Abstract
Naturally occurring vitamin E, comprised of four forms each of tocopherols and tocotrienols, are synthesized solely by photosynthetic organisms and function primarily as antioxidants. These different forms vary in their biological availability and in their physiological and chemical activities. Tocopherols and tocotrienols play important roles in the oxidative stability of vegetable oils and in the nutritional quality of crop plants for human and livestock diets. The isolation of genes for nearly all the steps in tocopherol and tocotrienol biosynthesis has facilitated efforts to alter metabolic flux through these pathways in plant cells. Herein we review the recent work done in the field, focusing on branch points and metabolic engineering to enhance and alter vitamin E content and composition in oilseed crops.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
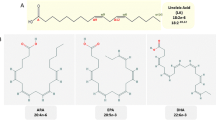
Tocopherols and tocotrienols comprise the vitamin E family of antioxidants and are synthesized by plants and other photosynthetic organisms. These molecules consist of a chromanol head group linked to an isoprenoid-derived hydrophobic tail. The aliphatic tail of tocopherols is fully saturated, while the side-chain of tocotrienols contains three trans double bonds (Fig. 1). Four different forms of tocopherols and tocotrienols occur in nature and differ by the numbers and positions of methyl groups on the aromatic portion of the chromanol head group (Fig. 1). The α form of tocotrienols and tocopherols has three methyl groups, the β and γ forms have two methyl groups, and the δ form has one methyl group on the aromatic ring. In the case of the β and γ forms, the methyl groups are at positions 5 and 8 or 7 and 8, respectively, of the chromanol head group. The α, β, γ, and δ forms of tocopherols and tocotrienols are often referred to collectively as “tocochromanols.”
Naturally occurring vitamin E molecules (or tocochromanols) have one of eight forms, varying in the saturation of the prenyl group and methylation pattern on the chromanol ring. Tocopherols contain fully saturated tails, while tocotrienol tails contain three trans double bonds. The hydrophilic head group has one, two, or three methyl groups. The hydrophobicity of the tail is greater than the hydrophilicity of the head group, thus vitamin E molecules are extracted with other lipids. Differences in methylation patterns are denoted by circles
Tocopherols occur widely in plants, but the form of tocopherol often differs in the leaves and seeds of the same species. α-Tocopherol is typically the primary form of tocopherols in leaves of plants and in photosynthetic prokaryotes such as Synechocystis. Seeds of different plant species, however, can be enriched in any of the four forms of tocopherol (Fig. 2). Soybean seeds, for example, contain primarily γ-tocopherol and lesser amounts of δ- and α-tocopherol. The occurrence of tocotrienols is more limited in plants. Tocotrienols are the principal tocochromanol of the seed endosperm of monocots, including important cereal grains such as wheat, rice, and barley (Fig. 2). Tocotrienols are also the major tocochromanol class in seeds of some dicots, including species of the Apiaceae (or Umbelliferae) family (e.g., coriander, celery) [1, 32]. Given their lipid soluble nature, tocopherols and tocotrienols are readily extracted as components of vegetable oils during the commercial processing of oilseeds. Soybean oil and palm oil, for example, are major commercial sources of tocopherols and tocotrienols, respectively.
C18-reverse phase HPLC analysis of tocochromanols in extracts from soybean seeds (a), sunflower seeds (b), rice seeds (c), and coconut endosperm (d). Tocochromanols were detected in crude lipid extracts by fluorescence (292 nm excitation/330 nm emission). C18-reverse phase HPLC columns are typically unable to resolve β and γ forms of tocotrienols and tocopherols. These molecules can be resolved by normal phase (e.g., silica) HPLC columns. In the soybean seed extract (a), the β/γ tocopherol peak is composed almost entirely of γ-tocopherol, and in the rice seed extract (c), the β/γ tocotrienol peak is composed almost entirely of γ-tocotrienol
Tocochromanols are potent lipid soluble antioxidants that protect plant cells against oxidative stresses. Tocopherols, for example, contribute to the maintenance of photosynthesis by mitigating photooxidative damage to photosystem II through the quenching of singlet oxygen [19]. In addition, seeds from Arabidopsis mutants that lack tocopherol display reduced storage life and accumulate increased amounts of compounds derived from the oxidation of stored oils [45]. The growth of these mutants at low temperatures is also reduced, suggesting that tocopherols contribute to the cold-adaptation of plants [31]. It is likely that tocotrienols are also important for reducing oxidative stresses in plants, but the physiological role of these compounds in plants has yet to be established.
Tocopherols and tocotrienols are classified as vitamin E based on their ability to prevent the resorption of rat fetuses maintained on defined diets, the classical measure of “vitamin E activity.” In this assay, α-tocopherol displays the greatest efficacy of the eight natural tocochromanols because of its ability to be more readily absorbed and retained by cells of the body [24]. As such, α-tocopherol is considered to have the highest nutritional value of the different forms of tocopherols and tocotrienols, and has thus received the greatest attention for the vitamin E biofortification of crop plants. By comparison, α-tocotrienol has about one-third of the vitamin E activity displayed by α-tocopherol, while γ-tocopherol has only about one-tenth of the vitamin E activity of α-tocopherol [24]. Regardless, all forms of tocochromanols are able to reduce free-radical damage to membrane lipids, and in model membrane studies, tocotrienols are better antioxidants than tocopherols [57, 48]. In addition, diverse health-promoting properties have been attributed to the various forms of tocochromanols. For example, low levels of γ-tocopherol, but not α-tocopherol, in serum correlate with the incidence of coronary heart disease [34]. Likewise, tocotrienols have physiological properties that are distinct from those of α-tocopherol. Tocotrienols inhibit cholesterol synthesis [39], and γ-tocotrienol is the most active inhibitor in studies with human hepatoma HepG2 cells [36]. Tocotrienols also reduce the in vitro growth of breast cancer cells [33], and α-tocotrienol provides the greatest protection against oxidative damage to neuronal cells of any of the tocochromanols [35].
From a commercial standpoint, the oxidative stability that tocochromanols confer to vegetable oils is perhaps their most important functional property. This property is especially valuable for reducing fatty acid oxidation and the formation of off-flavor compounds such as hexanals in foods fried or processed using vegetable oils [65, 67]. Of the different forms of tocopherol, δ- and γ-tocopherols are the most effective at reducing the oxidative breakdown of vegetable oils in frying applications [65, 66]. In addition, δ- and γ-tocotrienols have a slightly greater ability than the corresponding tocopherols to reduce the formation of fatty acid oxidation products during the frying of foods [66]. As interest in the use of vegetable oils as high-temperature lubricants grows [12], the ability to genetically enhance the tocochromanol content of oilseeds, particularly the δ and γ forms, to improve oxidative stability will likely become increasingly important. Recent efforts to engineer oilseeds to produce long-chain ω-3 polyunsaturated fatty acids (e.g., eicosapentaenoic acid), which are prone to oxidation, will also likely benefit from genetic enhancement of tocochromanol content [11]. In addition to their contributions to the oxidative stability of vegetable oils, tocochromanols are used commercially in cosmetics and sunscreens, and have potential value as livestock feed supplements to improve the quality and shelf-life of meats [30, 68].
Tocochromanols are synthesized in plastids of plants from precursors that derive from the shikimate and methylerythritol phosphate pathways. The biosynthetic pathway for tocopherols and many of the associated enzymes were determined in the 1980s. Research during the past 8 years has uncovered nearly all of the genes that are required for the synthesis and modification of tocopherols and tocotrienols. This review summarizes recent advances in our understanding of the biochemistry and genetics of tocopherol and tocotrienol biosynthesis in plants and describes how this research has been applied for the biotechnological enhancement of the vitamin E content and composition of oilseed crops.
Vitamin E Biosynthetic Enzymes
Vitamin E biosynthetic enzymes are found in chloroplasts [3, 21, 52, 55, 66] and chromoplasts [2], thus, presumably all plastids. Total vitamin E content increases during senescence [42], during chloroplast to chromoplast transition [2], and during seed development [15]. Although many of the enzymatic steps have been characterized for over 20 years [52], recent genetic work in dissecting the pathway has been facilitated by the availability of Arabidopsis mutants and the use of transgenic plants. Table 1 shows many of the available plant mutants and cloned genes. The generally accepted pathway for vitamin E biosynthesis is shown in Fig. 3 and major steps are summarized in Table 2. Only two steps are required to make a tocochromanol: the prenylation of homogentisate (HGA) and a subsequent cyclization step. In this case, the vitamin E will be either δ-tocopherol or δ-tocotrienol. Additional methylation steps produce α, β, and γ forms of vitamin E.
Biosynthesis of vitamin E occurs in plastids. The substrates, HGA and either GGDP (for tocotrienols) or PDP (for tocopherols) come from the shikimate and the deoxy-D-xylulose phosphate pathways, respectively. Enzymes in bold are specific to the synthesis of tocochromanols. Enzymes denoted in italics, while vital for vitamin E biosynthesis, are also used in other pathways. Neither methyltransferase step is required for tocochromanol synthesis. The figure shows the production of vitamin E molecules with α- and γ-head groups. δ-Head groups are produced when both methyltransferase steps are bypassed. β-Tocochromanols result when the first methylation step is omitted, but the second methylation step is included. The production of HPP varies. The pathway for plant synthesis is shown with solid arrows. Shunts, shown with dashed lines, have been used to manipulate vitamin E production in plants. In E. coli HPP is produced directly from chorismate by a bifunctional enzyme. In yeast, a prephenate dehydrogenase produces HPP
Two substrates are required for vitamin E biosynthesis: HGA and a C20 prenyldiphosphate (PrDP). HGA supplies the aromatic ring of the chromanol head group and is derived from tyrosine via the shikimate pathway. Tyrosine is deaminated to p-hydroxyphenylpyruvate (HPP), which in turn is oxygenated to HGA. HGA is also used in plastoquinone synthesis. The second required substrate is a prenyldiphosphate (PrDP), either phytyldiphosphate (PDP) or geranylgeranyldiphosphate (GGDP). These prenyl groups are supplied by the 1-deoxy-d-xyulose-5-phosphate (DXP) pathway, reviewed by Lichtenthaler [29]. GGDP can be used directly in tocotrienol synthesis or reduced to PDP for tocopherol biosynthesis. The proposed route for phytol synthesis has recently been revised [23, 61]. In the revised path, GGDP is incorporated into chlorophyll where the geranylgeranyl moiety is reduced to a phytyl moiety. Free phytol is released during chlorophyll breakdown [22]. One fate of this phytol may be phosphorylation and incorporation into tocopherols [23, 54, 61]. In addition to the two required substrates HGA and PrDP, tocochromanols are mono-, di-, or tri-methylated. The methyl donor is S-adenosyl-methionine [53]. The methylation pattern affects the bioavailability of the vitamin E as well as the antioxidant capabilities [24].
Homogentisate Prenyltransferases
The first step of vitamin E biosynthesis is the transfer of a prenyl group to HGA. This transfer, a condensation and decarboxylation [52], occurs via either homogentisate phytyltransferase (HPT) or homogentisate geranylgeranyl transferase (HGGT). The substrate specificity of the prenyltransferase determines whether the final tocochromanol will be a tocopherol or a tocotrienol. Tocopherols, with a phytyl tail, are more common in most plant tissues than tocotrienols, with a geranylgeranyl tail. Prenyltransferase activity was reported by Soll et al. [53] in 1980, working with spinach chloroplasts. To date, homogentisate prenyltransferase genes for vitamin E have been cloned from Synechocystis [7, 46, 47], Arabidopsis [7, 46], and the monocots barley, wheat, and rice [5]. The prenyltransferases are part of the ubiA prenyltransferase family and are predicted to be integral membrane proteins. Activity has been localized to the inner membranes of both chloroplasts [55] and chromoplasts [2] using cell fractionation methods.
The Arabidopsis HPT has been expressed in E. coli, where it has a strong substrate preference for PDP over GGDP [7, 43]. It shows no activity towards solanesyldiphosphate (SDP) [7, 43], a prenyltransferase activity required for phylloquinone biosynthesis. The Synechocystis enzyme has also been expressed in E. coli [7] and insect Sf-9 cells using a baculovirus system [46]. The Synechocystis HPT has activity for both PDP and GGDP, although in vitro GGDP activity is less than 20% of in vitro PDP activity [7]. No reports of HGGT expression have been made, and attempts to express the enzyme in E. coli have been unsuccessful [5]. However, HGGT is presumed to have higher activity with GGDP than with PDP (as indicated in Fig. 3), based on the ability of the barley, wheat, and rice enzymes to confer tocotrienol biosynthetic ability to plant cells upon transgenic expression [5]. An additional prenyltransferase (referred to in Table 1 as “homogentisate prenyldiphosphate transferase”) has recently been identified [43, 65]. The recombinant enzyme expressed in E. coli shows strongest activity toward SDP, with minor activity for GGDP and PDP [43], however, the in vivo function of this prenyltransferase is not yet clear.
HPTs and HGGTs have been transgenically expressed in plants. Overexpression of HPT increases tocopherol content of Arabidopsis leaves [8], and antisense suppression of HPT expression in Arabidopsis seeds reduces levels up to ten-fold compared to seeds from non-transformed plants [46]. Expression of the barley HGGT in Arabidopsis leaves shifts tocochromanol biosynthesis toward the production of tocotrienols rather than tocopherols and increases total tocochromanol content [5]. This result clearly indicates that homogentisate prenyltransferases alone can dictate whether tocopherols or tocotrienols are produced by plant cells. In addition, the enhanced levels of tocochromanols that accompany HPT or HGGT overexpression suggest that homogentisate prenyltransferases can assert flux control through the vitamin E biosynthetic pathway. As such, these enzymes have been major targets for biotechnological efforts to fortify the tocochromanol content of oilseeds, as described below.
2-Methyl-6-Prenylbenzoquinol Methyltransferase
In order to have γ- or α-tocochromanols, the number 3 position on the benzoquinol ring must be methylated. This methylation step is omitted when β- and δ-tocochromanols are produced. Methyltransferase activity was identified in 1980 [53] and a gene for 2-methyl-6-phytylbenzoquinol methyltransferase (PrBQMT) was cloned from Synechocystis in 2002 [51]. Since the production of γ- and α-tocotrienols can be accomplished with the addition of the HGGT gene alone, PrBQMT appears to methylate either tocopherols or tocotrienols [5]. The enzyme has been localized to the chloroplast inner membrane [55] and the Arabidopsis gene has a putative transit peptide [6]. The Synechocystis gene was cloned via homology to the Arabidopsis γ-tocopherol methyltransferase (TMT) gene. The Arabidopsis PrBQMT gene has only 18% amino acid sequence identity to the Synechocystis gene [6], and was identified by forward genetics using screens for altered tocopherol composition [6, 62]. The Arabidopsis and cyanobacterial genes have been expressed in E. coli. In this system, the transferase activity does not appear to depend on the prenyl tail; both phytylbenzoquinols and solanesylbenzoquinols are used [6, 51, 62]. Although Soll and Schultz [53] reported methyltransferase activity with β-tocopherol, recent work has not duplicated that activity, activity assayed from E. coli expression experiments shows no activity toward β- and δ-tocopherols [6, 51], thus, if the 3 position is to be methylated for either γ- or α-tocochromanols, it appears that this methylation must happen before the cyclization. The main effect of overexpression of PrBQMT is to alter the composition, but not the total content of vitamin E. For example, overexpression in soybean seeds converts the pools of δ- and β-tocopherols to γ- and α-tocopherols but does not affect the total tocopherol content [62].
Tocopherol/Tocotrienol Cyclase
Tocopherol/tocotrienol cyclase (TC) is the second enzyme required for vitamin E biosynthesis (Fig. 3). It forms the chromanol head group from a benzoquinol intermediate. The product is either γ-tocochromanol or δ-tocochromanol, depending on the methylation status of the chromanol head group. Unlike the other enzymes in the vitamin E pathway, TC has been localized to plastoglobules in chloroplasts [3, 64], but not to the inner membrane [52]. Since both δ- and γ- tocochromanols are found in plants it is presumed that TC cyclizes both mono- and di-methylated prenylbenzoquinols. Cyclase genes from Arabidopsis [38, 44], potato [21], and maize [44] as well as cyanobacteria [56] have been cloned and expressed in E. coli. The activity shows little, if any, preference for phytylated substrates over geranylgeranylated substrates [21, 38]. Thus, TC produces both γ- and δ-tocopherols and γ- and δ-tocotrienols.
Although the cyclase gene was first identified as a vitamin E biosynthetic gene in 2002 [38], the Arabidopsis VTE1 gene is homologous to a maize gene SXD1. The maize mutant, sxd1, is deficient in sucrose transport [37]. Although further work has established that the maize gene complements the Synechocystis slr1737 knockout mutant [44] and affects tocopherol production in potato and Arabidopsis as well as maize and Synechocystis [21, 44], the Arabidopsis mutants do not share the maize phenotype [38]. The link between vitamin E and sucrose transport is still under investigation.
When TC is constitutively overexpressed in Arabidopsis, the total leaf tocopherol content increases seven-fold, suggesting that TC, as well as the prenyltransferases, may play a role in flux control through the pathway [25].
Tocopherol/Tocotrienol Methyltransferase
Tocopherol/tocotrienol methyltransferase (TMT) catalyzes the final step in synthesis of α- or β-tocochromanols: the methylation of the number 5 carbon on the chromanol ring (Fig. 3). This enzyme is often referred to as γ-tocopherol methyltransferase, but it also has activity with δ-tocopherol, δ-tocotrienol, and γ-tocotrienol. This methyltransferase activity was identified in 1980 and localized to the inner membrane of spinach chloroplasts [53], and the enzyme was purified from pepper chromoplasts [10]. The TMT gene was isolated from Synechocystis using homology to previously identified genes involved in vitamin E biosynthesis and looking for candidate genes on the same operon [50]. The Arabidopsis gene was then cloned via homology to the newly identified Synechocystis TMT [50]. E. coli-expressed Arabidopsis TMT methylates both γ- and δ-tocopherol but has almost a three-fold preference for γ-tocopherol [50]. Overexpression of Arabidopsis TMT also converts nearly the entire content of γ-tocopherol in Arabidopsis seeds to α-tocopherol [50]. Because most dicot seeds are enriched in γ-tocopherol, this result provides clear evidence that TMT activity is limiting for α-tocopherol synthesis in dicot seeds, including major oilseeds such as canola and soybean.
Phytol Kinase and Phytylmonophosphate Kinase
While a phytol kinase (PK) activity had been reported in early work [54], conventional pathways had no source for PDP other than direct reduction of GGDP by geranylgeranyl reductase (GGR). However, recent work has demonstrated that a pathway from phytol to PDP exists [23] and a gene encoding PK has been identified and cloned [61]. Two separate kinase activities are required to synthesize PDP from phytol [23]. Free phytol increases during senescence as the phytyl moiety from chlorophyll is released by chlorophyllase [22]. Phytol from chlorophyll is phosphorylated by CTP or UTP [61, 23]. Using an Arabidopsis mutant with only 20% of wild type levels of total tocopherol Valentin et al. [61] cloned VTE5, which, when expressed in E. coli, phosphorylates free phytol. The Arabidopsis phytol kinase gene contains a putative transit peptide and is predicted to be an integral membrane protein. A separate kinase activity phosphorylates phytylmonophosphate (PMP) to PDP using any of the four nucleotide triphosphates [23]. However, the gene that codes for PMP kinase activity has not yet been reported. Thus, phytol from chlorophyll breakdown can be used in tocopherol synthesis via direct phosphorylation of phytol and phytylmonophosphate.
Substrate Supply: Homogentisate and Prenyldiphosphates
The HGA used in tocochromanol biosynthesis is derived from HPP via a decarboxylation by hydroxyphenylpyruvate dioxygenase (HPPD) [17]. HPP, in turn, is derived from tyrosine via the shikimate pathway. HPP is synthesized differently in plants, bacteria, and yeast. In plants, chorismate, the end product of the shikimate pathway, is isomerized to prephenate. An aminotransferase creates arogenate from prephenate, and the arogenate is dehydrogenated to tyrosine. This arogenate-to-tyrosine step is subject to strong feedback regulation by tyrosine [9]. In E. coli and yeast, bifunctional enzymes bypass some or all of these steps. In E. coli, the precursor for tyrosine is chorismate [20], and in yeast the precursor is prephenate [41].
Overexpression of HPPD by itself has a negligible or only modest effect on increasing total tocopherol content in leaves and seeds of tobacco and Arabidopsis [14, 41]. Unlike the rest of the enzymes involved in vitamin E biosynthesis, HPPD is a cytosolic enzyme [17]. However, redirecting HPPD to plastids by addition of a transit peptide does not enhance tocopherol content over levels achieved by cytosolic expression [16]. In spite of this, feeding experiments in soybean suspension cultures indicate that HGA may limit vitamin E production, since addition of exogenous HGA doubles tocochromanol levels [26]. Thus, while the supply of HGA may be limiting, overexpression of HPPD alone does not result in large increases of tocochromanol content.
The prenyldiphosphate is supplied by the DXP pathway (also called the methylerythritol phosphate pathway), reviewed in [29]. Products of the DXP pathway are used in carotenoid, phylloquinone, terpenoid, and gibberellin biosyntheses a well as vitamin E biosynthesis [13]. Increased flux through the DXP pathway can increase vitamin E content. When 1-deoxy-d-xylulose-5-phosphate synthase, the gene for the first step in the DXP path, is overexpressed, vitamin E content increases 1.5- to 2-fold [13]. Conversely, in deoxy-d-xylulose-5-phosphate synthase antisense plants, vitamin E content slightly decreases [13]. Additionally, competition for isoprenoid substrates between the vitamin E and other pathways can also decrease tocopherol content. For example, transgenic expression of a bacterial phytoene synthase, which uses GGDP as its substrate, results in a 50-fold increase in carotenoid levels in canola seeds, but the tocopherol content of these seeds is reduced by two-fold [49].
Four five-carbon isoprenoid moieties are required for the synthesis of the prenyldiphosphate moiety of tocochromanols. GGDP can be used directly in tocotrienol synthesis or reduced to PDP for tocopherol biosynthesis. Two paths for PDP synthesis have been proposed. In one, GGR reduces GGDP directly, and this PDP is available for incorporation into tocopherols [27, 55]. However, new evidence posits a second path for phytyl synthesis through chlorophyll [23, 61]. In this pathway, GGDP is incorporated into chlorophyll, then reduced to phytylated chlorophyll. During chlorophyll breakdown, free phytol is released. This free phytol is then phosphorylated, creating PDP for incorporation into tocopherols. Kinase activity has been discovered as discussed above [23, 54, 61]. It is likely that both routes are used in vivo, since the phytol kinase mutant still has tocopherol [61], and many seeds that produce tocopherols lack detectable chlorophyll to support flux through the second pathway.
Tocotrienol Biosynthesis
Tocotrienols occur principally in the endosperm of monocot seeds, and for the most part, tocotrienol biosynthesis differs little from tocopherol synthesis. The isolation of HGGT genes from barley, rice, and wheat provide a biochemical and genetic explanation for the occurrence of tocotrienols in monocot seeds [5]. The monocot HGGT identified to date share only 40–50% amino acid sequence identity with HPT, and the expression of the HGGT gene in barley was shown to be seed-specific, consistent with the location of tocotrienol accumulation in this plant [5]. Transgenic expression of the barley HGGT in tobacco callus and Arabidopsis leaves, which normally produce only tocopherols, results in the production of tocotrienols [5]. This result demonstrates that HGGT expression alone is sufficient to confer tocotrienol biosynthetic ability to plant cells. Seeds from some dicot species such as those of the Apiaceae family are also enriched in tocotrienols [1, 32]. Based on the monocot example, the simplest biosynthetic route that can account for tocotrienols in these seeds is the activity of a yet to be identified HGGT-like enzyme.
The other enzymes of tocopherol biosynthesis have activity for both phytylated and geranylgeranylated compounds (Fig. 3). The Arabidopsis TC has about equal activity towards both forms of benzoquinols [38]. When tobacco callus is transformed with barley HGGT, all four forms of tocotrienols are detected [5], hence methyltransferase can use geranylgeranyl-derived substrates to produce different methylated forms of tocotrienols. However, it remains to be determined if variant forms of TC, PrBQMT, and TMT have evolved for the more efficient synthesis of tocotrienols in the endosperm of monocot seeds.
HGGT-Independent Tocotrienol Synthesis
Several recent experiments in transgenic plants have uncovered an alternative route for tocotrienol synthesis. In these experiments, HGA synthesis is strongly upregulated by co-expression of HPPD with a yeast or bacterial enzyme that produces the HPP substrate for HPPD directly from the shikimate pathway. The yeast enzyme used in these studies was the prephenate dehydrogenase [41], and the bacterial enzyme used was the bifunctional chorismate mutase/prephenate dehydrogenase (encoded by the tyrA gene) [20, 26]. HPP is typically synthesized in two steps from tyrosine in plants (Fig. 3). The yeast and bacterial enzymes, however, shunt flux upstream of tyrosine (from prephenate or chorismate) toward the synthesis of HPP. This effectively bypasses steps in tyrosine synthesis that are normally negatively regulated by pool sizes of this amino acid [9, 20]. Through this strategy, high levels of tocotrienols were produced in tobacco leaves [20, 41], Synechocystis [26], and canola and soybean seeds [26], which normally accumulate only trace amounts of tocotrienols. In studies with soybean, HGA is increased to amounts high enough to alter the color of seeds [26]. Based on our current knowledge of vitamin E biosynthesis, it is unclear how greatly enhanced production of HGA results in tocotrienol synthesis. The Arabidopsis HPT, for example, displays relatively low activity with GGDP, the isoprenoid substrate for tocotrienol synthesis [7, 43]. Regardless, the production of tocotrienols, rather than tocopherols, through this alternative route suggests that available GGDP pools must greatly exceed those of PDP for tocochromanol synthesis. Although this transgenic method for production of tocotrienols is biochemically intriguing, it remains to be determined if this metabolic route normally contributes to tocotrienol synthesis in non-engineered plant cells.
Genetic Enhancement of Vitamin E Composition and Content in Oilseeds
Improvement of Tocochromanol Composition
The identification of vitamin E biosynthetic genes has facilitated biotechnological efforts to improve the nutritional value and antioxidant content of crop plants. One focus of this research has involved increasing the expression of methyltransferase genes to convert tocopherol in leaves or seeds into the more nutritious α form. This approach has been of particular interest for the nutritional enhancement of seed oils from dicotyledonous grain crops (e.g., soybean and canola), which are typically enriched in γ-tocopherol. The ability to genetically convert the bulk of the tocopherol in plant organs from the γ to α form was first demonstrated using the model plant Arabidopsis thaliana [50]. In this study, the tocopherol composition of A. thaliana seeds was shifted from 97% γ-tocopherol to 95% α-tocopherol by genetic transformation with a cDNA for the A. thaliana TMT under control of a strong seed-specific promoter. Given that α-tocopherol has ten-times greater vitamin E activity than γ-tocopherol, the net effect is an approximately nine-fold increase in the vitamin E activity of Arabidopsis seeds. Notably, this alteration in tocopherol composition is not accompanied by an increase in total tocochromanol content. The ability to increase relative amounts of α-tocopherol by increased expression of TMT has subsequently been shown in seeds of crop plants including soybean [26, 59, 62] and Brassica juncea [69].
Soybean seeds offer an additional challenge for the generation of high levels of α-tocopherol. The tocopherols of soybean seeds are composed of approximately 20% δ-tocopherol and 65% γ-tocopherol. This is in contrast to the tocopherols of A. thaliana seeds, which are almost exclusively in the γ form. The conversion of δ-tocopherol to α-tocopherol requires two sequential enzymatic steps: (a) PrBQMT to convert δ-tocopherol to γ-tocopherol and (b) TMT to convert the resulting γ-tocopherol to α-tocopherol. Consistent with this, seed-specific co-expression of PrBQMT and TMT transgenes in soybean yields seeds with >90% α-tocopherol [62]. In this study, enhanced expression of only PrBQMT results in seeds that contain exclusively the γ and α forms of tocopherol, with a ratio of 80% γ-tocopherol and 20% α-tocopherol. Increased expression of TMT alone yields seeds with approximately 75% α-tocopherol and 25% β-tocopherol. These results demonstrate the utility of altering flux through PrBQMT and TMT to generate novel tocochromanol compositions.
Given that δ- and γ-tocopherol confer greater oxidative stability to frying oils than α-tocopherol [65, 66], it can be envisioned that suppression of methyltransferase expression may be of commercial value for generating improved vegetable oils for food processing and high-temperature lubricant applications. This is of particular significance with regard to sunflower. The tocopherols in seeds of this crop are composed of >90% α-tocopherol. By mutational breeding, sunflower varieties have been generated with seeds that contain exclusively δ- and γ-tocopherol [18, 58]. This composition was achieved by crossing lines with lesions in genes for PrBQMT and TMT [18, 58].
Enhancement of Tocochromanol Content
A second focus of biotechnological efforts for vitamin E enhancement has been the production of increased total amounts of tocochromanols in seeds of crop species. As described in the “Introduction”, increasing the content of tocochromanols in seeds may be useful for enhancing the antioxidant capacity of vegetable oils. This, in turn, may improve the oxidative stability of vegetable oils in frying and other food processing applications and in high-temperature lubricants. This research to date has been successful in generating seeds with increased amounts of tocotrienols, but less successful in the enhancement of tocopherol levels in seeds. Attempts to increase the tocopherol content of seeds by use of a single transgene have centered on (a) increasing flux in the tocopherol biosynthetic pathway by overexpression of HPT or TC or (b) by increasing the supply of the HGA head group by overexpression of cDNAs for HPPD. Based on results obtained from overexpression studies in leaves, HPT and TC would appear to be logical targets for the enhancement of tocopherol levels in seeds. For example, overexpression of HPT and TC in leaves of A. thaliana results in a four- and seven-fold increase, respectively, in tocopherol content [7, 38]. However, expression of these enzymes using transgenes containing strong seed-specific promoters yields only modest increases (less than 1.5-fold) in the tocopherol content of seeds of A. thaliana [46], B. napus [28, 40], and soybean [26]. Similar results were also obtained with the overexpression of HPPD in seeds of these plants [26, 40, 60]. Only by co-expressing cDNAs for HPT, TC, and HPPD have increases in tocopherol content in the range 2- to 2.5-fold been achieved in B. napus seeds [40], which is the largest enhancement of tocopherol levels reported to date in an oilseed.
In contrast to the relatively small increases in tocopherol content attained by overexpression of HPT, TC, or HPPD, seed-specific expression of an HGGT transgene resulted in up to a six-fold enhancement of the tocochromanol content of corn seeds, largely in the form of tocotrienols [5]. This is currently the largest increase in tocochromanol content in an engineered seed obtained by expression of only one transgene. A possible explanation for the success of this approach compared to HPT overexpression is that GGDP pools that support tocotrienol synthesis may greatly exceed those of PDP that are available for tocopherol synthesis.
The largest increase in total tocochromanol content achieved to date in genetically enhanced seeds was reported by Karunanandaa et al. [26]. In this study, co-expression of transgenes for HPPD, GGR, HPT, and a bacterial bifunctional chorismate mutase/prephenate dehydrogenase (encoded by the tyrA gene) increases total tocochromanol content in soybean seeds 10- to 15-fold. Although the goal of this experiment was to enhance tocopherol levels, the increase in tocochromanol content was largely due to the production of tocotrienols. In the highest tocochromanol-producing seeds, tocotrienols account for over 90% of the tocochromanols. Notably, tocotrienols are only trace components of the tocochromanols of non-engineered soybean seeds. The production of tocotrienols in vast preference to tocopherols in these seeds (in the apparent absence of HGGT) is also consistent with the existence of GGDP pools that greatly exceed those of PDP in seed plastids. Presumably under the metabolic conditions created in these seeds, HPT is able to use GGDP as a substrate despite its relatively low in vitro activity with this substrate [43].
The inability to engineer large increases in the tocopherol content of oilseeds points to a lack of understanding of the biosynthesis of PDP in seed plastids. The studies by Karunanandaa et al. [26] suggest that the availability of PDP limits the amounts of tocopherol synthesized. As described above, recent evidence points to a route of PDP synthesis involving the modification of geranylgeranyl-chlorophyll by the activity of GGR (Fig. 3) [61]. Developing metabolic engineering strategies to increase flux through this pathway may be necessary for increasing PDP pool sizes in plastids to support high levels of flux toward the synthesis of tocopherols rather than tocotrienols.
In conclusion, the isolation of genes for PrBQMT and TMT has facilitated biotechnological efforts to improve the nutritional value of vegetable oils. By increasing the expression of genes for one or both of these enzymes, it has been possible to shift the tocopherol content of oilseeds from the δ, β, and γ forms to the α form, which has the highest vitamin E activity of any of the naturally occurring tocochromanols. Efforts to engineer increased total tocopherol content in oilseeds, however, have met with only modest success. This appears to be due to the inability to generate enhanced pool sizes of the PDP substrate, because of gaps in our understanding of its synthesis from GGDP in seeds. The genetic enhancement of total tocotrienol content by expression of HGGT or by strong upregulation of HGA synthesis has proven to be a more successful strategy for generating seeds with increased amounts of vitamin E antioxidants. Through these approaches, corn and soybean seeds with 6- to 10-fold increases in total tocochromanol content have been produced. The availability of tocotrienol-enriched corn and soybean seeds should allow for functionality testing of the extracted oil to evaluate its performance in food processing, lubricants, and other commercial applications.
Abbreviations
- DXP:
-
1-Deoxy-d-xyulose-5-phosphate
- GGDP:
-
Geranylgeranyldiphosphate
- GGR:
-
Geranylgeranyl reductase
- HGA:
-
Homogentisate
- HGGT:
-
Homogentisate geranylgeranyl transferase
- HPP:
-
p-Hydroxyphenylpyruvate
- HPPD:
-
Hydroxyphenylpyruvate dioxygenase
- HPT:
-
Homogentisate phytyltransferase
- PDP:
-
Phytyldiphosphate
- PK:
-
Phytol kinase
- PMP:
-
Phytylmonophosphate
- PrBQMT:
-
2-Methyl-6-prenylbenzoquinol methyltransferase
- PrDP:
-
Prenyldiphosphate
- SDP:
-
Solanesyldiphosphate
- TC:
-
Tocopherol/tocotrienol cyclase
- TMT:
-
Tocopherol/tocotrienol methyltransferase
References
Aizetmüller K (1997) Antioxidative effects of Carum seeds. J Am Oil Chem Soc 74:185
Arango Y, Heise K (1998) Short communication. Localization of α-tocopherol synthesis in chromoplast envelope membranes of Capsicum annuum L. Fruits J Exp Bot 49:1259–1262
Austin JR II, Frost E, Vidi PA, Kessler F, Staehelin LA (2006) Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18:1693–1703
Bergmüller E, Porfirova S, Dörmann P (2003) Characterization of an Arabidopsis mutant deficient in γ-tocopherol methyltransferase. Plant Mol Biol 52:1181–1190
Cahoon EB, Hall SE, Ripp KG, Ganzke TS, Hitz WD, Coughlan SJ (2003) Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nat Biotechnol 21:1082–1087
Cheng Z, Sattler S, Maeda H, Sakuragi Y, Bryant DA, DellaPenna D (2003) Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15:2343–2356
Collakova E, DellaPenna D (2001) Isolation and functional analysis of homogentisate phytyltransferase from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol 127:1113–1124
Collakova E, DellaPenna D (2003) Homogentisate phytyltransferase activity is limiting for tocopherol biosynthesis in Arabidopsis. Plant Physiol. 131:632–642
Coruzzi G, Last RL (2000) Amino acids (Chapter 8). In: Buchanan BB, Gruissem W, Jones RL (eds) Biochemistry and molecular biology of plants. American Society of Plant Physiologists, Rockville, pp 358–410
d’Harlingue A, Camara B (1985) Plastid enzymes of terpenoid biosynthesis. Purification and characterization of γ-tocopherol methyltransferase from Capsicum chromoplasts. J Biol Chem 260:15200–15203
Domergue F, Abbadi A, Heinz E (2005) Relief for fish stocks: oceanic fatty acids in transgenic oilseeds. Trends Plant Sci 10:112–116
Erhan SZ, Asadauskas S (2000) Lubricant basestocks from vegetable oils. Ind Crops Prod 11:277–282
Estévez JM, Cantero A, Reindl A, Reichler S, León P (2001) 1-Deoxy-d-xylulose-5-phosphate synthase, a limiting enzyme for plastidic isoprenoid biosynthesis in plants. J Biol Chem 276:22901–22909
Falk J, Andersen G, Kernebeck B, Krupinska K (2003) Constitutive overexpression of barley 4-hydroxyphenylpyruvate dioxygenase in tobacco results in elevation of the vitamin E content in seeds but not in leaves. FEBS Lett 540:35–40
Falk J, Krahnstöver A, van der Kooij TA, Schlensog M, Krupinska K (2004) Tocopherol and tocotrienol accumulation during development of caryopses from barley (Hordeum vulgare L.) Phytochemistry 65:2977–2985
Falk J, Brosch M, Schäfer A, Braun S, Krupinska K (2005) Characterization of transplastomic tobacco plants with a plastid localized barley 4-hydroxyphenylpyruvate dioxygenase. J Plant Physiol 162:738–742
Garcia I, Rodgers M, Pepin R, Hsieh TF, Matringe M (1999) Characterization and subcellular compartmentation of recombinant 4-hydroxyphenylpyruvate dioxygenase from Arabidopsis in transgenic tobacco. Plant Physiol 119:1507–1516
Hass CG, Tang S, Leonard S, Traber MG, Miller JF, Knapp SJ (2006) Three non-allelic epistatically interacting methyltransferase mutations produce novel tocopherol (vitamin E) profiles in Sunflower. Theor Appl Genet 113:767–782
Havaux M, Eymery F, Porfirova S, Rey P, Dörmann P (2005) Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17:3451–3469
Herbers K (2003) Vitamin production in transgenic plants. J Plant Physiol 160:821–829
Hofius D, Hajirezaei MR, Geiger M, Tschiersch H, Melzer M, Sonnewald U (2004) RNAi-mediated tocopherol deficiency impairs photoassimilate export in transgenic potato plants. Plant Physiol 135:1256–1268
Hörtensteiner S (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57:55–77
Ischebeck T, Zbierzak AM, Kanwischer M, Dörmann P (2006) A salvage pathway for phytol metabolism in Arabidopsis. J Biol Chem 281:2470–2477
Kamal-Eldin A, Appelqvist LA (1996) The chemistry and antioxidant properties of tocopherols and tocotrienols. Lipids 31:671–701
Kanwischer M, Porfirova S, Bergmüller E, Dörmann P (2005) Alterations in tocopherol cyclase activity in transgenic and mutant plants of Arabidopsis affect tocopherol content, tocopherol composition, and oxidative stress. Plant Physiol 137:713–723
Karunanandaa B, Qi Q, Hao M, Baszis SR, Jensen PK, Wong YH, Jiang J, Venkatramesh M, Gruys KJ, Moshiri F, Post-Beittenmiller D, Weiss JD, Valentin HE (2005) Metabolically engineered oilseed crops with enhanced seed tocopherol. Metab Eng 7:384–400
Keller Y, Bouvier F, d’Harlingue A, Camara B (1998) Metabolic compartmentation of plastid prenyllipid biosynthesis. Evidence for the involvement of a multifunctional geranylgeranyl reductase. Eur J Biochem 251:413–417
Kumar R, Raclaru M, Schüßeler T, Gruber J, Sadre R, Lühs W, Zarhloul KM, Friedt W, Enders D, Frentzen M, Weier D (2005) Characterisation of plant tocopherol cyclases and their overexpression in transgenic Brassica napus seeds. FEBS Lett 579:1357–1364
Lichtenthaler HK (1999) The 1-deoxy-d-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 50:47–65
Liu Q, Lanari MC, Schaefer DM (1995) A review of dietary vitamin E supplementation for improvement of beef quality. J Anim Sci 73:3131–3140
Maeda H, Song W, Sage TL, DellaPenna D (2006) Tocopherols play a crucial role in low-temperature adaptation and phloem loading in Arabidopsis. Plant Cell 18:2710–2732
Matthaus B, Vosmann K, Pham LQ, Aizetmüller K (2003) FA and tocopherol composition of Vietnamese oilseeds. J Am Oil Chem Soc 80:1013–1020
Nesaretnam K, Stephen R, Dils R, Darbre P (1998) Tocotrienols inhibit the growth of human breast cancer cells irrespective of estrogen receptor status. Lipids 33:461–469
Öhrvall M, Sundlöf G, Vessby B (1996) Gamma, but not alpha, tocopherol levels in serum are reduced in coronary heart disease patients. J Intern Med 239:111–117
Osakada F, Hashino A, Kume T, Katsuki H, Kaneko S, Akaike A (2004) α-Tocotrienol provides the most potent neuroprotection among vitamin E analogs on cultured striatal neurons. Neuropharmacology 47:904–915
Pearce BC, Parker RA, Deason ME, Qureshi AA, Wright JJ (1992) Hypocholesterolemic activity of synthetic and natural tocotrienols. J Med Chem 35:3595–3606
Porfirova S, Bergmüller E, Tropf S, Lemke R, Dörmann P (2002) Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc Natl Acad Sci USA 99:12495–12500
Provencher LM, Miao L, Sinha N, Lucas WJ (2001) Sucrose export defective1 encodes a novel protein implicated in chloroplast-to-nucleus signaling. Plant Cell 13:1127–1141
Qureshi AA, Burger WC, Peterson DM, Elson CE (1986) The structure of an inhibitor of cholesterol biosynthesis isolated from barley. J Biol Chem 261:10544–10550
Raclaru M, Gruber J, Kumar R, Sadre R, Lühs W, Zarhloul MK, Friedt W, Frentzen M, Weier D (2006) Increase of the tocochromanol content in transgenic Brassica napus seeds by overexpression of key enzymes involved in prenylquinone biosynthesis. Mol Breed 18:93–107
Rippert P, Scimemi C, Dubald M, Matringe M (2004) Engineering plant shikimate pathway for production of tocotrienol and improving herbicide resistance. Plant Physiol 134:92–100
Rise M, Cojocaru M, Gottlieb HE, Goldschmidt EE (1989) Accumulation of α-tocopherol in senescing organs as related to chlorophyll degradation. Plant Physiol 89:1028–1030
Sadre R, Gruber J, Frentzen M (2006) Characterization of homogentisate prenyltransferases involved in plastoquinone-9 and tocochromanol biosynthesis. FEBS Lett 580:5357–5362
Sattler SE, Cahoon EB, Coughlan SJ, DellaPenna D (2003) Characterization of tocopherol cyclases from higher plants and cyanobacteria. Evolutionary implications for tocopherol synthesis and function. Plant Physiol 132:2184–2195
Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16:1419–1432
Savidge B, Weiss JD, Wong YH, Lassner MW, Mitsky TA, Shewmaker CK, Post-Beittenmiller D, Valentin HE (2002) Isolation and characterization of homogentisate phytyltransferase genes from Synechocystis sp. PCC 6803 and Arabidopsis. Plant Physiol 129:321–332
Schledz M, Seidler A, Beyer P, Neuhaus G (2001) A novel phytyltransferase from Synechocystis Sp. PCC 6803 involved in tocopherol biosynthesis. FEBS Lett 499:15–20
Serbinova EA, Packer L (1994) Antioxidant properties of α-tocopherol and α-tocotrienol. Meth Enzymol 234:354–366
Shewmaker CK, Sheehy JA, Daley M, Colburn S, Ke DY (1999) Seed-specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J 20:401–412
Shintani D, DellaPenna D (1998) Elevating the vitamin E content of plants through metabolic engineering. Science 282:2098–2100
Shintani DK, Cheng Z, DellaPenna D (2002) The role of 2-methyl-6-phytylbenzoquinone methyltransferase in determining tocopherol composition in Synechocystis sp. PCC6803. FEBS Lett 511:1–5
Soll J (1987) α-Tocopherol and plastoquinone synthesis in chloroplast membranes. Meth Enzymol 148:383–392
Soll J, Schultz G (1980) 2-Methyl-6-phytylquinol and 2,3-dimethyl-5-phytylquinol as precursors of tocopherol synthesis in spinach chloroplasts. Phytochemistry 19:215–218
Soll J, Schultz G (1981) Phytol synthesis from geranylgeraniol in spinach chloroplasts. Biochem Biophys Res Commun 99:907–912
Soll J, Kemmerling M, Schultz G (1980) Tocopherol and plastoquinone synthesis in spinach chloroplasts subfractions. Arch Biochem Biophys 204:544–550
Stocker A, Fretz H, Frick H, Rüttimann A, Woggon WD (1996) The substrate specificity of tocopherol cyclase. Bioorg Med Chem 4:1129–1134
Suzuki YJ, Tsuchiya M, Wassall SR, Choo YM, Govil G, Kagan VE, Packer L (1993) Structural and dynamic membrane properties of α-tocopherol and α-tocotrienol: implication to the molecular mechanism of their antioxidant potency. Biochemistry 32:10692–10699
Tang S, Hass CG, Knapp SJ (2006) Ty3/gypsy-like retrotransposon knockout of a 2-methyl-6-phytyl-1,4-benzoquinone methyltransferase is non-lethal, uncovers a cryptic paralogous mutation, and produces novel tocopherol (vitamin E) profiles in sunflower. Theor Appl Genet 113:783–799
Tavva VS, Kim YH, Kagan IA, Dinkins RD, Kim KH, Collins GB (2007) Increased α-tocopherol content in soybean seed overexpressing the Perilla frutescens γ-tocopherol methyltransferase gene. Plant Cell Rep 26:61–70
Tsegaye Y, Shintani DK, DellaPenna D (2002) Overexpression of the enzyme p-hydroxyphenylpyruvate dioxygenase in Arabidopsis and its relation to tocopherol biosynthesis. Plant Physiol Biochem 40:913–920
Valentin HE, Lincoln K, Moshiri F, Jensen PK, Qi Q, Venkatesh TV, Karunanandaa B, Baszis SR, Norris SR, Savidge B, Gruys KJ, Last RL (2006) The Arabidopsis vitamin E pathway gene5–1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell 18:212–224
Van Eenennaam AL, Lincoln K, Durrett TP, Valentin HE, Shewmaker CK, Thorne GM, Jiang J, Baszis SR, Levering CK, Aasen ED, Hao M, Stein JC, Norris SR, Last RL (2003) Engineering vitamin E content: from Arabidopsis mutant to soy oil. Plant Cell 15:3007–3019
Venkatesh TV, Karunanandaa B, Free DL, Rottnek JM, Baszis SR, Valentin HE (2006) Identification and characterization of an Arabidopsis homogentisate phytyltransferase paralog. Planta 223:1134–1144
Vidi PA, Kanwischer M, Baginsky S, Austin JR, Csucs G, Dörmann P, Kessler F, Bréhélin C (2006) Tocopherol cyclase (VTE1) localization and vitamin E accumulation in chloroplast plastoglobule lipoprotein particles. J Biol Chem 281:11225–11234
Wagner KH, Elmadfa I (2000) Effects of tocopherols and their mixtures on the oxidative stability of olive oil and linseed oil under heating. Eur J Lipid Sci Tech 102:624–629
Wagner KH, Wotruba F, Elmadfa I (2001) Antioxidative potential of tocotrienols and tocopherols in coconut fat at different oxidation temperatures. Eur J Lipid Sci Tech 103:746–751
Warner K, Neff WE, Eller FJ (2003) Enhancing quality and oxidative stability of aged fried food with γ-tocopherol. J Agric Food Chem 51:623–627
Waylan AT, O’Quinn PR, Unruh JA, Nelssen JL, Goodband RD, Woodworth JC, Tokach MD, Koo SI (2002) Effects of modified tall oil and vitamin E on growth performance, carcass characteristics, and meat quality of growing-finishing pigs. J Anim Sci 80:1575–1585
Yusuf MA, Sarin NB (2007) Antioxidant value addition in human diets: genetic transformation of Brassica juncea with γ-TMT gene for increased α-tocopherol content. Transgenic Res 16:109–113
Acknowledgments
The authors’ work is supported by the National Research Initiative of the USDA Co-operative State Research, Education and Extension Service, grant number 2004-35318-14887.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Hunter, S.C., Cahoon, E.B. Enhancing Vitamin E in Oilseeds: Unraveling Tocopherol and Tocotrienol Biosynthesis. Lipids 42, 97–108 (2007). https://doi.org/10.1007/s11745-007-3028-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11745-007-3028-6