Abstract
Purpose of Review
Clinically available biomarkers for hepatocellular carcinoma (HCC) early diagnosis and prognostication have limited utility. Further lack of routine biopsy in hepatocellular carcinoma limits the availability of molecular information to guide drug development. Recent studies investigating liquid biopsy using circulating tumor cells (CTCs) and cell-free deoxyribonucleic acid (cfDNA) have yielded promising data that could address both of these limitations.
Recent Findings
For early HCC diagnosis, CTCs have modest sensitivity but high specificity. CfDNA methylation scores have shown high sensitivity and specificity in two large phase II studies. Presence of CTCs has been associated with poorer prognosis in numerous studies, particularly increased cancer recurrence following curative therapy, while the literature on cfDNA and prognosis is less robust.
Summary
Liquid biopsy using CTCs and cfDNA has shown promise in prognostication and early diagnosis in HCC. Further robust validation of this liquid biopsy is required for routine clinical use.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer and a leading cause of cancer death [1]. HCC incidence in the USA is rising, primarily due to increasing non-alcoholic fatty liver disease prevalence and peaking hepatitis C-related complications, and is one of few malignancies in the USA whose attributable mortality is increasing [2, 3]. HCC carries a poor prognosis with median survival under two years and is one of few malignancies in the USA. Several major challenges in HCC care exist. First, most patients are diagnosed at an advanced stage, partly because the current surveillance method of ultrasound and alpha-fetoprotein (AFP) has limited sensitivity and poor compliance [4]. Second, in patients with advanced-stage disease, currently available systemic therapy is ineffective, with objective response rates of only 10–20% [5, 6].
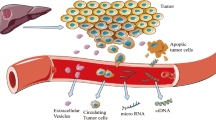
One factor limiting advances in HCC care and development of targeted systemic therapies is lack of biopsy tissue: HCC can be diagnosed and treated based on imaging alone and biopsy is not generally obtained [7]. Partly due to this, our understanding of HCC cancer biology lags behind that of other cancer types [8]. “Liquid biopsy,” i.e., analysis of circulating tumor-derived molecules, may address this limitation by offering molecular insights into established cancer and serving as an early diagnostic biomarker for surveillance.
Circulating tumor cells (CTCs) and cell-free deoxyribonucleic acid (cfDNA) for early cancer detection, prognostication, and guiding choice of therapy will be the focus of this review. CTCs are believed to represent an intermediate stage between localized disease and distant metastasis and have been detected in virtually all major solid tumors [9]. CfDNA is released into the circulation by death of and secretion from cells and can be quantified, characterized for integrity, and sequenced to detect mutations, methylation, and insertion-deletions [10]. (The term “circulating tumor DNA” is frequently used in the literature and refers to the subset of cfDNA that originates from tumors.) Non-coding ribonucleic acid can be detected in serum of patients with HCC or chronic liver disease, but we will focus on CTCs and cfDNA these non-coding RNA have been recently reviewed [11, 12].
Early Diagnosis
Circulating Tumor Cells
CTCs can be isolated from whole blood using a number of methods. The most commonly used and only Food and Drugs Administration–cleared method is CellSearch, which detects cells with an epithelial phenotype, i.e., cells which express EPCAM and cytokeratins [9]. Other methods exploit the fact that CTCs are typically larger than white blood cells: for example, CanPatrol utilizes size- and shape-based filtration, generally followed by fluoresecent labeling for cell-surface markers of interest [13], while other methods use microfluidics [14].
Several studies have investigated the utility of CTC in early diagnosis of HCC. Overall, CTC detection has moderate sensitivity (60–70%) and high specificity (> 70%) for distinguishing HCC from chronic liver disease or healthy controls [15]. One study of 296 HCC and 39 benign liver disease patients found a sensitivity of 65% using CanPatrol, a method based on filtration followed by fluoresecent labeling [16]. Studies using the commercially available CellSearch system have generally had lower sensitivities ranging from 31 to 68% [17, 18••].
High-throughput genetic analysis of CTCs or liver tissue can be used to develop predictive genetic scores for HCC [19•]. One study performed whole transcriptomic sequencing of circulating cells with epithelial phenotypes in patients with HCC vs. non-malignant chronic liver disease and found that cells from HCC patients had different transcriptomic profiles [20•]. They then created a score based on expression of specific genes to distinguish between HCC and chronic liver disease controls. Another study identified liver-specific transcripts and developed a serum risk score based on expression of these transcripts to distinguish between HCC and non-HCC controls [21]. While these genetic risk scores have not been externally validated, they may have greater sensitivity and specificity than those of CTCs alone.
Other attempts to develop more sensitive surrogate markers for CTCs have used targeted sequencing of genes such as EPCAM, whose corresponding protein has been used for CTC detection, or AFP, the protein product of which is clinically used as a noninvasive marker for HCC. One study of EPCAM mRNA found that EPCAM had lower sensitivity than AFP protein for HCC detection overall [22]. In two studies using AFP mRNA as a marker for HCC, sensitivity ranged from 54 to 100% depending on cancer stage, but specificity was modest at 56–86% [23, 24]. Use of multiple mRNAs may result in superior test characteristics: one such study used a combination of EPCAM, CD133, CD90, and CK19 mRNA expression and reported sensitivity 73–82% and specificity > 90% (vs. chronic liver disease patients), with similar performance in early stage and AFP-negative HCC [25•]. Additional prospective studies on sequencing of CTC-associated genes are needed.
Cell-Free DNA
Various properties of cfDNA have been evaluated for early HCC diagnosis (Table 1). Early studies in this field investigated mutations in TP53 [43, 47] or methylation of P16 [39, 41], which generally had high specificity but low sensitivity around 26–55% depending on the control group. Similarly, other hotspot mutations in TERT promoter and CTNNB1 are present in < 50% of patients with HCC [48, 50]. More recently, some studies have investigated total cfDNA amount or cfDNA integrity (defined as ratio of short- vs. long-circulating DNA strands), which generally have higher sensitivity around 70–80% and 80–90%, respectively [28,29,30, 34].
More recently, there has been interest in cfDNA methylation scores, which have yielded substantially higher sensitivity and specificity. One recent study from the USA identified a panel of six cfDNA differentially methylation regions that was tested in a cohort of 21 patients with HCC and 30 with cirrhosis, then validated in another with 95 HCC patients, 51 cirrhosis patients, and 98 healthy controls [51•]. On cross-validation, the sensitivity was 85% and specificity 91%, with sensitivity 75% for BCLC stage 0 and 93% for BCLC stage A HCC. Another larger study from China identified a set of ten cfDNA methylation markers on a training set of 715 HCC and 560 healthy controls, then validated these markers on 383 HCC patients and 275 controls [52••]. The authors reported sensitivity 83–86% and specificity 91–94% for distinguishing HCC from healthy controls. The authors also reported that this methylation score could also distinguish between HCC and non-malignant chronic liver disease (viral hepatitis and fatty liver), but sensitivity/specificity and numbers of the chronic liver disease patients were not reported.
Limitations
The existing literature on early HCC diagnosis using CTCs and cfDNA suffer from several major limitations. First, many of the studies detailed above, including the two large studies on methylation scores, included healthy individuals in their control arms, which could have resulted in inflated sensitivity/specificity estimates and is not a clinically relevant comparison. Also, some studies included patients with advanced-stage HCC, but generally the more clinically relevant question is comparing early stage HCC to chronic liver disease controls, in order to achieve early detection. Most cfDNA studies utilized post hoc cutoffs of mRNA expression and cross-cohort validity remain to be determined; further, cross-ancestry validity of cfDNA scores remains to be determined.
Prognosis
Circulating Tumor Cells
Table 2 is a partial list of studies on the prognostic significance of CTCs in HCC. The literature on CTCs in prognosis in HCC is most robust in patients undergoing partial hepatectomy with curative intent [15]. In this population, unadjusted hazard ratio for recurrence after resection was 2.7 (95% confidence interval 2.1–3.4). Overall survival as an outcome has been less consistently reported in patients undergoing resection, but presence of CTCs is usually associated with poorer overall survival as well [17, 57].
The utility of CTCs for prognostication is less clear in patients with intermediate- or advanced-stage disease. In patients receiving locoregional therapy, presence of CTCs has been associated with progression [22] and poorer overall survival [63, 68], but other studies showed no significant associations [69, 70]. Notably, the sizes of the cohorts including intermediate- and advanced-stage HCC are far smaller than those of the resection studies. Among patients receiving systemic therapy, there are very limited data on the association between CTCs and prognosis. Two small studies in patients with advanced-stage disease, one of the patients receiving sorafenib and temsirolimus and the other of patients not on systemic therapy at time of enrollment, showed no difference in overall survival based on presence or absence of CTCs [71, 72]. Another study showed a significant association between CTC presence and progression-free survival, but the definition of CTCs used here (phosphorylated ERK or Akt) has not been separately validated in HCC [56].
CTCs may be in a sense more relevant in patients with early stage disease as they indicate micrometastatic disease that may not be clinically apparent. In contrast, patients with advanced-stage HCC have by definition overt portal vein involvement or extrahepatic metastasis. However, further studies using larger cohorts of patients with intermediate- or advanced-stage disease will be required to better evaluate the significance of CTCs in this population.
CTCs can have different phenotypes, namely an epithelial phenotype, a mesenchymal phenotype, or a mixed phenotype; the epithelial-mesenchymal transition (EMT) is believed to be an important step in carcinogenesis that facilitates invasion into the circulation and, thus, metastasis [73]. As with other cancer types, in HCC primary tumors, expression of EMT genes is associated with poorer prognosis [74]. Likewise, CTCs with mesenchymal phenotypes appear to be associated with a poorer prognosis in HCC. CTC expression of the EMT proteins Slug, Snail, Twist, ZEB1, or Vimentin has been associated with the presence of tumor thrombus and metastatic disease [18••, 75]. A more recent study found that in patients undergoing curative resection, post-operative presence of mesenchymal CTCs has been associated with increased risk of early HCC recurrence, while presence of epithelial or mixed CTCs was not [76]. Whole transcriptome sequencing of CTCs has also been performed in limited studies; however, whether transcriptomic analysis of HCC CTCs yields information beyond that of targeted sequencing of candidate genes is not known [19•].
Location of CTCs may also have clinical significance. One study measured CTC counts in different vascular spaces of patients undergoing curative resection for HCC, including peripheral veins and arteries, portal vein, hepatic vein, and infrahepatic inferior vena cava [25•]. As might be predicted, CTC counts were highest in the hepatic vein, then in the peripheral vein, then lowest from the other vascular territories. Further, these vascular spaces had different clinical significance. Patients who subsequently developed intrahepatic recurrence had higher numbers of CTCs in the systemic circulation but similar numbers of CTCs in the hepatic circulation, compared with patients without intrahepatic recurrence. In contrast, those who developed lung metastasis had more CTCs in the hepatic vein but similar numbers of systemic circulation CTCs. While it is not practical to routinely sample the hepatic vein in all patients, it may be useful for risk stratification in patients undergoing resection or transplantation.
Circulating tumor clusters consisting of CTCs and, possibly, white blood cells and stromal cells may have special clinical significance. Several recent studies in breast cancer showed that circulating tumor clusters are more tumorigenic, perhaps because clusters are enriched for cancer stem cells whereas single circulating tumor cells are not [77]. Circulating tumor clusters are not well-studied in HCC, but one study suggested that they may portend an even poorer prognosis than single CTCs alone [18••].
Cell-Free DNA
Studies on cfDNA have evaluated various properties for their effect on prognosis. However, these studies are typically less standardized in terms of tumor stage and treatment type than studies on CTCs, and there is insufficient evidence to comment on differential utility of cfDNA in early stage vs. advanced-stage disease. While some studies on total cfDNA amount showed poorer overall survival and earlier time to progression with higher cfDNA amounts [31, 78], other studies across a range of HCC stages found no association between cfDNA amount and overall survival [28, 29]. Timing of cfDNA collection may be significant: one study in patients receiving radiotherapy found that while pre-therapy cfDNA amount correlated with tumor size, post-therapy cfDNA amount was more prognostically significant and was associated with intrahepatic recurrence [79]. CfDNA integrity has not been well-studied in the setting of prognosis but may be associated with poorer overall survival [28]. Similarly, mutations in or methylation of candidate genes have been variably associated with poorer overall survival [80] or recurrence [40, 48], and requires further characterization.
As with early diagnosis, cfDNA methylation scores may have greater predictive power than individual hotspot mutations or cfDNA amount/integrity. One large study found that the same methylation score used for early diagnosis was associated with poorer survival in both derivation (N = 680) and validation (N = 369) cohorts, with primarily advanced-stage HCC (64% TNM stage III/IV) [52••]. This score added prognostic information beyond that of TNM stage alone. Another study found that having ≥ 3 of 6 methylation markers in RASSF1A, CCND2, CFTR, SPINT2, SRD5A2, and/or BASP1 was associated with poorer adjusted disease-free survival and overall survival, in a mix of early and advanced-stage HCC (54% TNM stage III/IV) [81].
Limitations
Several limitations exist in the literature on liquid biopsy in for HCC prognostication. First, outcomes are inconsistently reported: usually studies in patients receiving curative therapy report recurrence or recurrence-free survival while those in patients receiving non-curative therapy report progression or progression-free survival, and overall survival is frequently not mentioned at all. In addition, some studies report hazard ratios while others describe mean/median overall survival, progression-free survival, etc. More consistent reporting will facilitate an improved understanding of the use of CTCs for predicting prognosis in HCC. Finally, cfDNA studies are prone to ad hoc cutoffs in amount, integrity, or methylation proportion.
Liquid Biopsy to Guide Therapy
In several cancer types, genetic data are used to guide systemic therapy decisions. For example, erlotinib and alectinib are approved for lung adenocarcinoma with selected EGFR mutations and EML4-ALK fusions, respectively, while pembrolizumab is approved as second-line therapy for microsatellite instability-high or mismatch repair-deficient cancer regardless of primary site [82, 83]. In HCC, biomarkers to guide treatment in the setting of systemic disease are much more limited, and no therapies are approved for use specifically in tumors with specific genetic alterations. The SHARP study identified no biomarker as predictive of treatment response to sorafenib [84], and neither AFP nor c-Met is associated with response to regorafenib treatment [85]. Until recently, AFP was the only biomarker associated with response to certain treatments in HCC: ramucirumab improved overall survival in advanced-stage HCC only in patients with AFP concentration > 400 ng/mL [86], and survival benefit to cabozantinib was greater in HCC patients with serum AFP ≥ 200 ng/mL [87]. A recent study identified a set of micro-RNAs and plasma proteins associated with response to regorafenib, though this requires further validation [88]. No studies have identified biomarkers predicting response to checkpoint inhibitor therapy, which are currently approved as second-line agents in patients with unresectable HCC.
In order for liquid biopsy to be useful for guiding clinical decision-making, it should ideally reflect the biology of the primary tumor. In HCC, genetic alterations in cfDNA are inconsistently concordant with those of the primary tumor. In one study of paired tumor-plasma samples, 89% of patients with TP53 R249S mutations in the primary tumor also had them in cfDNA [89], but in another study, only 69% of patients with GSTP1 promoter hypermethylation in the primary tumor had this in cfDNA [90]. Several studies have systematically sequenced selected cancer-related genes in both cfDNA and primary tumors: while cfDNA mutations are typically detected in primary tumors, the sensitivity of cfDNA for primary tumor mutations is widely variable from 30 to > 90% and may vary based on the specific gene [91, 92]. It is not known whether HCC CTC mutations correlate with those of the primary tumor as to our knowledge DNA sequencing in HCC CTCs has not been reported. However, studies in prostate cancer [93] and multiple myeloma [94] suggest that mutations are usually concordant between CTCs and primary tumors. It is difficult to determine whether transcriptomes are concordant between primary tumor and CTCs due to tumor heterogeneity and small numbers of CTCs, but in prostate cancer, CTCs more closely resemble their corresponding primary tumor than primary tumors from other individuals [93].
Liquid biopsy could hypothetically guide therapy in a few ways. One would be to use liquid biopsy to guide choice of initial therapy. Using cfDNA to identify specific mutations is unlikely, at present, to be an adequate tool to achieve this, since most patients with HCC do not have targetable mutations [95]. Other properties of cfDNA have been explored as well: for instance, genomic instability and total amount have been associated with poorer response to sorafenib [96]. CTCs may provide an alternate method of identifying response to therapy: they can be characterized by transcriptomic analysis or proteomics, which would potentially allow for expression profiles (rather than mutations only) that are associated with response to one drug or another. While whole transcriptome sequencing of HCC CTCs has been performed [19•, 20•], whether it has implications for treatment response remains to be determined.
Another potential application of liquid biopsy is to monitor patients on therapy and determine whether to continue treatment or switch to another option. RNA-based scores (as an approximation of CTCs) have been used to monitor response to treatment in HCC [21] but to our knowledge, there are no published studies showing that liquid biopsy for monitoring is superior or comparable to imaging studies. Literature in other cancer types may provide a roadmap to liquid biopsy-guided HCC treatment. In prostate cancer, cfDNA mutational profiles may predict development of enzalutamide resistance [97]. In breast cancer, cfDNA correlates with tumor burden and may be an early predictor of disease progression or recurrence [98]. Monitoring may not result in clinically relevant improvement in outcomes though. One study of 319 patients with metastatic breast cancer and detectable CTCs at baseline who were receiving standard first-line chemotherapy followed CTC counts longitudinally; patients with persistently increased CTCs were randomized to either change to an alternative chemotherapy regimen or continue current treatment, with the idea that persistently increased CTC count indicated treatment failure [99]. This was a negative study: there was no change in overall survival in the patients who underwent CTC-guided change in therapy compared with those who did not. Prospective studies on this topic in HCC are required to further clarify any potential role of liquid biopsy for monitoring.
Currently, no major professional societies endorse use of CTCs or cfDNA to guide treatment or for monitoring across all cancers. We agree that it would be premature to use liquid biopsy as reviewed in this article to guide therapy at this time. However, studies on CTCs and cfDNA in other cancer types suggest that in the future it may be possible to apply these methods to HCC as well.
Conclusions
Liquid biopsy has the potential to address two of the major limitations in clinical HCC management, namely inadequate HCC surveillance tools and providing molecular data, that will, hopefully, one day help guide choice of systemic therapy in HCC. We believe that liquid biopsy using CTCs and cfDNA is promising in early diagnosis and prognostication of HCC. CfDNA has shown the most potential for early diagnosis, especially if polygenic scores are used; in contrast, CTCs have only modest sensitivity for early HCC detection. In contrast, the data for CTCs and HCC prognosis are more robust, and CTCs also offer the opportunity to characterize the cancer transcriptome noninvasively. However, the field is still immature and there are inadequate data to recommend using liquid biopsy to determine choice of initial treatment or for monitoring on treatment. Further research is required to further evaluate potential clinical applications of liquid biopsy in HCC.
Abbreviations
- AFP:
-
Alpha-fetoprotein
- cfDNA:
-
Cell-free deoxyribonucleic acid
- CTC:
-
Circulating tumor cell
- EMT:
-
Epithelial-mesenchymal transition
- HCC:
-
Hepatocellular carcinomas
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Bertuccio P, Turati F, Carioli G, Rodriguez T, La Vecchia C, Malvezzi M, et al. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67(2):302–9.
Tandon P, Garcia-Tsao G. Prognostic indicators in hepatocellular carcinoma: a systematic review of 72 studies. Liver Int. 2009;29(4):502–10.
Tapper EB, Parikh ND. Mortality due to cirrhosis and liver cancer in the United States, 1999-2016: observational study. BMJ. 2018;362:k2817.
Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154(6):1706–18.e1.
El-Khoueiry AB, Sangro B, Yau T, Crocenzi TS, Kudo M, Hsu C, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492–502.
Bruix J, Qin S, Merle P, Granito A, Huang Y-H, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389(10064):56–66.
Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53(3):1020–2.
Tapper EB, Lok AS. Use of liver imaging and biopsy in clinical practice. N Engl J Med. 2017;377(8):756–68.
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–904.
Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–37.
Tricoli L, Niture S, Chimeh U, Ressom H, Kumar D. Role of microRNAs in the development of hepatocellular carcinoma and drug resistance. Front Biosci (Landmark Ed). 2019;24:382–91.
Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15(3):137–51.
Wu S, Liu S, Liu Z, Huang J, Pu X, Li J, et al. Classification of circulating tumor cells by epithelial-mesenchymal transition markers. PLoS One. 2015;10(4):e0123976.
Lin E, Rivera-Baez L, Fouladdel S, Yoon HJ, Guthrie S, Wieger J, et al. High-throughput microfluidic labyrinth for the label-free isolation of circulating tumor cells. Cell Syst. 2017;5(3):295–304 e4.
Chen VL, Xu D, Harouaka R, Wicha M, Lok A, Parikh ND. Mo1451–liquid biopsy for prognosis in hepatocellular carcinoma (HCC) using circulating tumor cells (CTCS): a systematic review and meta-analysis. Gastroenterology. 2019;156(6):S-1311.
Guo BT, Liu XC, Huang Y, Ou HH, Li XH, Yang DH. Positive circulating tumor cells in the peripheral blood may indicate a poor prognosis in patients with hepatocellular carcinoma. Nan Fang Yi Ke Da Xue Xue Bao. 2016;36(8):1134–9.
Yu JJ, Xiao W, Dong SL, Liang HF, Zhang ZW, Zhang BX, et al. Effect of surgical liver resection on circulating tumor cells in patients with hepatocellular carcinoma. BMC Cancer. 2018;18(1):835.
•• Sun YF, Guo W, Xu Y, Shi YH, Gong ZJ, Ji Y, et al. Circulating tumor cells from different vascular sites exhibit spatial heterogeneity in epithelial and mesenchymal composition and distinct clinical significance in hepatocellular carcinoma. Clin Cancer Res. 2018;24(3):547–59 Identified that vascular territories have unique prognostic significance, and also discussed circulating tumor clusters.
• D'Avola D, Villacorta-Martin C, Martins-Filho SN, Craig A, Labgaa I, von Felden J, et al. High-density single cell mRNA sequencing to characterize circulating tumor cells in hepatocellular carcinoma. Sci Rep. 2018;8(1):11570 Transcriptomic sequencing of circulating tumor cells in hepatocellular carcinoma.
• Bhan I, Mosesso K, Goyal L, Philipp J, Kalinich M, Franses JW, et al. Detection and analysis of circulating epithelial cells in liquid biopsies from patients with liver disease. Gastroenterology. 2018. Transcriptomic sequencing of circulating tumor cells in hepatocellular carcinoma.
Kalinich M, Bhan I, Kwan TT, Miyamoto DT, Javaid S, LiCausi JA, et al. An RNA-based signature enables high specificity detection of circulating tumor cells in hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2017;114(5):1123–8.
Guo W, Yang XR, Sun YF, Shen MN, Ma XL, Wu J, et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin Cancer Res. 2014;20(18):4794–805.
Yang SZ, Dong JH, Li K, Zhang Y, Zhu J. Detection of AFPmRNA and melanoma antigen gene-1mRNA as markers of disseminated hepatocellular carcinoma cells in blood. Hepatobiliary Pancreat Dis Int. 2005;4(2):227–33.
Yao F, Guo JM, Xu CF, Lou YL, Xiao BX, Zhou WH, et al. Detecting AFP mRNA in peripheral blood of the patients with hepatocellular carcinoma, liver cirrhosis and hepatitis. Clin Chim Acta. 2005;361(1–2):119–27.
• Guo W, Sun YF, Shen MN, Ma XL, Wu J, Zhang CY, et al. Circulating tumor cells with stem-like phenotypes for diagnosis, prognosis, and therapeutic response evaluation in hepatocellular carcinoma. Clin Cancer Res. 2018;24(9):2203–13.
Chen H, Sun LY, Zheng HQ, Zhang QF, Jin XM. Total serum DNA and DNA integrity: diagnostic value in patients with hepatitis B virus-related hepatocellular carcinoma. Pathology. 2012;44(4):318–24.
Chen K, Zhang H, Zhang LN, Ju SQ, Qi J, Huang DF, et al. Value of circulating cell-free DNA in diagnosis of hepatocelluar carcinoma. World J Gastroenterol. 2013;19(20):3143–9.
El-Shazly SF, Eid MA, El-Sourogy HA, Attia GF, Ezzat SA. Evaluation of serum DNA integrity as a screening and prognostic tool in patients with hepatitis C virus-related hepatocellular carcinoma. Int J Biol Markers. 2010;25(2):79–86.
Huang Z, Hua D, Hu Y, Cheng Z, Zhou X, Xie Q, et al. Quantitation of plasma circulating DNA using quantitative PCR for the detection of hepatocellular carcinoma. Pathol Oncol Res. 2012;18(2):271–6.
Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, Stark M, et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer Res. 2006;26(6c):4713–9.
Piciocchi M, Cardin R, Vitale A, Vanin V, Giacomin A, Pozzan C, et al. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol Int. 2013;7(4):1050–7.
Ren N, Qin LX, Tu H, Liu YK, Zhang BH, Tang ZY. Quantitative analysis of circulating DNA level in plasma from patients with hepatocellular carcinoma and its potential clinical value. Fudan Univ J Med Sci. 2005;32(2):134–8.
Yan L, Chen Y, Zhou J, Zhao H, Zhang H, Wang G. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int J Infect Dis. 2018;67:92–7.
Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, et al. Plasma circulating cell-free DNA integrity as a promising biomarker for diagnosis and surveillance in patients with hepatocellular carcinoma. J Cancer. 2016;7(13):1798–803.
Chang H, Yi B, Li L, Zhang HY, Sun F, Dong SQ, et al. Methylation of tumor associated genes in tissue and plasma samples from liver disease patients. Exp Mol Pathol. 2008;85(2):96–100.
Chu HJ, Heo J, Seo SB, Kim GH, Kang DH, Song GA, et al. Detection of aberrant p16INK4A methylation in sera of patients with liver cirrhosis and hepatocellular carcinoma. J Korean Med Sci. 2004;19(1):83–6.
Tan SH, Ida H, Lau QC, Goh BC, Chieng WS, Loh M, et al. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol Rep. 2007;18(5):1225–30.
Wong IHN, Dennis Lo YM, Zhang J, Liew C-T, Ng MHL, Wong N, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59(1):71.
Wong IH, Lo YM, Yeo W, Lau WY, Johnson PJ. Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin Cancer Res. 2000;6(9):3516–21.
Wong IHN, Johnson PJ, Lai PBS, Lau WY, Lo YMD. Tumor-derived epigenetic changes in the plasma and serum of liver cancer patients: implications for cancer detection and monitoring. Ann N Y Acad Sci. 2000;906(1):102–5.
Wong IH, Zhang J, Lai PB, Lau WY, Lo YM. Quantitative analysis of tumor-derived methylated p16INK4a sequences in plasma, serum, and blood cells of hepatocellular carcinoma patients. Clin Cancer Res. 2003;9(3):1047–52.
Zhang YJ, Rossner P Jr, Chen Y, Agrawal M, Wang Q, Wang L, et al. Aflatoxin B1 and polycyclic aromatic hydrocarbon adducts, p53 mutations and p16 methylation in liver tissue and plasma of hepatocellular carcinoma patients. Int J Cancer. 2006;119(5):985–91.
Huang XH, Sun LH, Lu DD, Sun Y, Ma LJ, Zhang XR, et al. Codon 249 mutation in exon 7 of p53 gene in plasma DNA: maybe a new early diagnostic marker of hepatocellular carcinoma in Qidong risk area, China. World J Gastroenterol. 2003;9(4):692–5.
Igetei R, Otegbayo JA, Ndububa DA, Lesi OA, Anumudu CI, Hainaut P, et al. Detection of p53 codon 249 mutation in Nigerian patients with hepatocellular carcinoma using a novel evaluation of cell-free DNA. Ann Hepatol. 2008;7(4):339–44.
Jackson PE, Qian G-S, Friesen MD, Zhu Y-R, Lu P, Wang J-B, et al. Specific p53 mutations detected in plasma and tumors of hepatocellular carcinoma patients by electrospray ionization mass spectrometry. Cancer Res. 2001;61(1):33–5.
Jackson PE, Kuang S-Y, Wang J-B, Strickland PT, Muñoz A, Kensler TW, et al. Prospective detection of codon 249 mutations in plasma of hepatocellular carcinoma patients. Carcinogenesis. 2003;24(10):1657–63.
Kirk GD, Lesi OA, Mendy M, Szymañska K, Whittle H, Goedert JJ, et al. 249ser TP53 mutation in plasma DNA, hepatitis B viral infection, and risk of hepatocellular carcinoma. Oncogene. 2005;24(38):5858–67.
Liao W, Yang H, Xu H, Wang Y, Ge P, Ren J, et al. Noninvasive detection of tumor-associated mutations from circulating cell-free DNA in hepatocellular carcinoma patients by targeted deep sequencing. Oncotarget. 2016;7(26):40481–90.
Marchio A, Amougou Atsama M, Bere A, Komas NP, Noah Noah D, Atangana PJA, et al. Droplet digital PCR detects high rate of TP53 R249S mutants in cell-free DNA of middle African patients with hepatocellular carcinoma. Clin Exp Med. 2018;18:421–31.
Jiao J, Watt GP, Stevenson HL, Calderone TL, Fisher-Hoch SP, Ye Y, et al. Telomerase reverse transcriptase mutations in plasma DNA in patients with hepatocellular carcinoma or cirrhosis: prevalence and risk factors. Hepatol Commun. 2018;2(6):718–31.
• Kisiel JB, Dukek BA, Kanipakam R.V.S.R, Ghoz HM, Yab TC, Berger CK, et al. Hepatocellular carcinoma detection by plasma methylated DNA: discovery, phase I pilot, and phase II clinical validation. Hepatology. 2018;0(0). Large study of circulating tumor DNA for early detection in hepatocellular carcinoma.
•• Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155–61 Largest study to date of circulating tumor DNA for early diagnosis and prognosis in hepatocellular carcinoma.
Court CM, Hou S, Winograd P, Segel NH, Li QW, Zhu Y, et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 2018;24(7):946–60.
Fan ST, Yang ZF, Ho DW, Ng MN, Yu WC, Wong J. Prediction of posthepatectomy recurrence of hepatocellular carcinoma by circulating cancer stem cells: a prospective study. Ann Surg. 2011;254(4):569–76.
Lai HC, Yeh CC, Jeng LB, Huang SF, Liao PY, Lei FJ, et al. Androgen receptor mitigates postoperative disease progression of hepatocellular carcinoma by suppressing CD90+ populations and cell migration and by promoting anoikis in circulating tumor cells. Oncotarget. 2016;7(29):46448–65.
Li J, Shi L, Zhang X, Sun B, Yang Y, Ge N, et al. pERK/pAkt phenotyping in circulating tumor cells as a biomarker for sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncotarget. 2016;7(3):2646–59.
Liu S, Li N, Yu X, Xiao X, Cheng K, Hu J, et al. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology. 2013;144(5):1031–41 e10.
Nel I, Baba HA, Ertle J, Weber F, Sitek B, Eisenacher M, et al. Individual profiling of circulating tumor cell composition and therapeutic outcome in patients with hepatocellular carcinoma. Transl Oncol. 2013;6(4):420–8.
Ogle LF, Orr JG, Willoughby CE, Hutton C, McPherson S, Plummer R, et al. Imagestream detection and characterisation of circulating tumour cells - a liquid biopsy for hepatocellular carcinoma? J Hepatol. 2016;65(2):305–13.
Ou H, Huang Y, Xiang L, Chen Z, Fang Y, Lin Y, et al. Circulating tumor cell phenotype indicates poor survival and recurrence after surgery for hepatocellular carcinoma. Dig Dis Sci. 2018;63(9):2373–80.
Qi LN, Xiang BD, Wu FX, Ye JZ, Zhong JH, Wang YY, et al. Circulating tumor cells undergoing EMT provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res. 2018;78:4731–44.
Schulze K, Von Felden J, Krech T, Ewald F, Nashan B, Lohse AW, et al. EpCAM-positive circulating tumor cells as liquid biomarker for early micrometastases and HCC recurrence risk. J Hepatol. 2017;66(1):S457.
Shen J, Wang W, Zhu X, Ni C. EpCAM-positive circulating tumor cells independently predict poor outcomes of transcatheter arterial chemoembolization in patients with unresectable hepatocellular carcinoma. J Vasc Interv Radiol. 2018;29(4):S119.
Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ, et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013;57(4):1458–68.
von Felden J, Schulze K, Krech T, Ewald F, Nashan B, Pantel K, et al. Circulating tumor cells as liquid biomarker for high HCC recurrence risk after curative liver resection. Oncotarget. 2017;8(52):89978–87.
Wang S, Zheng Y, Liu J, Huo F, Zhou J. Analysis of circulating tumor cells in patients with hepatocellular carcinoma recurrence following liver transplantation. J Investig Med. 2018;66(5):1–6.
Xue F, Shi S, Zhang Z, Xu C, Zheng J, Qin T, et al. Application of a novel liquid biopsy in patients with hepatocellular carcinoma undergoing liver transplantation. Oncol Lett. 2018;15(4):5481–8.
Zhou Y, Wang B, Wu J, Zhang C, Zhou Y, Yang X, et al. Association of preoperative EpCAM circulating tumor cells and peripheral Treg cell levels with early recurrence of hepatocellular carcinoma following radical hepatic resection. BMC Cancer. 2016;16:506.
Yin LC, Luo ZC, Gao YX, Li Y, Peng Q, Gao Y. Twist expression in circulating hepatocellular carcinoma cells predicts metastasis and prognoses. Biomed Res Int. 2018;2018:1–12.
Fang ZT, Zhang W, Wang GZ, Zhou B, Yang GW, Qu XD, et al. Circulating tumor cells in the central and peripheral venous compartment - assessing hematogenous dissemination after transarterial chemoembolization of hepatocellular carcinoma. OncoTargets Ther. 2014;7:1311–8.
Kelley RK, Lee MR, Hwang J, Gordan JD, Nimeiri HS, Bocobo AG, et al. Detection of circulating tumor cells (CTC) using a non-EpCAM-based, high-definition, single-cell assay in advanced hepatocellular carcinoma (HCC) for patients enrolled on phase I and II trials of sorafenib plus temsirolimus. J Clin Oncol. 2017;35:4.
Kelley RK, Magbanua MJM, Butler TM, Collisson EA, Hwang J, Sidiropoulos N, et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer. 2015;15(1):206.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–96.
Li P, Chen P, Peng X, Ma C, Zhang W, Dai X. HOXC6 predicts invasion and poor survival in hepatocellular carcinoma by driving epithelial-mesenchymal transition. Aging. 2018;10:2570–84.
Lee HM, Joh JW, Seo SR, Kim WT, Kim MK, Choi HS, et al. Cell-surface major vault protein promotes cancer progression through harboring mesenchymal and intermediate circulating tumor cells in hepatocellular carcinomas. Sci Rep. 2017;7(1):13201.
Wang Z, Luo L, Cheng Y, He G, Peng B, Gao Y, et al. Correlation between postoperative early recurrence of hepatocellular carcinoma and mesenchymal circulating tumor cells in peripheral blood. J Gastrointest Surg. 2018;22(4):633–9.
Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176(1–2):98–112 e14.
Park S, Lee EJ, Rim CH, Seong J. Cell-free DNA as a predictive marker after radiation therapy for hepatocellular carcinoma. Int J Radiat Oncol Biol Phys. 2017;99(2):S89–90.
Park S, Lee EJ, Rim CH, Seong J. Plasma cell-free DNA as a predictive marker after radiotherapy for hepatocellular carcinoma. Yonsei Med J. 2018;59(4):470–9.
Huang ZH, Hu Y, Hua D, Wu YY, Song MX, Cheng ZH. Quantitative analysis of multiple methylated genes in plasma for the diagnosis and prognosis of hepatocellular carcinoma. Exp Mol Pathol. 2011;91(3):702–7.
Kanekiyo S, Iizuka N, Tsunedomi R, Tokumitsu Y, Hashimoto N, Tokuhisa Y, et al. Preoperative serum methylation signature as prognostic tool after curative hepatectomy in patients with hepatocellular carcinoma. Anticancer Res. 2015;35(2):997–1007.
Peters S, Camidge DR, Shaw AT, Gadgeel S, Ahn JS, Kim D-W, et al. Alectinib versus Crizotinib in untreated ALK-positive non–small-cell lung cancer. N Engl J Med. 2017;377(9):829–38.
Diaz LA, Marabelle A, Delord J-P, Shapira-Frommer R, Geva R, Peled N, et al. Pembrolizumab therapy for microsatellite instability high (MSI-H) colorectal cancer (CRC) and non-CRC. J Clin Oncol. 2017;35(15_suppl):3071.
Llovet JM, Pena CE, Lathia CD, Shan M, Meinhardt G, Bruix J, et al. Plasma biomarkers as predictors of outcome in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2012;18(8):2290–300.
Teufel M, Köchert K, Meinhardt G, Bruix J. Efficacy of regorafenib (REG) in patients with hepatocellular carcinoma (HCC) in the phase III RESORCE trial according to alpha-fetoprotein (AFP) and c-Met levels as predictors of poor prognosis. J Clin Oncol. 2017;35(15_suppl):4078.
Zhu AX, Kang Y-K, Yen C-J, Finn RS, Galle PR, Llovet JM, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019;20(2):282–96.
Abou-Alfa GK, El-Khoueiry AB, Meyer T, Rimassa L, Merle P, Chan SL, et al., editors. Outcomes by baseline alpha-fetoprotein (AFP) levels in the phase 3 celestial trial of cabozantinib (C) versus placebo (P) in previously treated advanced hepatocellular carcinoma (HCC)2018: WILEY 111 RIVER ST, HOBOKEN 07030–5774, NJ USA.
Teufel M, Seidel H, Kochert K, Meinhardt G, Finn RS, Llovet JM, et al. Biomarkers associated with response to Regorafenib in patients with hepatocellular carcinoma. Gastroenterology. 2019;156(6):1731–41.
Szymańska K, Lesi OA, Kirk GD, Sam O, Taniere P, Scoazec J-Y, et al. Ser-249TP53 mutation in tumour and plasma DNA of hepatocellular carcinoma patients from a high incidence area in the Gambia, West Africa. Int J Cancer. 2004;110(3):374–9.
Wang J, Qin Y, Li B, Sun Z, Yang B. Detection of aberrant promoter methylation of GSTP1 in the tumor and serum of Chinese human primary hepatocellular carcinoma patients. Clin Biochem. 2006;39(4):344–8.
Howell J, Atkinson SR, Pinato DJ, Knapp S, Ward C, Minisini R, et al. Identification of mutations in circulating cell-free tumour DNA as a biomarker in hepatocellular carcinoma. Eur J Cancer. 2019;116:56–66.
Riviere P, Fanta PT, Ikeda S, Baumgartner J, Heestand GM, Kurzrock R. The mutational landscape of gastrointestinal malignancies as reflected by circulating tumor DNA. Mol Cancer Ther. 2018;17(1):297–305.
Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351–6.
Lohr JG, Kim S, Gould J, Knoechel B, Drier Y, Cotton MJ, et al. Genetic interrogation of circulating multiple myeloma cells at single-cell resolution. Sci Transl Med. 2016;8(363):–363ra147.
Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–11.
Park SR, Oh CR, Kong SY, Kim MK, Yoon KA, Cho EH, et al. Biomarker analysis in circulating cell-free DNA in patients treated with sorafenib for advanced hepatocellular carcinoma. Cancer Res. 2018;78:13.
Wyatt AW, Azad AA, Volik SV, Annala M, Beja K, McConeghy B, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016;2(12):1598–606.
Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7(302):302ra133.
Smerage JB, Barlow WE, Hortobagyi GN, Winer EP, Leyland-Jones B, Srkalovic G, et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol. 2014;32(31):3483–9.
Funding
Vincent Chen was supported in part by a University of Michigan Training in Basic and Translational Digestive Sciences T32 grant (5T32DK094775).
Author information
Authors and Affiliations
Contributions
Vincent Chen: drafting of the manuscript
Neehar Parikh: critical review of manuscript
All authors identified above have critically reviewed the paper and approve the final version of this paper, including the authorship statement.
Corresponding author
Ethics declarations
Conflict of Interest
Vincent Chen declares no potential conflicts of interest.
Neehar Parikh: Consultant: Bristol-Myers Squibb, Exelixis, Freenome; Advisory Board: Eisai, Bayer, Exelixis, Wako Diagnostics; Research Grants: Bayer, Target Pharmasolutions, Exact Sciences.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Hepatic Cancer
Rights and permissions
About this article
Cite this article
Chen, V.L., Parikh, N.D. Liquid Biopsy for Hepatocellular Carcinoma. Curr Hepatology Rep 18, 390–399 (2019). https://doi.org/10.1007/s11901-019-00491-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11901-019-00491-z