phytase-containing transgenic seeds as a nove1 feed additive for improved phosphorus utilization. Biotechnology, 11, 811–814.
Rao, D. E. C. S., Rao, K. V., & Reddy, V. D. (2008). Cloning and expression of Bacillus phytase gene (phy) in Escherichia coli and recovery of active enzyme from the inclusion bodies. Journal of Applied Microbiology, 4, 1128–1137.
Elkhalil, E. A. I., Manner, K., Borriss, R., & Simon, O. (2007). In vitro and in vivo characteristics of bacterial phytases and their efficacy in broiler chickens. British Poultry Science, 48, 64–70.
Oh, B. C., Chang, B. S., & Park, K. H. (2001). Calcium-dependent catalytic activity of a novel phytase from Bacillus amyloliquefaciens DS11. Biochemistry, 40, 9669–9676.
Esteve-Garcia, E., Perez-Vendrell, A. M., & Broz, J. (2005). Phosphorus equivalence of a Consensus phytase produced by Hansenula polymorpha in diets for young turkeys. Archives of Animal Nutrition, 59, 53–59.
Kim, D. H., Oh, B. C., Choi, W. C., Lee, J. K., & Oh, T. K. (1999). Enzymatic evaluation of Bacillus amyloliquefaciens phytase as a feed additive. Biotechnology Letters, 21, 925–927.
Acknowledgments
The authors are thankful to the ICGEB, New Delhi, for providing support and facilities to carry out the research. The authors acknowledge the award of Senior Research Fellowship to M. Manna and Research Associateship to V. M. M. Achary by CSIR and DBT, India, respectively.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Reddy, C.S., Achary, V.M.M., Manna, M. et al. Isolation and Molecular Characterization of Thermostable Phytase from Bacillus subtilis (BSPhyARRMK33). Appl Biochem Biotechnol 175, 3058–3067 (2015). https://doi.org/10.1007/s12010-015-1487-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1487-4