Abstract
Purpose
We summarized the recent findings of liquid biopsy in cancer field and discussed its potential utility in hepatocellular carcinoma.
Methods
Literature published in MEDLINE, EMBASE, and Science Direct electronic databases was searched and reviewed.
Results
Liquid biopsy specially referred to the detection of nucleic acids (circulating cell-free DNA, cfDNA) and circulating tumor cells (CTCs) in the blood of cancer patients. Compared to conventional single-site sampling or biopsy method, liquid biopsy had the advantages such as non-invasiveness, dynamic monitoring, and the most important of all, overcoming the limit of spatial and temporal heterogeneity. The genomic information of cancer could be profiled by genotyping cfDNA/CTC and subsequently applied to make molecular classification, targeted therapy guidance, and unveil drug resistance mechanisms. The serial sampling feature of liquid biopsy made it possible to monitor treatment response in a real-time manner and predict tumor metastasis/recurrence in advance.
Conclusions
Liquid biopsy is a non-invasive, dynamic, and informative sampling method with important clinical translational significance in cancer research and practice. Much work needs to be done before it is used in the management of HCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Technological advancements in the past decade have greatly enriched our knowledge of cancer genomics. The diagnostic and therapeutic significance of genetic aberrations that initiate tumorigenesis and promote disease progression has been increasingly recognized, and as the cost of next-generation sequencing (NGS) falls and its accuracy improves, genomic information has become a powerful tool for aiding clinicians in the management of malignancies [1]. However, the limitations of traditional sampling/biopsy modalities, such as their invasiveness and inability to account for tumor heterogeneity, have curtailed the utility of genetic tools for cancer management. Therefore, some researchers have advocated for liquid biopsy, a novel sampling approach that has wide clinical utility [2]. Herein, we will focus on the concept, implementation, and application of liquid biopsy in cancer research and will discuss the current status and future prospects of liquid biopsy in hepatocellular carcinoma (HCC) in detail.
The Concept of Liquid Biopsy and Rationale for its Use in Cancer Management
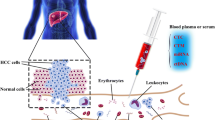
Liquid biopsy is a modality that samples bodily fluids instead of solid tissue for pathophysiological or sequencing analysis; it has been introduced in the fields of prenatal testing, transplantation rejection, diabetes mellitus, and cancer research. Generally, any bodily fluids, including blood, urine, sputum, pleural effusion, ascites, and cerebrospinal fluid, can be used as potential samples for liquid biopsy. A blood sample is most frequently used because of several advantages, including availability, its representation of whole-body condition, non-invasiveness, and the opportunity to repeatedly sample it for dynamic evaluation of health/disease status. Once blood is drawn, cells, proteins, molecules, nucleic acids, and vesicles can be isolated and utilized by different analytical approaches to gather as much information as possible to make an early diagnosis, evaluate treatment response, and/or monitor disease progression. Currently, tumor-derived cells and DNA in the blood, namely circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA), are the markers most widely sought in liquid biopsy for cancer research.
Surgery, being the most probable therapy to achieve curative effect, has reached its peak in improving the survival of patients with resectable tumors and postoperative recurrence and/or metastases aggravate such conditions. For patients with unresectable solid tumors and hematological malignancies, chemo-radiotherapy and targeted agents are commonly prescribed. Since the identification of mutations responsible for tumor initiation, genotype-based treatments, such as EGFR and BRAF kinase inhibitors, have become the focus of targeted therapeutic strategies. Thus, uncovering the genetic background of disease to tailor personalized drugs and treatments in real-time can improve the clinical outcomes of cancer patients.
Traditionally, genomic information is obtained by sequencing tumor tissue that has been either surgically resected or biopsied. However, this method has some intrinsic limits. First, for patients with unresectable metastatic diseases, tumor tissue is unavailable; though biopsy might be conducted in some of these patients, it is not without complications and has weaknesses. The biopsy procedure is painful and bears the risk of bleeding and iatrogenic tumor inoculation; furthermore, in cases where tumors are located deep inside the body and/or close to important blood vessels, biopsy is anatomically challenging and highly risky. Should a biopsy fail, repeated sampling aggravates these situations. Most importantly, tumor sampling and biopsy are both confined to regional sites within a tumor and are thus easily subject to selection bias resulting from tumor heterogeneity.
Morphological tumor heterogeneity has long been recognized, but only recently have researchers begun to realize the importance of genetic heterogeneity, which in turn consists of spatial and temporal heterogeneity. Spatial heterogeneity (intratumor heterogeneity, ITH) is caused by subclonal cellular populations harboring heterogeneous genetic, epigenetic, and phenotypic features among different regions of a tumor or between the primary and metastatic sites. ITH has been reported in a variety of cancers, including renal cancer [3, 4], breast cancer [5], lung cancer [6, 7], glioma [8], ovarian cancer [9], HCC [10], and hematological malignancies [11–13]. The common observation that only some oncogenetic mutations are shared by all cellular populations within a tumor [14] has led the medical community to question the use of biopsy, which can be likened to a snapshot of the whole tumor or a pathological slide that is only a cross-sectional image. Additionally, the differences in genetic backgrounds between primary and metastatic lesions make it less effective to use mutational profiles of primary tumors to guide targeted therapy for metastasis.
Temporal heterogeneity confers even greater difficulties for solid tumor biopsy because the changes in a tumor’s genome over time cannot be detected by a single test. The development of cancer can be compared to the evolution of species: somatic mutations that occur as early events during tumorigenesis tend to propagate in many or all clones, whereas later mutations exist only in some clones, thus forming heterogeneous populations within tumor lesions. As time goes on, the initiation of treatment (selective pressure), ITH in the tumor microenvironment, the emergence of drug-resistant subclones, selection of rare mutants by treatment, and the seeding of metastatic cells from subclones collectively contribute to the hierarchical tumor model [15], which is characterized by different genetic backgrounds at different times. Thus, a tumor genome in the later course of the disease might differ significantly from its initial status, and this difference cannot be recognized unless serial sampling is performed. However, repeated biopsy is rarely feasible, and without knowledge of genetic changes, the complete personalization of treatment and targeted therapy is impossible.
Liquid biopsy, conversely, is a non-invasive, dynamic means of capturing spatial and temporal tumor heterogeneity in a wide range of cancer types. For metastatic cancers, increasing evidence demonstrates that distant metastases harbor genomic aberrations different from those of the primary tumor, and that such aberrations might provide specific information for systemic or targeted therapy [16]. Moreover, recent work has demonstrated that different metastatic sites harbor different genomic aberrations, and biopsy of one or two accessible metastases may not be representative of every mutant profile [17]. Thus, liquid biopsy surveillance for ctDNA has been proposed as an alternative to metastasis biopsy for non-invasive genotyping across all tumor types [18] and dynamic sequencing of ctDNA suggests that it could be a novel, non-invasive cancer biomarker [19]. Indeed, sequencing liquid biopsies of plasma provides information comparable with that of metastatic biopsies [20], and, in non-small-cell lung carcinoma (NSCLC) patients, rare EGFR mutants absent in tumor samples can be detected in plasma [21]. The applications of liquid biopsy in clinical research and practice will be discussed thoroughly in a later part of this chapter.
Biomarkers Measured by Liquid Biopsy
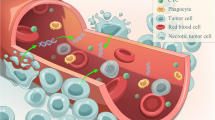
CTCs
CTCs are rare tumor cells shed from either the primary or metastatic tumor that circulates in the blood of cancer patients. The CTC research has flourished over the past decade, spanning fields including CTC capture, development of technology for characterization and isolation, identification of its prognostic significance for disease surveillance, and evaluation of treatment response and therapy choices [22]. CTCs have been detected in different cancers; the number of CTCs varies among malignancies but correlates significantly with tumor characteristics, patient prognosis, and disease progression [23]. More recently, the isolation of CTCs has become an alternative sampling method because these cells are shed randomly from multiple sites within primary and/or metastatic lesions and could, therefore, better represent tumor heterogeneity than any single tumor biopsy, providing additional opportunities to monitor therapeutic efficacy and predict metastatic potential [24]. The molecular analysis of CTCs, especially the genomic profiling of single CTCs, could provide information on the mutational landscape and clonal dominance, which might benefit patients in the form of targeted drugs. Moreover, longitudinal studies of clonal changes in mutant status during chemo- or targeted therapy that successively genotype CTCs demonstrate the utility of CTCs for monitoring the treatment response and discovering drug resistance mechanisms [25, 26].
Circulating Cell-Free Nucleic Acids
Circulating cell-free nucleic acids include circulating cell-free DNA (cfDNA) and microRNA (miRNA). The existence of cfDNA in bodily fluids, such as blood, urine, and cerebrospinal fluid, has long been recognized. cfDNA can readily be quantified and correlates with health status: patients with malignancies tend to have higher levels of cfDNA than normal individuals, and concentration of cfDNA is positively associated with tumor burden, suggesting the potential use of cfDNA concentration as a biomarker [27–29]. The size distribution of cfDNA also varies accordingly: due to more DNAs originated from apoptotic and necrotic cells, cfDNA molecules from cancer patients are often damaged and therefore much shorter than those of healthy patients or those with benign diseases, indicating that, with proper cutoff value, it is possible to use cfDNA length to identify malignancies [30]. Recently, extensive researches have focused on ctDNA. ctDNA belongs to cfDNAs and they differ in that while cfDNA contains copies released by normal cells, ctDNAs are DNA fragments originated only from tumor cells and could be detected by tumor-specific mutants [31]. ctDNA is currently gaining extensive attention [32] and has demonstrated itself useful as liquid biopsy in the early detection, surveillance, and personalized treatment of cancer [33].
Since the discovery of circulating cell-free miRNA, its use in liquid biopsy has received wide research interest. Although the precise origin of circulating miRNA remains incompletely understood, a number of studies have explored its practical utility, and these studies suggest the possibility of using circulating miRNA to detect cancers. The expression pattern of circulating miRNA differs between cancerous and normal cells, as well as among tumor types. Specific panels consisting of different miRNA combinations can be used as alternative diagnostic tools in a variety of cancers, such as breast cancer [34], NSCLC [35], and colorectal cancer (CRC) [36]. Furthermore, the concentration of circulating miRNAs in cancer patients can be calculated according to stable endogenous control of the miRNA miR-1228 [37], the close correlation between the concentration of circulating miRNA and tumor status enables non-invasive and dynamic monitoring of disease progression [38], and clinical outcomes can be predicted by circulating miRNA [39]. Thus, miRNA is a promising diagnostic tool, and further research on the mechanism of its release into bodily fluids will elucidate cancer biology.
Exosomes and Microvesicles
Extracellular vesicles (EVs) are vesicles widely present in bodily fluids. Exosomes are membrane-enclosed EVs formed during the inward budding of late endosomes that are subsequently released through exocytosis. Upon release, exosomes enter circulation and can be detected based on vesicular contents such as nucleic acids, enzymes, cytokines, and various soluble factors expressed during different pathophysiological statuses. Research on the use of a panel of several proteins and/or miRNAs to identify cancer cell-derived exosomes for diagnosis of different cancers indicates that the identification of exosomes and exosomal contents is a feasible and efficacious tool [40]. The similarity in gene expression between miRNA from circulating exosomes and lung tumor tissue suggests that circulating exosomes might be a useful means of detecting lung cancers [41]. Exosomal miRNA panels have also been reported for early diagnosis of other malignancies including acute myeloid leukemia [42], CRC [43], glioma [44], thyroid cancer [45], and esophageal adenocarcinoma [46]. Tumor-associated proteins may be enriched in tumor-derived exosomes, making it a precise biomarker for cancer detection [47]. Urinary exosomes differentially express proteins in bladder cancer patients, and a panel combining transmembrane protein 256 and late endosomal/lysosomaladaptor MAPK and MTOR activator 1 (LAMTOR1) reaches a detection sensitivity of 100 % [48]. In addition, exosomes have also been used for prognosis prediction [47, 49] and progression surveillance [44]. Unpublished data from our research group indicate that exosome may be a useful drug-delivery tool for liver cancer treatment: autologous exosomes assembled with synthetic small interfering RNA (siRNA) could inhibit the growth and metastasis of HCC in vivo and in vitro.
Technological Advances in Liquid Biopsy
The biomarkers for liquid biopsy are generally present in low levels, which cause the majority of them to be overwhelmed by normal physiological background noise. Indeed, CTCs are estimated to occur once among every 107 white blood cells, and the majority of cfDNA is released by normal blood cells without mutations, resulting in ctDNA prevalence as low as 0.1 % of all cfDNA. Furthermore, miRNAs and exosomes originate from both normal and malignant cells. Thus, effective separation and enrichment methods with high specificity and practicability are of primary importance in liquid biopsy.
Isolation of CTCs
CTCs are admixed with billions of blood cells; this extreme rarity is the key technical challenge in the research on their implementation as biomarkers. A vast array of technology has been developed to isolate CTCs, mostly by jointly using the principles of enrichment and detection. Because controversy exists around the numbers, molecular and biological properties, and significance of CTCs in the natural history of cancer, no gold standard has yet been established to measure the efficiency of different approaches in isolating CTCs.
However, immunomagnetic capture is a successful and widely-accepted CTC enrichment approach. Based on the hypothesis that CTCs express tumor-specific cell surface markers and morphological characteristics but lack leukocyte marker CD45, the CellSearch system (Janssen Diagnostics), the only Food and Drug Administration-approved CTC device, uses ferrofluids loaded with an epithelial cell adhesion molecule (EpCAM) antibody to capture and define CTCs [50]. This platform has undergone a full validation for reproducibility and performance characteristics in large-scale multicenter trials and is currently used for prognostics in breast, prostate, and CRC [51]. Other enrichment tools include the CTC-iChip, Herringbone chip, MagSweeper, and IsoFlux [52]. The CTC-iChip, which captures viable CTCs not by cell surface markers but by simply removing the blood components, appears to have the broadest applications for cancer patients [53]. Considering that the morphology and marker signature of CTCs might change upon release into circulation, technology platforms that use marker-independent enrichment methods have received increasing attention. For example, physical properties of CTCs, namely the difference in buoyant density relative to red blood cells and larger size than leukocytes, have been exploited to isolate CTCs using gradient centrifugation and filtration-based approaches [54, 55]. Additional innovative approaches include quantitative real-time PCR (qRT-PCR)-based platforms [56], fiber-optic array scanning technology [57], microfluidic technology [58], etc. Because wild-type DNA and RNA from leukocytes isolated during CTC enrichment represent a significant technical hurdle for the molecular profiling of rare cells, novel methods such as the DEPArray (Silicon Biosystems) employ physical micromanipulation to trap and move single or groups of CTCs into a separate container for further analysis [59].
Extraction and Pre-Analytical Procedures of Circulating Nucleic Acids and Vesicles
Cell-free nucleic acids and exosomes circulate in blood at low levels, necessitating the standardization of extraction procedures. Plasma is a better source for cfDNA than serum because contamination from blood cells during the clotting process in serum can increase the background noise of wild-type DNA and lower the detection positivity of ctDNA. Similarly, the intercellular miRNA trafficking during the coagulation process also leads to large differences in circulating miRNA profiles between serum and plasma. Although the impact of the coagulation process on cell-free miRNAs is unknown, serum is currently more commonly used for cell-free miRNA and vesicle research [60].
The pre-analytical procedures also significantly influence the performance of these plasma-based biomarkers [61, 62], including time between blood sampling and isolation procedure, centrifugation conditions, time between plasma cryopreservation and extraction, and kits or methods used for extraction. Variation among quantification methods, including spectrophotometric methods, fluorescent dyes, or quantitative PCR-based methods, also affects the performance of nucleic acids and vesicles as biomarkers due to different measurement ranges and operating principles. This variation makes results difficult to compare across studies. Moreover, little is known about the origins, release, and degradation of nucleic acids and exosomes. Thus, their utility for liquid biopsy is complicated by potential bias from confounding events, such as nonmalignant diseases, inflammation, heavy smoking, pregnancy, exercise, and blood volume.
NGS
The development of NGS technology has illuminated the interpretation of cancer genomes and, more recently, accelerated the implementation of liquid biopsy. Although traditional Sanger sequencing is not suitable for liquid biopsy due to its low mutation detection threshold and throughput, the sequencing of whole genomes, whole exomes, or targeted regions using deep sequencing of plasma DNA has identified mutations from cfDNA that represent those of tumor tissues. Although whole genome and whole exome sequencing could provide more comprehensive information regarding the mutant status of ctDNA, targeted region deep sequencing, which focuses on a fewer gene loci with ultra-deep sequencing, has gained popularity. The sequencing region can be customized according to sequencing purpose, cancer types, costs, and turnaround time [63, 64]. Single-cell sequencing of CTCs with NGS is now increasingly performed, which allows rapid, sensitive, and non-invasive assessment of tumor genotype in CRC [65], NSCLC [66], and breast cancer [67] patients.
Digital PCR (dPCR)
The concept of absolute quantification of nucleic acids at the single-molecule level was first introduced in 1992 by Sykes et al. [68] forming the basic principle of dPCR: templates are diluted and distributed into individual reactions so that only one target molecule per reaction partition is examined. Although this is a theoretical goal, in practice, some partitions could contain more than one template, requiring Poisson statistics to correct the distortion [69]. Compared to qRT-PCR, dPCR does not rely on external calibrants, has a higher tolerance for enzyme-inhibiting substances, and has significantly higher sensitivity [70]. Compartmentalization of templates reduces noise from background DNA, thus offering superior precision and reproducibility to qPCR. Furthermore, the development of multiplex dPCR enables the simultaneous detection of several targets in one reaction, which is useful when limited samples are available. dPCR has wide applicability in the liquid biopsy of cancer, especially in combination with circulating nucleic acids: it permits the detection of rare mutations within ctDNA to guide targeted therapy, absolute quantification of copy number variations to predict disease progression, measurement of changes in gene expression to diagnosis of cancer early, and assessment of methylation status to identify epigenetic dysregulation during carcinogenesis [71].
Clinical Utility of Liquid Biopsy
An Alternative to Traditional Sampling Methods
Although the standard currently used for profiling cancer genomes is the sequencing of tumor tissues, obtaining this tissue can be difficult or impossible, especially in cases of metastatic cancers, because the genomics of primary tumors and metastases are not always concordant. Even if a tumor can be biopsied, some inherent limitations are inevitable: a tissue sample only represents a single snapshot in time and is subjected to spatial selection bias of ITH. Moreover, chemotherapeutic agents and targeted drugs, acting as selective pressures, alter and complicate the molecular landscape during treatment [72]. We know a little of these dynamics because repeated sampling is impossible, and, thus, we cannot accurately monitor disease progression and treatment response. Ultimately, the absence of available tumor tissues and the resulting lack of mutant information can lead to targeted treatment failure.
Fortunately, liquid biopsy can solve these issues inherent to solid tumor biopsy, including spatial and temporal heterogeneity. For example, in a metastatic melanoma patient, who was ineligible for BRAF inhibitor-based therapy because no viable tumor cells from a biopsy could be found for mutation testing [73], examination of ctDNA detected a BRAF V600E mutation. Such findings buttress the notion that directly investigating plasma, instead of the primary tumor, can identify actionable mutations. For patients with pancreatic and biliary carcinomas, personalized treatment options are few, in part because adequate biopsies cannot be obtained for molecular characterization. In this setting, cfDNA sequencing reliably and accurately detects tumor-derived mutations with 97.7 % diagnostic accuracy, 92.3 % sensitivity, and 100 % specificity [74]. In CRC patients, liquid biopsy has the potential to replace tumor sampling; analysis of plasma DNA identified mutations that were not detected in matched tumor tissue, indicating that ctDNA can more comprehensively capture intra-patient disease heterogeneity [75]. Similarly, among NSCLC patients whose tumor tissues were EGFR-negative, cfDNA samples indicated EGFR mutations that made them eligible for TKI treatment [76].
Circulating miRNAs are another non-invasive sampling tool that can be used in lieu of biopsied tissues. Indeed, reduced or increased levels of circulating miRNAs have been reported in cancers, and combinations of specific miRNAs have differential significance [77]. Combining several miRNAs in a panel can effectively distinguish colitis-associated cancer from ulcerative colitis [78] and a targeted miRNAs panel (miR-19a, −98, −146b, −186, −191, −222, −331-5p, −452, −625, −664, and −1247) shows significant expression differences among normal patients versus those with polyps or cancer [36]. miRNA panels are also superior to carcinoembryonic antigen (CEA) and carbohydrate antigen 19–9 (CA19-9) for the diagnosis of early-stage CRC [79]. Similar findings using circulating miRNA panels as tools for early disease diagnosis have also been reported in other malignancies, such as breast cancer [28], gastric cancer [80], pancreatic cancer [81], and lung cancer [82]. We have previously identified a microRNA panel (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801) with high diagnostic accuracy for HCC; this panel could differentiate HCC from healthy patients, those with chronic hepatitis B and those with cirrhosis [83].
Early Detection and Prediction of Tumor Progression
Compared to traditional imaging and serum-based biomarkers, liquid biopsy shows higher sensitivity in the early detection of tumor progression. ctDNA-based detection preceded the clinical detection of metastasis in 86 % of patients, with an average lead time of 11 months. A recent study revealed that detection of glypican-1 positive exosomes non-invasively identifies late-stage pancreatic cancer with 100 % sensitivity and distinguishes patients with precancerous pancreatic lesions from those with benign pancreatic diseases [47]. For malignancies such as gastric cancer, effective modalities for disease progression surveillance are rare: serological biomarkers, such as CA19-9 and CEA, are not sensitive, and CT has difficulty detecting minimal residual disease and peritoneal dissemination, the latter of which is the most frequent recurrent pattern in gastric cancer. However, Hamakawa et al. found that concentrations of ctDNA decrease after surgical resection and correlate with the disease status of gastric cancer patients, suggesting the utility of this approach for detection of residual disease and monitoring for recurrence [84]. Similarly, diffuse large B-cell lymphoma, though curable, results in poor outcomes upon treatment failure, but imaging-based surveillance is imprecise and radiation exposure is potentially risky. ctDNA can identify patients’ risk of recurrence at a median of 3–5 months before clinical disease is evident, which enhances the likelihood of reducing disease burden through chemotherapy and improving survival [85]. Similarly, the detection of ctDNA by liquid biopsy in pancreatic cancer patients after curative resection predicts recurrence 6.5 months earlier than CT imaging [86]; in metastatic CRC (mCRC) treated by chemotherapy, a reduction in circulating ctDNA precedes tumor shrinkage visible by CT imaging, which indicates the utility of ctDNA as a biomarker to evaluate the treatment response in a timely fashion in order to make therapeutic decisions [87].
Unveiling Drug Resistance and Guiding Treatment Choice
For advanced diseases, chemotherapy and targeted monoclonal antibodies or inhibitors are commonly used as first-line treatment; however, drug resistance often occurs, leading to treatment failure. The mechanisms of oncological drug resistance are largely unknown because repeated biopsies to study genomic evolution following therapy are difficult, invasive, and limited by heterogeneity. However, liquid biopsy provides some information that solid tumor tissue cannot. In NSCLC patients harboring activating somatic mutations in EGFR, TKIs such as gefitinib and erlotinib are effective at initial treatment; however, acquired resistance usually develops after a median of 9–14 months [88, 89]. Though a few EGFR mutations in plasma DNA have been detected prior to treatment [76], the presence of which can predict tumor response in NSCLC patients [90], de novo mutations of KRAS, NRAS, and BRAF and amplification of Erb-B2 receptor tyrosine kinase 2 (ERBB2) and MET might drive resistance to anti-EGFR treatment. Sequencing plasma DNA before and after the development of drug resistance indicates that, upon developing TKI resistance, new mutations in EGFR, such as a C797S mutation, emerge, and cause treatment failure [91]. Similarly, the detection of T790M mutation in CTCs from patients with EGFR mutations who have received TKIs is predictive of reduced progression-free survival [92].
Despite the prevalence of newly-acquired mutations that confer resistance to treatment, pre-existing rare clones within the primary tumor can also result in cancer refractory to anti-EGFR treatment. Measurement of KRAS and EGFR mutation status in plasma DNA of posttreatment mCRC patients indicates that drug resistance-associated mutants are indeed derived from rare, pre-existing clones in the primary tumors [93]. Though these mutants occur at very low frequencies prior to EGFR blockade therapy, they become evident 22 weeks after initiation of treatment, the time interval required for the mutant subclone to repopulate the lesion and lead to recurrence [94]. As a result, evaluating the serial plasma DNA mutation status during monoclonal antibody treatment suggests that combination therapies targeting multiple pathways and timely adjustments of therapeutic regimens can delay or prevent disease progression [95].
Furthermore, quantitative and genetic characterization of CTCs can also guide treatment choice. A decrease in total CTCs suggests response to therapy and predicts longer overall survival in lung cancer patients [96], whereas an increase in CTCs after chemotherapy is associated with shorter overall survival in patients with breast cancer and CRC [97, 98]. The positive HER2 status of CTCs in breast cancer patients whose primary tumors were HER2-negative indicates a benefit from trastuzumab therapy [99]. Moreover, the results from the SWOG 0500 trial found that for breast cancer patients with persistently increased CTCs after 21 days of first-line chemotherapy, an early switch to an alternative cytotoxic therapy was not effective in prolonging overall survival, indicating that this population needs more effective treatment than standard chemotherapy [100].
Confronting Issues of Liquid Biopsy
Several issues must be solved before liquid biopsy can be widely implemented. First, the pre-analytical conditions of sampling should be defined, including the markers used for CTC enrichment in different cancers, optimization and standardization of circulating DNA and miRNA extraction and quantification, and selection of the proper method of mutation analysis. Second, the high costs of NGS and low throughput of dPCR must be taken into account when selecting either method. Although NGS can provide broader mutation detection than dPCR, the high cost, long turnover time, and need for professional operators limit its popularity. In contrast, the benefit of high sensitivity, low cost, and wide applicability of dPCR is discounted by its low throughput and inability to discover unknown mutants. Moreover, the ultimate goal of liquid biopsy is to aid clinicians in making efficient treatment decisions; thus, clinicians using liquid biopsy must be capable of accurate interpretation of testing results and its translational significance. Collaboration between industry, research institutes, and clinical centers will be critical for solving such issues.
Liquid Biopsy in HCC
Although the role of liquid biopsy in HCC has not been widely and thoroughly evaluated, it has utility for the early diagnosis and prognosis of HCC patients. The presence of CTCs in HCC patients correlates with tumor invasion and is a predictor of reduced survival [101]. A variety of enrichment and detection methods can detect CTCs. Using the CellSearch platform, Sun et al. found that 66.67 % of HCC patients have detectable EpCAM-positive CTCs prior to surgery, which decreases significantly 1 month after surgery; moreover, preoperative CTC ≥ 2 per 7.5 ml blood was a positive predictor of recurrence [102]. Intercellular adhesion molecule 1 (ICAM-1) can also be used as a biomarker to detect CTC in HCC; elevated numbers of CD45 (−) ICAM-1 (+) cells is predictive of poorer clinical outcomes [103]. In a highly metastatic orthotopic nude mouse model, the number of CTCs strongly correlates with disease progression and treatment response to sorafenib [104]. Furthermore, a recent study demonstrated that pretreatment CTC level has prognostic significance not only in HCC patients who have undergone surgery, but also in those who have received transcatheter arterial chemoembolization and radiotherapy [105].
Apart from the quantification of CTCs, recent research has paid more attention to their genetic characterization. Sequencing of CTC DNA could serve as an alternative to solid tumor biopsy to identify known HCC mutations, especially for low-frequency variants [106]. Furthermore, single-cell sequencing of CTCs provides genomic profiling for copy number or mutations, which might better describe the landscape of a single-cell genome. This information could enrich our knowledge about the genomic amplifications of oncogenes and deletions of tumor suppressors, supplying clinicians with the information necessary to direct therapy or monitor disease following treatment [107].
In addition to the quantification and characterization of CTCs, circulating cell-free nucleic acids are promising biomarkers for diagnosis and prognosis in HCC [108]. Methylation of p15 and p16 is present and concordant in plasma and tumor tissue, but not in controls with chronic hepatitis/cirrhosis or healthy subjects [109, 110]. Furthermore, p16INK4a [111], RASSF1A [112], and INK4A [113] are hypermethylated in plasma and useful for HCC screening, detection, and prognostication. Thus, sequencing of plasma cfDNA to identify genomic copy variations could help to identify patients at high risk for HCC among individuals with chronic liver diseases [114]. The 249 (Ser) p53 mutation, a “hotspot” mutation in HCC patients exposed to aflatoxin B1 and hepatitis B virus, can be readily identified in plasma, facilitating the early diagnosis of HCC [115–117]. For patients undergoing liver transplantation, genetic variations of rs894151 and rs12438080 in pre-transplant circulating cfDNA effectively predict tumor recurrence and identify a subgroup of patients with low risk of post-transplant recurrence despite exceeding the Milan criteria [118]. Beyond the presence or absence of such mutations, cfDNA concentration is also prognostic in HCC. cfDNA concentration significantly differs between patients with HCC and liver cirrhosis, and higher cfDNA levels predict advanced tumor status and poor survival of HCC patients [119, 120]. Furthermore, increased levels of cfDNA are observed in hepatitis C virus-associated HCC [121] and predict inflammatory status prior to curative surgery [122] and distant metastasis afterwards [123].
The quantity of circulating miRNA is another aspect of liquid biopsy that facilitates early diagnosis of HCC [124]. A plasma miRNA panel consisting of miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801 accurately differentiates HCC from healthy, chronic hepatitis B, and cirrhosis patients [83]. Similarly, the combination of circulating miR-122 and let-7b differentiates early HCC from dysplastic nodules in chronic hepatitis B patients [125], and the panel of miR-19a, miR-195, miR-192, and miR-146a has high accuracy in the early diagnosis of hepatitis C virus-related HCC [126]. Other circulating miRNAs with diagnostic or prognostic value include miRNA-16 [127], miRNA-101 [128], and miRNA-21 and 199-a [129, 130] are also reported. Finally, plasma miRNA predicts the success of liver transplantation in HCC patients [131].
In addition, miRNA extracted from circulating exosomes also has diagnostic value [132] and is predictive of HCC recurrence after liver transplantation [133]. The combination of miRNA with established markers, such as AFP, has better diagnostic performance than conventional markers alone [127]. Our research group has found that low expression of miR-26a in HCC tissues predicts sensitivity to interferon after hepatectomy [134], and an ongoing study is evaluating the ability of miR-26a expression in serum to guide interferon therapy for the prevention of postoperative recurrence and metastasis. However, caution should be taken when these results are applied clinically because many factors influence circulating miRNA levels [135].
Conclusion
Liquid biopsy, an application of blood-based analysis for cancer diagnosis, surveillance, and prognosis, has broad utility in the management of cancer. The major strengths of liquid biopsy include genotyping a tumor’s mutant landscape, the feasibility of serial sampling to generate a dynamic profile of disease progression, and the practicability of monitoring treatment response, unveiling drug resistance, and making therapeutic choice in real-time. The non-invasiveness and high specificity of liquid biopsy, as well as the homogeneous nature of blood, make liquid biopsy an ideal tool for modern oncological research and practice. The widespread use of liquid biopsy in HCC is on the horizon in this era of precision medicine.
References
Kruglyak KM, Lin E, Ong FS. Next-generation sequencing in precision oncology: challenges and opportunities. Expert Rev Mol Diagn. 2014;14(6):635–7.
Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10(8):472–84.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92.
Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46(3):225–33.
Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–9.
de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science (New York, NY). 2014;346(6206):251–6.
Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science (New York, NY). 2014;346(6206):256–9.
Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5):458–68.
Schwarz RF, Ng CK, Cooke SL, Newman S, Temple J, Piskorz AM, et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: a phylogenetic analysis. PLoS Med. 2015;12(2):e1001789.
Friemel J, Rechsteiner M, Frick L, Bohm F, Struckmann K, Egger M, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res: Off J Am Assoc Cancer Res. 2015;21(8):1951–61.
Gawad C, Koh W, Quake SR. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci U S A. 2014;111(50):17947–52.
Landau DA, Clement K, Ziller MJ, Boyle P, Fan J, Gu H, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26(6):813–25.
Oakes CC, Claus R, Gu L, Assenov Y, Hullein J, Zucknick M, et al. Evolution of DNA methylation is linked to genetic aberrations in chronic lymphocytic leukemia. Cancer Discov. 2014;4(3):348–61.
Jamal-Hanjani M, Quezada SA, Larkin J, Swanton C. Translational implications of tumor heterogeneity. Clin Cancer Res: Off J Am Assoc Cancer Res. 2015;21(6):1258–66.
McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27(1):15–26.
McCreery MQ, Halliwill KD, Chin D, Delrosario R, Hirst G, Vuong P, et al. Evolution of metastasis revealed by mutational landscapes of chemically induced skin cancers. Nature Med. 2015;21(12):1514–20.
Alix-Panabieres C, Pantel K. Real-time liquid biopsy: circulating tumor cells versus circulating tumor DNA. Ann Transl Med. 2013;1(2):18.
Lebofsky R, Decraene C, Bernard V, Kamal M, Blin A, Leroy Q, et al. Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol. 2015;9(4):783–90.
Lianos GD, Mangano A, Kouraklis G, Roukos DH. Dynamic sequencing of circulating tumor DNA: novel noninvasive cancer biomarker. Biomark Med. 2014;8(5):629–32.
Rothe F, Laes JF, Lambrechts D, Smeets D, Vincent D, Maetens M, et al. Plasma circulating tumor DNA as an alternative to metastatic biopsies for mutational analysis in breast cancer. Ann Oncol: Off J Eur Soc Med Oncol / ESMO. 2014;25(10):1959–65.
Zhu G, Ye X, Dong Z, Lu Y, Sun Y, Liu Y, et al. Highly sensitive droplet digital PCR method for detection of EGFR activating mutations in plasma cell-free DNA from patients with advanced Non-small cell lung cancer. J Mol Diagn: JMD. 2015;17(3):265–72.
Pantel K, Speicher MR. The biology of circulating tumor cells. Oncogene. 2015. doi:10.1038/onc.2015.192.
Yap TA, Lorente D, Omlin A, Olmos D, de Bono JS. Circulating tumor cells: a multifunctional biomarker. Clin Cancer Res: Off J Am Assoc Cancer Res. 2014;20(10):2553–68.
Caceres G, Puskas JA, Magliocco AM. Circulating tumor cells: a window into tumor development and therapeutic effectiveness. Cancer Control: J Moffitt Cancer Cent. 2015;22(2):167–76.
Schneck H, Blassl C, Meier-Stiegen F, Neves RP, Janni W, Fehm T, et al. Analysing the mutational status of PIK3CA in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. 2013;7(5):976–86.
Polzer B, Medoro G, Pasch S, Fontana F, Zorzino L, Pestka A, et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol Med. 2014;6(11):1371–86.
Hao TB, Shi W, Shen XJ, Qi J, Wu XH, Wu Y, et al. Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer. 2014;111(8):1482–9.
Shin VY, Siu JM, Cheuk I, Ng EK, Kwong A. Circulating cell-free miRNAs as biomarker for triple-negative breast cancer. Br J Cancer. 2015;112(11):1751–9.
Singh N, Gupta S, Pandey RM, Chauhan SS, Saraya A. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Investig. 2015;33(3):78–85.
Zonta E, Nizard P, Taly V. Assessment of DNA integrity, applications for cancer research. Adv Clin Chem. 2015;70:197–246.
Sausen M, Parpart S, Diaz Jr LA. Circulating tumor DNA moves further into the spotlight. Genome Med. 2014;6(5):35.
Bidard FC, Weigelt B, Reis-Filho JS. Going with the flow: from circulating tumor cells to DNA. Sci Transl Med. 2013;5(207):207ps14.
Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61(1):112–23.
Matamala N, Vargas MT, Gonzalez-Campora R, Minambres R, Arias JI, Menendez P, et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61(8):1098–106.
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol: Off J Am Soc Clin Oncol. 2010;28(10):1721–6.
Verma AM, Patel M, Aslam MI, Jameson J, Pringle JH, Wurm P, et al. Circulating plasma microRNAs as a screening method for detection of colorectal adenomas. Lancet. 2015;385 Suppl 1:S100.
Hu J, Wang Z, Liao BY, Yu L, Gao X, Lu S, et al. Human miR-1228 as a stable endogenous control for the quantification of circulating microRNAs in cancer patients. Int J Cancer. 2014;135(5):1187–94.
Aushev VN, Zborovskaya IB, Laktionov KK, Girard N, Cros MP, Herceg Z, et al. Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS One. 2013;8(10):e78649.
Nair VS, Maeda LS, Ioannidis JP. Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Inst. 2012;104(7):528–40.
Thery C. Cancer: diagnosis by extracellular vesicles. Nature. 2015;523(7559):161–2.
Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–6.
Hornick NI, Huan J, Doron B, Goloviznina NA, Lapidus J, Chang BH, et al. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci Rep. 2015;5:11295.
Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113(2):275–81.
Shi R, Wang PY, Li XY, Chen JX, Li Y, Zhang XZ, et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget. 2015;6(29):26971–81.
Lee JC, Zhao JT, Gundara J, Serpell J, Bach LA, Sidhu S. Papillary thyroid cancer-derived exosomes contain miRNA-146b and miRNA-222. J Surg Res. 2015;196(1):39–48.
Chiam K, Wang T, Watson DI, Mayne GC, Irvine TS, Bright T, et al. Circulating serum exosomal miRNAs as potential biomarkers for esophageal adenocarcinoma. J Gastrointest Surg: Off J Soc Surg Aliment Tract. 2015;19(7):1208–15.
Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–82.
Overbye A, Skotland T, Koehler CJ, Thiede B, Seierstad T, Berge V, et al. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget. 2015;6(30):30357–76.
Tokuhisa M, Ichikawa Y, Kosaka N, Ochiya T, Yashiro M, Hirakawa K, et al. Exosomal miRNAs from peritoneum lavage fluid as potential prognostic biomarkers of peritoneal metastasis in gastric cancer. PLoS One. 2015;10(7):e0130472.
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res: Off J Am Assoc Cancer Res. 2004;10(20):6897–904.
Coumans F, Terstappen L. Detection and characterization of circulating tumor cells by the CellSearch approach. Methods Mol Biol (Clifton, NJ). 2015;1347:263–78.
Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells-biology and biomarkers. Nat Rev Clin Oncol. 2014;11(3):129–44.
Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5(179):179ra47.
Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450(7173):1235–9.
Hou HW, Warkiani ME, Khoo BL, Li ZR, Soo RA, Tan DS, et al. Isolation and retrieval of circulating tumor cells using centrifugal forces. Sci Rep. 2013;3:1259.
He W, Wang H, Hartmann LC, Cheng JX, Low PS. In vivo quantitation of rare circulating tumor cells by multiphoton intravital flow cytometry. Proc Natl Acad Sci U S A. 2007;104(28):11760–5.
Somlo G, Lau SK, Frankel P, Hsieh HB, Liu X, Yang L, et al. Multiple biomarker expression on circulating tumor cells in comparison to tumor tissues from primary and metastatic sites in patients with locally advanced/inflammatory, and stage IV breast cancer, using a novel detection technology. Breast Cancer Res Treat. 2011;128(1):155–63.
Sarioglu AF, Aceto N, Kojic N, Donaldson MC, Zeinali M, Hamza B, et al. A microfluidic device for label-free, physical capture of circulating tumor cell clusters. Nat Methods. 2015;12(7):685–91.
Fabbri F, Carloni S, Zoli W, Ulivi P, Gallerani G, Fici P, et al. Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs. Cancer Lett. 2013;335(1):225–31.
He Y, Lin J, Kong D, Huang M, Xu C, Kim TK, et al. Current state of circulating microRNAs as cancer biomarkers. Clin Chem. 2015;61(9):1138–55.
McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57(6):833–40.
El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: preanalytical considerations. Clin Chim Acta; Int J Clin Chem. 2013;424:222–30.
Martinez P, McGranahan N, Birkbak NJ, Gerlinger M, Swanton C. Computational optimisation of targeted DNA sequencing for cancer detection. Sci Rep. 2013;3:3309.
Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20):20ra14.
Mohamed Suhaimi NA, Foong YM, Lee DY, Phyo WM, Cima I, Lee EX, et al. Non-invasive sensitive detection of KRAS and BRAF mutation in circulating tumor cells of colorectal cancer patients. Mol Oncol. 2015;9(4):850–60.
Marchetti A, Del Grammastro M, Felicioni L, Malatesta S, Filice G, Centi I, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One. 2014;9(8):e103883.
Pestrin M, Salvianti F, Galardi F, De Luca F, Turner N, Malorni L, et al. Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients. Mol Oncol. 2015;9(4):749–57.
Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13(3):444–9.
Hudecova I. Digital PCR, analysis of circulating nucleic acids. Clin Biochem. 2015;48(15):948–56.
Zhang BO, Xu CW, Shao Y, Wang HT, Wu YF, Song YY, et al. Comparison of droplet digital PCR and conventional quantitative PCR for measuring gene mutation. Exp Ther Med. 2015;9(4):1383–8.
Huggett JF, Cowen S, Foy CA. Considerations for digital PCR as an accurate molecular diagnostic tool. Clin Chem. 2015;61(1):79–88.
Stewart GD, O’Mahony F, Laird A, Eory L, Lubbock A, Mackay A, et al. Sunitinib treatment exacerbates intratumoral heterogeneity in metastatic renal cancer. Clin Cancer Res: Off J Am Assoc Cancer Res. 2015;21(18):4212–23.
Tsao SC, Weiss J, Hudson C, Christophi C, Cebon J, Behren A, et al. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci Rep. 2015;5:11198.
Zill OA, Greene C, Sebisanovic D, Siew L, Leng J, Vu M, et al. Cell-free DNA next-generation sequencing in pancreatobiliary carcinomas. Cancer Discov. 2015;5(10):1040–8.
Siravegna G, Mussolin B, Buscarino M, Corti G, Cassingena A, Crisafulli G, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21(7):795–801.
Watanabe M, Kawaguchi T, Isa SI, Ando M, Tamiya A, Kubo A, et al. Ultra-sensitive detection of the pretreatment EGFR T790M mutation in non-small-cell lung cancer patients with an EGFR-activating mutation using droplet digital PCR. Clin Cancer Res: Off J Am Assoc Cancer Res. 2015;21(15):3552–60.
Mishra PJ. MicroRNAs as promising biomarkers in cancer diagnostics. Biomarker Res. 2014;2:19.
Patel M, Verma A, Aslam I, Pringle H, Singh B. Novel plasma microRNA biomarkers for the identification of colitis-associated carcinoma. Lancet. 2015;385 Suppl 1:S78.
Fang Z, Tang J, Bai Y, Lin H, You H, Jin H, et al. Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res: CR. 2015;34(1):86.
Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang F, et al. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Sci Rep. 2015;5:11251.
Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311(4):392–404.
Li N, Ma J, Guarnera MA, Fang H, Cai L, Jiang F. Digital PCR quantification of miRNAs in sputum for diagnosis of lung cancer. J Cancer Res Clin Oncol. 2014;140(1):145–50.
Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol: Off J Am Soc Clin Oncol. 2011;29(36):4781–8.
Hamakawa T, Kukita Y, Kurokawa Y, Miyazaki Y, Takahashi T, Yamasaki M, et al. Monitoring gastric cancer progression with circulating tumour DNA. Br J Cancer. 2015;112(2):352–6.
Roschewski M, Dunleavy K, Pittaluga S, Moorhead M, Pepin F, Kong K, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 2015;16(5):541–9.
Sausen M, Phallen J, Adleff V, Jones S, Leary RJ, Barrett MT, et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat Commun. 2015;6:7686.
Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol: Off J Eur Soc Med Oncol / ESMO. 2015;26(8):1715–22.
Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361(10):958–67.
Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13(3):239–46.
Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol: Off J Am Soc Clin Oncol. 2009;27(16):2653–9.
Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21(6):560–2.
Maheswaran S, Sequist LV, Nagrath S, Ulkus L, Brannigan B, Collura CV, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359(4):366–77.
Morelli MP, Overman MJ, Dasari A, Kazmi SM, Mazard T, Vilar E, et al. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol: Off J Eur Soc Med Oncol / ESMO. 2015;26(4):731–6.
Diaz Jr LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–40.
Mohan S, Heitzer E, Ulz P, Lafer I, Lax S, Auer M, et al. Changes in colorectal carcinoma genomes under anti-EGFR therapy identified by whole-genome plasma DNA sequencing. PLoS Genet. 2014;10(3):e1004271.
Hiltermann TJ, Pore MM, van den Berg A, Timens W, Boezen HM, Liesker JJ, et al. Circulating tumor cells in small-cell lung cancer: a predictive and prognostic factor. Ann Oncol: Off J Eur Soc Med Oncol / ESMO. 2012;23(11):2937–42.
Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, et al. Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res: Off J Am Assoc Cancer Res. 2006;12(14 Pt 1):4218–24.
Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol: Off J Am Soc Clin Oncol. 2008;26(19):3213–21.
Liu Y, Liu Q, Wang T, Bian L, Zhang S, Hu H, et al. Circulating tumor cells in HER2-positive metastatic breast cancer patients: a valuable prognostic and predictive biomarker. BMC Cancer. 2013;13:202.
Smerage JB, Barlow WE, Hortobagyi GN, Winer EP, Leyland-Jones B, Srkalovic G, et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol: Off J Am Soc Clin Oncol. 2014;32(31):3483–9.
Zhang Y, Li J, Cao L, Xu W, Yin Z. Circulating tumor cells in hepatocellular carcinoma: detection techniques, clinical implications, and future perspectives. Semin Oncol. 2012;39(4):449–60.
Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ, et al. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology (Baltimore, Md). 2013;57(4):1458–68.
Liu S, Li N, Yu X, Xiao X, Cheng K, Hu J, et al. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology. 2013;144(5):1031–41.e10.
Yan J, Fan Z, Wu X, Xu M, Jiang J, Tan C, et al. Circulating tumor cells are correlated with disease progression and treatment response in an orthotopic hepatocellular carcinoma model. Cytometry A: J Int Soc Anal Cytol. 2015;87(11):1020–8.
Guo W, Yang XR, Sun YF, Shen MN, Ma XL, Wu J, et al. Clinical significance of EpCAM mRNA-positive circulating tumor cells in hepatocellular carcinoma by an optimized negative enrichment and qRT-PCR-based platform. Clin Cancer Res: Off J Am Assoc Cancer Res. 2014;20(18):4794–805.
Kelley RK, Magbanua MJ, Butler TM, Collisson EA, Hwang J, Sidiropoulos N, et al. Circulating tumor cells in hepatocellular carcinoma: a pilot study of detection, enumeration, and next-generation sequencing in cases and controls. BMC Cancer. 2015;15:206.
Azvolinsky A. Beyond counting: new way to use circulating tumor cells. J Natl Cancer Inst. 2014;106(10):dju343.
Zhou J, Shi YH, Fan J. Circulating cell-free nucleic acids: promising biomarkers of hepatocellular carcinoma. Semin Oncol. 2012;39(4):440–8.
Wong IH, Lo YM, Zhang J, Liew CT, Ng MH, Wong N, et al. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59(1):71–3.
Wong IH, Lo YM, Yeo W, Lau WY, Johnson PJ. Frequent p15 promoter methylation in tumor and peripheral blood from hepatocellular carcinoma patients. Clin Cancer Res: Off J Am Assoc Cancer Res. 2000;6(9):3516–21.
Wong IH, Zhang J, Lai PB, Lau WY, Lo YM. Quantitative analysis of tumor-derived methylated p16INK4a sequences in plasma, serum, and blood cells of hepatocellular carcinoma patients. Clin Cancer Res: Off J Am Assoc Cancer Res. 2003;9(3):1047–52.
Chan KC, Lai PB, Mok TS, Chan HL, Ding C, Yeung SW, et al. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem. 2008;54(9):1528–36.
Huang G, Krocker JD, Kirk JL, Merwat SN, Ju H, Soloway RD, et al. Evaluation of INK4A promoter methylation using pyrosequencing and circulating cell-free DNA from patients with hepatocellular carcinoma. Clin Chem Lab Med: CCLM / FESCC. 2014;52(6):899–909.
Xu H, Zhu X, Xu Z, Hu Y, Bo S, Xing T, et al. Non-invasive analysis of genomic copy number variation in patients with hepatocellular carcinoma by next generation DNA sequencing. J Cancer. 2015;6(3):247–53.
Huang XH, Sun LH, Lu DD, Sun Y, Ma LJ, Zhang XR, et al. Codon 249 mutation in exon 7 of p53 gene in plasma DNA: maybe a new early diagnostic marker of hepatocellular carcinoma in Qidong risk area, China. World J Gastroenterol: WJG. 2003;9(4):692–5.
Szymanska K, Lesi OA, Kirk GD, Sam O, Taniere P, Scoazec JY, et al. Ser-249TP53 mutation in tumour and plasma DNA of hepatocellular carcinoma patients from a high incidence area in the Gambia, West Africa. Int J Cancer J Int Cancer. 2004;110(3):374–9.
Kirk GD, Lesi OA, Mendy M, Szymanska K, Whittle H, Goedert JJ, et al. 249(ser) TP53 mutation in plasma DNA, hepatitis B viral infection, and risk of hepatocellular carcinoma. Oncogene. 2005;24(38):5858–67.
Hu J, Wang Z, Fan J, Dai Z, He YF, Qiu SJ, et al. Genetic variations in plasma circulating DNA of HBV-related hepatocellular carcinoma patients predict recurrence after liver transplantation. PLoS One. 2011;6(10):e26003.
Ren N, Qin LX, Tu H, Liu YK, Zhang BH, Tang ZY. The prognostic value of circulating plasma DNA level and its allelic imbalance on chromosome 8p in patients with hepatocellular carcinoma. J Cancer Res Clin Oncol. 2006;132(6):399–407.
Ren N, Ye QH, Qin LX, Zhang BH, Liu YK, Tang ZY. Circulating DNA level is negatively associated with the long-term survival of hepatocellular carcinoma patients. World J Gastroenterol: WJG. 2006;12(24):3911–4.
Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, Stark M, et al. Elevated levels of circulating cell-free DNA in the blood of patients with hepatitis C virus-associated hepatocellular carcinoma. Anticancer Res. 2006;26(6c):4713–9.
Iida M, Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, et al. Relation between serum levels of cell-free DNA and inflammation status in hepatitis C virus-related hepatocellular carcinoma. Oncol Rep. 2008;20(4):761–5.
Tokuhisa Y, Iizuka N, Sakaida I, Moribe T, Fujita N, Miura T, et al. Circulating cell-free DNA as a predictive marker for distant metastasis of hepatitis C virus-related hepatocellular carcinoma. Br J Cancer. 2007;97(10):1399–403.
Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology (Baltimore, Md). 2013;57(2):840–7.
Hung CH, Hu TH, Lu SN, Kuo FY, Chen CH, Wang JH, et al. Circulating microRNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int J Cancer J Int Cancer. 2015. doi:10.1002/ijc.29802.
Motawi TK, Shaker OG, El-Maraghy SA, Senousy MA. Serum microRNAs as potential biomarkers for early diagnosis of hepatitis C virus-related hepatocellular carcinoma in Egyptian patients. PLoS One. 2015;10(9):e0137706.
Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol. 2011;45(4):355–60.
Fu Y, Wei X, Tang C, Li J, Liu R, Shen A, et al. Circulating microRNA-101 as a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Oncol Lett. 2013;6(6):1811–5.
Wang H, Hou L, Li A, Duan Y, Gao H, Song X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. BioMed Res Int. 2014;2014:864894.
Amr KS, Ezzat WM, Elhosary YA, Hegazy AE, Fahim HH, Kamel RR. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene. 2015;575(1):66–70.
Hu J, Wang Z, Tan CJ, Liao BY, Zhang X, Xu M, et al. Plasma microRNA, a potential biomarker for acute rejection after liver transplantation. Transplantation. 2013;95(8):991–9.
Sohn W, Kim J, Kang SH, Yang SR, Cho JY, Cho HC, et al. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp Mol Med. 2015;47:e184.
Sugimachi K, Matsumura T, Hirata H, Uchi R, Ueda M, Ueo H, et al. Identification of a bona fide microRNA biomarker in serum exosomes that predicts hepatocellular carcinoma recurrence after liver transplantation. Br J Cancer. 2015;112(3):532–8.
Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361(15):1437–47.
Wang Y, Liang Z, Gao Y, Zhai D, Rao Q, Shi W, et al. Factors influencing circulating MicroRNA level in the studies of hepatocellular carcinoma biomarker. Neoplasma. 2015;62(5):798–804.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, J., Huang, A. & Yang, XR. Liquid Biopsy and its Potential for Management of Hepatocellular Carcinoma. J Gastrointest Canc 47, 157–167 (2016). https://doi.org/10.1007/s12029-016-9801-0
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12029-016-9801-0