Abstract
Wildtype sunflower (Helianthus annuus L.) seeds are a rich source of α-tocopherol (vitamin E). The g = Tph 2 mutation disrupts the synthesis of α-tocopherol, enhances the synthesis of γ-tocopherol, and was predicted to knock out a γ-tocopherol methyltransferase (γ-TMT) necessary for the synthesis of α-tocopherol in sunflower seeds—wildtype (g + g +) lines accumulated > 90% α-tocopherol, whereas mutant (g g) lines accumulated > 90% γ-tocopherol. We identified and isolated two γ-TMT paralogs (γ-TMT-1 and γ-TMT-2). Both mapped to linkage group 8, cosegregated with the g locus, and were transcribed in developing seeds of wildtype lines. The g mutation greatly decreased γ-TMT-1 transcription, caused alternative splicing of γ-TMT-1, disrupted γ-TMT-2 transcription, and knocked out one of two transcription initiation sites identified in the wildtype; γ-TMT transcription was 36 to 51-fold greater in developing seeds of wildtype (g + g +) than mutant (g g) lines. F2 populations (B109 × LG24 and R112 × LG24) developed for mapping the g locus segregated for a previously unidentified locus (d). B109, R112, and LG24 were homozygous for a null mutation (m = Tph 1) in MT-1, one of two 2-methyl-6-phytyl-1,4-benzoquinone/2-methyl-6-solanyl-1,4-benzoquinone methyltransferase (MPBQ/MSBQ-MT) paralogs identified in sunflower. The d mutations segregating in B109 × LG24 and R112 × LG24 were allelic to a cryptic mutation identified in the other MPBQ/MSBQ-MT paralog (MT-2) and disrupted the synthesis of α- and γ-tocopherol in F2 progeny carrying m or g mutations—m m g + g + d d homozygotes accumulated 41.5% α- and 58.5% β-T, whereas m m g g d d homozygotes accumulated 58.1% γ- and 41.9% δ-T. MT-2 cosegregated with d and mapped to linkage group 4. Hence, novel tocopherol profiles are produced in sunflower seed oil by three non-allelic epistatically interacting methyltransferase mutations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Vitamin E (tocol) antioxidants (α-, β-, γ-, and δ-tocopherol and tocotrienol) have diverse metabolic and biological functions in plants and animals and are essential in human and animal diets (Kamal-Eldin and Appelqvist 1996; Traber and Sies 1996; Brigelius-Flohe and Traber 1999; Bramley et al. 2000; Traber 2004a, b; Pfluger et al. 2004; Sattler et al. 2004; Havaux et al. 2005). The role of α-tocopherol (α-T) in human health and nutrition has been the primary focus of vitamin E research because the other forms are rapidly metabolized and have lower vitamin E activities in vivo (Sheppard et al. 1993; Traber and Sies 1996; Pfluger et al. 2004). However, a greater understanding of the roles and importance of other tocols is beginning to emerge and has been stimulated in part by the prevalence of α-T in European and γ-tocopherol (γ-T) in US diets (Ohrvall et al. 1996; Christen et al. 1997; Bramley et al. 2000; Jiang et al. 2000; Devaraj and Traber 2003; Hensley et al. 2004; Wagner et al. 2004). The dietary difference partly traces to the widespread consumption of α-T-rich sunflower (Helianthus annuus L.) oil in Europe and γ-T-rich soybean (Glycine max L.) oil in the USA.
Seeds are important natural sources of vitamin E compounds (Sheppard et al. 1993; Eitenmiller 1997; Cahoon et al. 2003). α- and γ-T accumulate in varying proportions in the seeds of most dicots. α-T percentages are commonly lower than γ-T percentages in oilseed crops, e.g., α- and γ-T percentages are < 10–20 and > 70–80%, respectively, in soybean and corn (Zea mays L.) oils (Sheppard et al. 1993; Eitenmiller 1997; Shintani and DellaPenna 1998; Dolde et al. 1999; Grusak and DellaPenna 1999; Rocheford et al. 2002; Wong et al. 2003). The proportions are closer to 50% α- and 50% γ-T in cottonseed (Gossypium hirsutum L.) and oil palm (Elaeis guineensis Jacq.). Wildtype sunflower produces > 90% α-T and is unique among commodity oilseeds other than safflower (Carthamus tinctorius L.), another α-T-rich oilseed in the Compositae. The natural vitamin E activity of sunflower seed oil was greater than the other seed oils, nuts, and seeds surveyed by Sheppard et al. (1993).
The α-T profile in physiologically mature seeds of wildtype sunflower is produced by naturally strong 2-methyl-6-phytyl-1,4-benzoquinone/2-methyl-6-solanyl-1,4-benzoquinone methyltransferase (MPBQ/MSBQ-MT) and γ-tocopherol methyltransferase (γ-TMT) activities in developing seeds (Shintani and DellaPenna 1998; Shintani et al. 2002; Bergmüller et al. 2003; Cheng et al. 2003; Van Eenennaam et al. 2003). MPBQ/MSBQ-MT and γ-TMT are necessary for the synthesis of α-T and, when mutated, disrupt α-T synthesis and enhance β-, γ-, and δ-T synthesis in characteristically and predictably different ways. The methylation of MPBQ by MPBQ/MSBQ-MT yields DMPBQ, the substrate for the synthesis of γ-T by tocopherol cyclase (TC) (Porfirova et al. 2002; Cheng et al. 2003; Van Eenennaam et al. 2003; Kanwischer et al. 2005); hence, MPBQ/MSBQ-MT mutations disrupt flow through the MPBQ ⇒ DMPBQ ⇒ γ-T ⇒ α-T branch and redirect flow through the MPBQ ⇒ δ-T ⇒ β-T branch of the tocopherol biosynthetic pathway (Cheng et al. 2003; Van Eenennaam et al. 2003). The methylation of δ- and γ-T by γ-TMT yields β- and α-T, respectively; hence, γ-TMT mutations disrupt the synthesis of β- and α-T and cause a build up of δ- and γ-T, with proportions of the latter hinging on the activity of MPBQ/MSBQ-MT (Shintani and DellaPenna 1998; Shintani et al. 2002; Bergmüller et al. 2003; Cheng et al. 2003; Van Eenennaam et al. 2003; Tang et al. 2006). Null TC mutations disrupt ring cyclization of MPBQ to δ-T and DMPBQ to γ-T and are lethal in Arabidopsis (Porfirova et al. 2002). Non-lethal TC mutations, however, affect tocopherol composition, in addition to tocopherol accumulation (Kanwischer et al. 2005). Null MPBQ/MSBQ-MT mutations, in addition to redirecting flow in the tocopherol biosynthetic pathway and modifying tocopherol composition, disrupt plastoquinone (PQ) biosynthesis and are seedling lethal in Arabidopsis (Cook and Miles 1992; Cheng et al. 2003).
Two non-lethal mutations (m = Tph 1 and g = Tph 2) are known to disrupt α-T synthesis and produce novel tocopherol profiles in sunflower seeds, primarily increased β- and γ-T (Demurin 1993; Demurin et al. 1996; Hass et al. 2003; Tang et al. 2005). The functions disrupted by the m and g mutations have not been identified. The tocopherol composition phenotypes produced by the m and g loci, however, are characteristic of MPBQ/MSBQ-MT and γ-TMT mutations, respectively (Shintani and DellaPenna 1998; Shintani et al. 2002; Bergmüller et al. 2003; Cheng et al. 2003; Van Eenennaam et al. 2003). We focused on isolating the g locus and identifying the activity disrupted by the g mutation in the present study, whereas Tang et al. (2006), in a companion study, focused on isolating the m locus and identifying the activity disrupted by the m mutation. Tang et al. (2006) identified two MPBQ/MSBQ-MT paralogs (MT-1 and MT-2). The m mutation partially disrupts the synthesis of α-T, increases β- and δ-T percentages, and was caused by the insertion of a 5.2 kb Ty3/gypsy-like retrotransposon in exon 1 of MT-1. MT-1 was not transcribed in mutant homozygotes (m m); hence, the spontaneous m = Tph 1 mutation discovered by Demurin (1993) is a knockout of one of two MPBQ/MSBQ-MT paralogs (Tang et al. 2005, 2006). MT-1 and m cosegregated and mapped to linkage group 1.
The g mutation disrupts the synthesis of α-T and greatly increases γ-T percentages in sunflower seeds—wildtype homozygotes (g + g+) produced > 90% α-T and < 10% γ-T, whereas mutant homozygotes (g g) produced < 10% α-T and > 90% γ-T (Demurin 1993; Demurin et al. 1996). The phenotype of the mutant is consistent with the disruption of γ-TMT activity in developing sunflower seeds (Shintani and DellaPenna 1998; Bergmüller et al. 2003; Van Eenennaam et al. 2003). Our initial goals were to isolate genes encoding γ-TMT activities in sunflower and test for associations between g locus phenotypes and γ-TMT locus genotypes. Fortuitously, a previously unidentified mutation segregated in F2 populations developed for mapping the g locus (B109 × LG24 and R112 × LG24) and were subsequently targeted for further study and analysis. The mystery mutations were discovered to be allelic to d, a cryptic MT-2 mutation identified in a population (NMS373 × SRA16) segregating for MT-1 and MT-2 mutations; the mutant MT-2 allele was epistatically masked by the wildtype MT-1 allele in the wildtype parent (NMS373) (Tang et al. 2006). Here, we describe the isolation and analysis of γ-TMT paralogs (γ-TMT-1 and γ-TMT-2), identify the methyltransferase activity disrupted by the g mutation, describe offtype tocopherol profiles produced by interactions among three non-allelic methyltransferase mutations (m, g, and d), and substantiate, through forward genetic analyses, the association between d and MT-2 found by Tang et al. (2006).
Materials and methods
Genetic stocks and segregating populations
Tocopherol phenotypes and m, g, and d locus genotypes of several of the sunflower inbred lines used in our study were not known a priori. Genotypes for the biochemical mutant loci were ascertained, as described herein and by Tang et al. (2006), from biochemical phenotypes, γ-TMT, MT-1, and MT-2 allele sequences and INDEL marker genotypes, and γ-TMT, MT-1, and MT-2 transcript profiles. Genetic analyses were performed on wildtype and offtype inbred lines described by Tang et al. (2006; RHA280, RHA801, NMS373, VHG8, HG81, HD55, SRA16, VHB18, VHB45, and MB17), in addition to the wildtype and offtype inbred lines described herein (HA341, HA821, RHA344, 2696-1, Krasnodar 917, B109, R112, and LG24). Hybrids were produced between B109 and LG24 and R112 and LG24 by manually emasculating B109 and R112 flowers and using LG24 as the pollen parent (R112, B109, and LG24 are inbred lines). B109 × LG24 and R112 × LG24 F2 seeds were produced by manually selfing one hybrid individual per cross. Demurin (1993) developed LG24 (m m g g d + d +). R112 and B109 (m m g + g + d d) were developed from three-way hybrids (HA341/HA821//Krasnodar 917 and RHA344/2696-1//Krasnodar 917, respectively). Krasnodar 917 (m m g + g + d d) is a single-cross hybrid (VK373/VK66) (Demurin et al. 1996). VK373 and VK66 were developed from crosses to LG15 (Demurin 1993). HA341 (PI 509051), HA821 (PI 599984), RHA344 (PI 509054), and 2696-1 are wildtype inbred lines (Roath et al. 1986; Miller et al. 1987; Miller and Vick 2001).
Genomic DNA was isolated from 4- to 6-week-old leaves using a modified CTAB method (Murray and Thompson 1980). Total RNA was isolated from leaves (60 days after germination; 60-DAG) and kernels (seeds) (25 days after flowering; 25-DAF) using TRIZOL reagent (Invitrogen Life Technology, Carlsbad, CA, USA). Total RNA samples were treated with DNase I using TURBO DNA-Free (Ambion Inc., Austin, TX, USA). RNA quality and quantity were checked using denatured agarose gels on Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA).
Tocopherol phenotyping
Total lipids were isolated from seeds of several inbred lines (RHA344, HA821, HA341, 2696-1, B109, R112, and LG24), a single-cross hybrid (Krasnodar 917), 190 B109 × LG24 F2 progeny, and 239 R112 × LG24 F2 progeny for tocopherol quantification by reverse-phase high-performance liquid chromatography (HPLC). Single-cross hybrids and inbred lines were phenotyped by sampling and bulking eight seeds per genotype, whereas F2 progeny were phenotyped by assaying half-seed samples (the upper half of each F2 seed, distal to the embryo, was sampled). Tocopherols were extracted from seeds using protocols described by Burton et al. (1985) and Podda et al. (1996). The hull (pericarp) was removed and discarded and whole or half seeds were weighed, added to 5 ml ethanol and 2.5 ml 1% w/v aqueous ascorbic acid in a 25 ml screw-top glass tube, and crushed with a Teflon rod. We added 1.5 ml of concentrated potassium hydroxide (KOH) to the ground samples. Samples were saponified at 70°C for 30 min, mixed every 10 min, and cooled on crushed ice before adding 2.5 ml of 1% w/v aqueous ascorbic acid and 50 μl 2,6-di-tert-butyl-4-methylphenol as preservatives. Lipids were extracted from the latter using 5 ml hexane. The supernatant (4 ml) was drawn off and dried under vacuum using a SpeedVac (Savant Instruments, Holbrook, NY, USA). The dried samples were resuspended in 150 μl of 1:1 methanol:ethanol, vortexed, and added to an HPLC autosampler tube (HPLC analyses were perform on 50 μl samples).
Tocopherols were separated on a Waters 2695 HPLC using a 996 photodiode array detector (Milford, MA, USA). F2 progeny were phenotyped using a Waters Nova-Pak C18 3.9 × 150 mm2 column, whereas inbreds and hybrids were phenotyped using a Phenomenex Develosil RP-Aqueous C30 150 mm × 4.6 mm column (Torrance, CA, USA). β- and γ-T could not be separated on the Waters column, whereas α-, β-, γ-, and δ-T were separated by the Phenomenex column and separately quantified. The mobile phase was 99% MeOH and 1% water run under isocratic conditions of 1 ml/min with a run time of 14 min according to the manufacturer’s directions (Phenomenex, Torrance). Samples were held at 5°C to prevent oxidation of the tocopherols, whereas the column temperature was held at 20°C. UV detection was recorded at 295 nm.
α-, β-, γ-, and δ-T standards were obtained from Matreya Inc. (Pleasant Gap, PA, USA). Standards were dried of all hexane, diluted in 100% ethanol, and stored at − 20°C. Concentrations of the standards were measured by absorbance at the λ max for each tocopherol on a spectrophotometer (Spectronic 21, Bausch & Lomb, Rochester, NY, USA) and calculated using standard E-value divisors. Standards were diluted to an equal concentration and mixed to create a standard mixture, and a standard curve was developed by injecting different quantities of the standard mixture (0.05, 0.1, 0.1, 0.5, 1.0, and 2.0 μg) onto the column, recording areas under response peaks, and graphing the injected quantities against response peak area.
The isolation of γ-TMT and TC
Arabidopsis γ-TMT (GeneBank accession nos. AF104220 and At1g64970; Shintani and DellaPenna 1998; Bergmüller et al. 2003) and TC (GenBank accession nos. AL022537 and AT4g32770; Porfirova et al. 2002; Sattler et al. 2003) cDNA sequences were used as query templates to search for sunflower γ-TMT and TC homologs in the Compositate Genome Program Database (CGPdb; http://cgpdb.ucdavis.edu) and NCBI GenBank (http://www.ncbi.nlm.nih.gov). Once putative homologs were identified, cDNA clones for selected CGPdb ESTs were isolated and sequenced. We used 5′ and 3′ RACE (Rapid Amplification of cDNA Ends; FirstChoice RLM-RACE Kits, Ambion Inc.) to amplify 5′- and 3′-UTRs from developing seed RNAs (25-DAF) of three genotypes (NMS373, R112, and LG24). Full-length γ-TMT and TC cDNAs were isolated from NMS373, R112, LG24, RHA280, and RHA801 by RT-PCR using developing seed RNA templates. Full-length γ-TMT and TC genomic DNA sequences were isolated from NMS373, R112, LG24, RHA280, and RHA801 by PCR using leaf DNA templates. DNA sequencing was performed on ABI 3730 and 3730 XL DNA Analyzers (Applied Biosystems, Foster City, CA, USA). cDNA, genomic DNA, and protein sequences of γ-TMT and TC were aligned using GeneDoc 2.6 (http://www.psc.edu/biomed/genedoc/).
Genotyping and genetic mapping
Goodness-of-fit of observed segregation ratios for the g and d loci among 239 R112 × LG24 and 190 B109 × LG24 F2 individuals were tested using χ 2-statistics (Sokal and Rohlf 1981). The two loci were predicted to segregate 9 g + _ d + _ : 3 g +_ d d : 3 g g d + _ : 1 g g d d in both populations, where g + and d + are dominant (wildtype) alleles and g and d are recessive (mutant) alleles and g and d locus genotypes were inferred from α-, β-/γ-, and δ-T phenotypes. DNA bulks were developed for g + g + d + d + and g g d d genotypes by selecting 10 individuals from the upper tail of the g + _ d + _ α-T distribution and 10 individuals from the lower tail of the g g d d β-/γ-T distribution in the R112 × LG24 F2 population (Fig. 1). We performed bulked segregant analyses (BSA) (Michelmore et al. 1991) by screening R112, LG24, and the two DNA bulks for polymorphisms among mapped SSR marker loci (Tang et al. 2003). Polymorphic SSR markers were genotyped in R112 × LG24 on an ABI Prism 377 (Applied Biosystems). Genetic mapping analyses were performed using MAPMAKER (Lander et al. 1987), as described by Tang et al. (2002).
Single strand conformational polymorphism (SSCP) and INDEL markers were developed for γ-TMT and INDEL markers were developed for TC and screened for polymorphisms among several mutant and wildtype inbred lines (NMS373, R112, LG24, RHA280, RHA801, SRA16, VHG8, VHB18, VHB45, HG81, MB17, and HD55) using previously described methods (Kolkman et al. 2004). γ-TMT and MT-2 INDEL markers were genotyped and mapped in R112 × LG24 F2 (n = 94) and RHA280 × RHA801 RIL (n = 94) mapping populations using MAPMAKER (Lander et al. 1987), as previously described (Tang et al. 2002).
γ-TMT and TC expression analyses
Quantitative RT-PCR (qRT-PCR) analyses of γ-TMT, TC, and actin (GenBank acc. no. AF282624) transcript accumulation were performed in triplicate on RNAs isolated from developing seeds (25-DAF) and leaves (60-DAG) on an MJ Opticon Detector (MJ Research Inc., Waltham, MA, USA) using Platinum SYBR Green qPCR Kits (Invitrogen Life Technology); DNA sequences for forward and reverse actin primer sequences were FA = GCAAAAAGCAGCTCGTCTGT and RA = AGCAGCTTCCATTCCAATCA, respectively. The reaction contained in a final volume of 25 μl, 10 or 20 ng of reverse-transcribed total RNA from seeds or leaves, respectively, 2.0 μM of the forward and reverse primers, and 12.5 μl of 2× Platinum SYBR Green qPCR SuperMix. The initial denaturation step was performed at 50°C for 2 min, then 95°C for 2 min, followed by 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s. The PCR reaction was held at 95°C for 1 min, followed by 50°C for 3 min. Reassociation curves were produced by ramping the temperature from 60 to 90°C and recording the fluorescence intensity every 0.2°C with 10 s between reads. Standard curves were produced for γ-TMT, TC, and actin using 32, 16, 8, 4, and 2 ng of total RNA isolated from developing seeds (25-DAF) and 64, 32, 16, 8, and 4 ng of total RNA isolated from leaves (60-DAG). γ-TMT and TC transcript accumulation was normalized to actin using comparative threshold (C t) values calculated using the Opticon Monitor Analysis Software.
Statistical analyses
The intragenic and intergenic effects of the g and d loci on α-, β-/γ-, and δ-T percentages among B109 × LG24 and R112 × LG24 F2 progeny (n = 190 and n = 239, respectively) were estimated using Type III mixed model analysis of variance (ANOVA) statistics, where the independent variables were g and d locus genotypes (g + _ d +_, g + _ d d, g g d + _, and g g d d) inferred from phenotypes. SAS PROC GLM (Statistical Analysis System, Cary, NC, USA; http://www.sas.com) programs were developed to perform statistical analyses and estimate least square means for each of the four g × d genotypic classes (the data were unbalanced with no missing cells). Type III F-tests (F = M G/M R:G) were used to estimate the statistical significance of intragenic and intergenic effects, where genotype (G) effects (g, d, and g × d) were fixed, F2 progeny nested in genotype (R:G) effects were random, M G is the mean square for g, d, or g × d, and M R:G is the residual (R:G) mean square. The coefficient of determination (R 2) was estimated for the complete 22 factorial (two-locus) model (three degrees of freedom). ESTIMATE statements were used to estimate intragenic (g and d) and intergenic (g × d) effects.
The additive (A), dominant (D), A × A, A × D, D × A, and D × D effects of γ-TMT and MT-2 on α-, β-/γ-, and δ-T percentages among R112 × LG24 F2 progeny (n = 94) were estimated using Type III ANOVA statistics and orthogonal contrasts among γ-TMT × MT-2 genotype means (Falconer and Mackay 1996; Lynch and Walsh 1998). SAS PROC GLM programs were developed to perform statistical analyses and estimate least square means for each of the nine γ-TMT × MT-2 genotypes; A, D, A × A, A × D, D × A, and D × D effects were estimated using ESTIMATE statements. The data were unbalanced with no empty cells (genotypic data were present for each of the nine γ-TMT × MT-2 genotypes). Type III F-tests (F = M G/M R:G) were used to estimated the statistical significance of each intragenic and intergenic effect, where DNA marker genotype (G) effects (γ-TMT, MT-2, and γ-TMT × MT-2) were fixed, F2 progeny nested in genotype (R:G) effects were random, M G is the mean square for γ-TMT, MT-2, or γ-TMT × MT-2, and M R:G is the residual (R:G) mean square. The coefficient of determination (R 2) was estimated for the complete 32 factorial (two-locus) model (eight degrees of freedom).
Results
Two mutant loci (g and d) segregated, epistatically interacted, and produced novel tocopherol profiles in sunflower seed oil
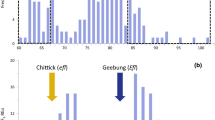
Tocopherol percentages were quantified in physiologically mature kernels (seeds) of wildtype and mutant inbred lines and F2 individuals by HPLC (Table 1). RHA344, 2696-1, HA821, and HA341 produced 92.0–96.7% α-T and had wildtype tocopherol profiles, whereas B109, R112, and Krasnodar 917 produced significantly less α-T (35.3–45.1%) and significantly more β-T (54.9–64.7%) than wildtype inbred lines (P < 0.0001). Four non-overlapping phenotypic classes were produced by the segregation of g and d among B109 × LG24 and R112 × LG24 F2 progeny (Fig. 1). The observed segregation ratios were not significantly different from 9 g + _ d + _ : 3 g +_ d d : 3 g g d + _ : 1 g g d d in B109 × LG24 (108 g + _ d + _ : 44 g +_ d d : 30 g g d + _ : 8 g g d d; χ 2 = 4.52; P = 0.21) or R112 × LG24 (139 g +_ d + _ : 42 g +_ d d : 46 g g d + _ : 12 g g d d; χ 2 = 1.09; P = 0.78). The intragenic and intergenic interaction effects of g and d and least square means for the four m × g × d genotypic classes (m m g +_ d + _, m m g + _ d d, m m g g d + _, and m m g g d d) were estimated among 190 B109 × LG24 and 239 R112 × LG24 F2 progeny (Tables 2, 3; Fig. 1). The segregation of g and d produced 91–96% of the phenotypic variability for α-, β-/γ-, and δ-T percentages (Tables 2, 3).
The g mutation completely disrupted the synthesis of α-T and caused the accumulation of γ-T in m m d + d + individuals and δ-T in m m d d individuals, whereas the d mutation partially disrupted the synthesis of α-T and caused the accumulation of β-T in m m g + g + individuals and δ-T in m m g g individuals (Fig, 1; Tables 1, 2, 3). The intragenic effect of g was significant for α-, β-/γ-, and δ-T percentages (P < 0.0001), the intragenic effect of d was significant for α- and δ-T percentages (P < 0.0001), and the two loci epistatically interacted—g × d effects were significant for α-, β-/γ-, and δ-T percentages (P < 0.0001) (Table 3).
Genetic mapping of tocopherol mutant loci
The mutant loci (g and d) were genetically mapped in the R112 × LG24 F2 population using dominant and recessive genotypes (g + _ d + _, g + _ d d, g g d + _, and g g d d) inferred from tocopherol phenotypes (Fig. 1). Bulked-segregant analyses (Michelmore et al. 1991) identified SSR markers linked to g and d and narrowed the search for additional linked SSR marker loci down to two linkage groups (4 and 8) identified by Hass et al. (2003). We screened 78 SSR markers for polymorphisms between the parents of the R112 × LG24, g + _ and g g F2 DNA bulks, and d + _ and d d F2 DNA bulks using the PCR-multiplexes described by Tang et al. (2003). Twenty-four of the 78 SSR markers were polymorphic between R112 and LG24 and one (ORS70) was polymorphic between g + _ and g g DNA bulks. None of the SSR markers in the multiplex set were polymorphic between d + _ and d d bulks, so 82 additional SSR markers were screened. The latter were dispersed across the 17 chromosomes in the haploid genome (x = 17) of sunflower and spanned gaps between SSR loci interrogated by the multiplexes (Tang et al. 2003). Twelve of the 82 SSR markers were polymorphic in R112 × LG24 and one (ORS676) was polymorphic between d + _ and d d DNA bulks.
ORS676 and ORS70 had been previously mapped to linkage groups 4 and 8, respectively (Tang et al. 2002, 2003; Yu et al. 2003), so 37 additional linkage group (LG) 4 and 43 additional LG 8 SSR markers were screened for polymorphisms between R112 and LG24. Three polymorphic SSR markers from LG 4 and 12 polymorphic SSR markers from LG 8 were genotyped and mapped in R112 × LG24 (Fig. 2). g was tightly linked to ORS70 and mapped to LG 8 and d was tightly linked to ORS676 and mapped to LG 4.
Genetic mapping of methyltransferase and tocopherol mutant loci. Genetic mapping of γ-tocopherol methyltransferase (γ-TMT-1 and γ-TMT-2), 2-methyl-6-phytyl-1,4-benzoquinone/2-methyl-6-solanyl-1,4-benzoquinone methyltransferase (MPBQ/MSBQ-MT-2), tocopherol cyclase (TC), and tocopherol mutant (g and d) loci on linkage groups 4 and 8 in the R112 × LG24 F2 (displayed on the left in each linkage group pair) and RHA280 × RHA801 recombinant inbred line (displayed on the right of each linkage group pair) mapping populations. Linkage groups were aligned using SSR marker loci (ORS and CRT prefixes) genotyped in both mapping populations (highlighted in bold)
DNA polymorphisms in γ-tocopherol methyltransferase (γ-TMT) alleles resequenced from mutant and wildtype inbred lines
We isolated full-length cDNA and genomic DNA sequences for γ-TMT and resequenced γ-TMT alleles from wildtype and mutant inbred lines to screen for mutations. GenBank (http://www.ncbi.nlm.nih.gov/) and the Compositae Genome Database (CGPdb; http://cgpdb.ucdavis.edu/) were searched for sunflower cDNA and genomic DNA sequences homologous to Arabidopsis γ-TMT (GeneBank acc. nos. AF104220 and At1g64970; Shintani and DellaPenna 1998; Bergmüller et al. 2003) and yielded an EST contig (CGPdb contig5094; 697 bp) and an EST (GeneBank acc. no. CD852146; 874 bp) with significant homology to Arabidopsis γ-TMT (e−60 and 4e−96, respectively); three ESTs (QHK3O09, QHM1I15, and QHM3O15) were found in the contig. cDNA clones for two ESTs (QHK3O09 and QHM1I15) were isolated, sequenced, and aligned with CD852146 and created a 1,291 bp long contig spanning the complete coding sequence (CDS). Complete UTR sequences were isolated by RACE-PCR using R10, R14, or F67 (Table 4).
Full-length cDNA alleles were resequenced from wildtype and mutant inbred lines (NMS373, R112, LG24, RHA280, and RHA801). Two γ-TMT transcripts were amplified from RHA801 (γ-TMT-1 and γ-TMT-2), whereas only one (γ-TMT-1) was amplified from NMS373, R112, LG24, and RHA280 (Supplemental Figure S1). The presence of two γ-TMT transcripts in developing seeds of RHA801 and polymorphisms in DNA sequences isolated from LG24 and RHA801 prompted us to screen for γ-TMT paralogs in other inbred lines. Genomic DNA fragments amplified from several inbred lines using the F1/R19 primer pair produced a single 571 bp band on agarose (the F1/R19 primer pair amplifies exon 1 and the first 207 bp of intron 1 in γ-TMT-1) (Fig. 3; Table 4). When separated by SSCP analysis, two loci (four strands) were observed in the wildtype (g + g +) inbred line RHA801 and four mutant (g g) inbred lines (LG24, VHG8, HG81, and HD55), whereas a single locus (two strands) was observed in other g + g + inbred lines (NMS373, R112, RHA280, SRA16, VHB18, VHB45, and MB17)—VHG8, HG81, and HD55 inherited the mutant g allele from LG24 (Fig. 3). The presence of a deletion in one of the paralogs (γ-TMT-1) facilitated the development of a codominant INDEL marker (F67/R78) to screen for the presence of both paralogs and a dominant INDEL marker (F69/R78) to screen for the presence of the second paralog only (Fig. 3; Table 4).
γ-TMT is duplicated in LG24 and RHA801. a The F1/R19 primer pair amplified two loci from genomic DNAs isolated from RHA801, LG24, VHG8, HG81, and HD55 and a single locus from genomic DNAs isolated from NMS373, R112, SRA16, VHB18, VHB45, MB17, and RHA280. Genomic DNA fragments amplified by F1/R19 were nearly identical in length and could not be separated on agarose (lower panel), but harbored several DNA polymorphisms and were separated by single strand confirmational polymorphism (SSCP) analysis (upper panel); complementary γ-TMT-1 and γ-TMT-2 strands are labeled 1 and 2, respectively. b Locations of F67 and F69 primers in the γ-TMT genomic DNA sequence alignment (nt 3,080 to 3,200). F67 is complementary to DNA sequences present in both paralogs, whereas F69 is complementary to DNA sequences present in the second paralog only (LG24-2 and RHA801-2). c Length polymorphisms for two γ-TMT INDEL markers (F67/R78 and F69/R78) among several inbred lines. F67/R78 amplified both paralogs (518, 638, or 665 bp), whereas F69/R78 only amplified γ-TMT-2 (567 bp) from LG24, VHG8, HG81, HD55, and RHA801. VHG8, HG81, and HD55 are homozygous for the mutant g allele transmitted by LG24. NMS373, R112, SRA16, VHB18, VHB45, MB17, and RHA280 either carry one paralog or two identical paralogs
Full-length genomic DNA sequences were subsequently isolated from wildtype and mutant inbred lines (GenBank acc. no. DQ229828-34; Supplemental Figure S1). One paralog (γ-TMT) was identified from NMS373, R112, and RHA280, whereas two paralogs (γ-TMT-1 and γ-TMT-2) were identified from LG24 and RHA801 (Fig. 3). Six exons encoding 314 amino acids were present in both paralogs (intron splice sites were conserved). The amino acid similarity between sunflower and Arabidopsis γ-TMT proteins was 68%. We identified 173 SNPs and 33 INDELs in coding and non-coding sequences. Five haplotypes were present: NMS373 (haplotype 1; 4,294 bp), R112 (haplotype 2; 4,029 bp), RHA280 (haplotype 3; 4,035 bp), and LG24-1 and RHA801-1 (haplotype 4; 4,151 bp), and LG24-2 and RHA801-2 (haplotype 5; 4,304 bp). LG24-1 and RHA801-1 genomic DNA sequences were 100% identical (haplotype 4), as were LG24-2 and RHA801-2 genomic DNA sequences (haplotype 5); hence, LG24-1 and RHA801-1 were allelic and assigned to paralog 1 and LG24-2 and RHA801-2 were allelic and assigned to paralog 2. Haplotypes 4 and 5 were distinguished by 96 SNPs, but 92% identical (Supplemental Figure S1). Genomic DNA sequences isolated from NMS373, R112, and RHA280 could not be unequivocally identified as one paralog or the other. Nucleotide identities were slightly greater between NMS373 (haplotype 1) and haplotype 5 (94%) than between NMS373 and haplotype 4 (92%), whereas nucleotide identities were slightly greater between R112 (haplotype 2) or RHA280 (haplotype 3) and haplotype 4 (93%) than between R112 or RHA280 and haplotype 5 (88%); the nucleotide identity between R112 and RHA280 was 99%.
We identified 12 synonymous and three non-synonymous SNPs in coding sequences (CDSs). The non-synonymous SNPs—TCT (Ser-84) to TAT (Tyr-84), CCG (Pro-89) to CTG (Leu-89), and CCT (Pro-157) to TCT (Ser-157)—were not associated with mutant phenotypes; SNP alleles identified in inbred lines homozygous for mutant g or d alleles were found in one or more inbred lines homozygous for wildtype g or d alleles (Supplemental Figures S1, S2).
The g mutation disrupts γ-TMT transcription
γ-TMT transcription was 10- to 13-fold greater in leaves and 36 to 51-fold greater in developing seeds of wildtype (g + g +) than mutant (g g) inbred lines (Fig. 4). γ-TMT-1 was alternatively spliced in LG24 (Supplemental Figure S3). The alternatively spliced transcript (LG24-1-AS) was identified by sequencing 132 and 251 bp cDNA fragments amplified by the F27/R35 primer pair (flanking intron 2) from RNAs isolated from leaves and developing seeds of LG24 (g g). The normally spliced (NS) transcripts isolated from wildtype and mutant inbred lines were identical (R112-NS and LG24-1-NS in Supplemental Figure S3). The alternatively spliced (AS) transcript (251 bp) isolated from LG24 harbored the first 102 bp and the last 17 bp of the intron 2 sequence (LG24-1-AS in Supplemental Figure S3). Sequences of the normally and alternatively spliced LG24 transcripts were identical to haplotype 4 (LG24-1) and, consequently, were produced by γ-TMT-1. γ-TMT-2 was not transcribed in LG24 (Fig. 5).
γ-Tocopherol methyltransferase is weakly transcribed and alternatively spliced in inbred lines homozygous for the g mutation. a RT-PCR analyses of RNAs isolated from developing seeds 25 days after flowering (25-DAF) and leaves 60 days after germination (60-DAG) of two wildtype (g + g + = +) inbred lines (NMS373 and R112) and two mutant (g g = −) inbred lines (LG24 and HG81) using a primer pair (F27/R35) flanking the second intron in γ-TMT amplified a 132 bp cDNA fragment from wildtype and 132 and 251 bp cDNA fragments from mutant inbred lines. The 132 bp amplicon was produced by normal splicing, whereas the longer amplicon was produced by alternative splicing of intron 2. F27/R35 amplified a 327 bp genomic DNA fragment from genomic DNA isolated from LG24. M is a DNA ladder. b Transcript accumulation in developing seeds (25-DAF) and leaves (60-DAG) of two wildtype (g + g +) inbred lines (R112 and NMS373) and two mutant (g g) inbred lines (LG24 and HG81) quantified by real-time RT-PCR using forward and reverse primers (F5/R7) complementary to conserved DNA sequences spanning a 158 bp fragment in the exon 1 of γ-TMT. The dependent (x-axis) variable was calculated using the ratio of the cycle threshold for each inbred line to the cycle threshold for LG24 (x = 1 for LG24) normalized using a sunflower actin (ACT) gene
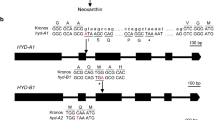
γ-Tocopherol methyltransferase-2 is not transcribed in LG24, an inbred line homozygous for the g mutation. a Transcription initiation sites T1 (nt 1) and T39 (nt 39) found in γ-TMT transcripts. Forward primers were placed upstream (F1; nt 1–22) and downstream (F2; nt 39–59) of the T39 transcription initiation site. b Ethidium bromide-stained agarose gel showing cDNA fragments amplified by nested 5′-RACE PCR from developing seed RNAs (25-DAF) using an inner primer in exon 1 (R10) and an outer primer in exon 2 (R24). Two cDNA fragments (343 and 381 bp) were amplified from wildtype (g + g + = +) inbred lines (NMS373 and R112), whereas a single cDNA fragment (343 bp) was amplified from a mutant (g g = −) inbred line (LG24). Two transcription initiation sites (bp 1 and bp 39) were found in wildtype transcripts, whereas only one (bp 39) was found in the mutant transcript. c cDNA fragments amplified by RT-PCR from RNAs isolated from developing seeds (25-DAF) using forward primers F1 or F2 and a reverse primer (R24) complementary to DNA sequences in exon 2 (nt 1,452 to 1,471). F1/R24 amplified a 411 bp cDNA fragment from wildtype (+) inbred lines (NMS373, R112, RHA280, and RHA801) and a very faint 411 bp cDNA fragment from the mutant (−) inbred line (LG24). F2/R24 amplified a 373 bp cDNA fragment from wildtype and mutant inbred lines
Two transcription initiation sites (nt 1 and 39) were identified in wildtype (g + g +) inbred lines, but only one (nt 39) was identified in mutant (g g) inbred lines—343 and 381 bp cDNA fragments were amplified from NMS373 and R112 (g + g +), whereas only the 343 bp cDNA fragment was amplified from LG24 (g g) (Fig. 5). The latter was identical to LG24-1 cDNA and genomic DNA sequences (Supplemental Figure S1). The absence of the nt 1 transcription site (T1) in the mutant was substantiated by RT-PCR analyses using forward primers upstream (F1) and downstream (F2) of the nt 39 transcription initiation site (T39) and a reverse primer (R24) complementary to conserved DNA sequences in exon 2 (nt 1,452-1,471) (Fig. 5; Table 4). F1/R24 amplified a 411 bp cDNA fragment from leaves and developing seeds of wildtype (NMS373, R112, RHA280, and RHA801) and mutant (LG24) inbred lines, although the LG24 cDNA fragment was barely perceptible (Fig. 5). F2/R24 amplified a single cDNA fragment (343 bp) from wildtype and mutant inbred lines. The signal was substantially weaker in LG24 than wildtype inbred lines (Fig. 5). Hence, both γ-TMT paralogs were transcribed in leaves and seeds of RHA801 (g + g +), whereas only γ-TMT-1 was transcribed, albeit weakly, in LG24 (g g) (Figs. 4, 5).
Cosegregation of methyltransferase and tocopherol mutant loci
We tested for an association between γ-TMT and g by developing and mapping a codominant γ-TMT INDEL marker (F9/R24) in R112 × LG24 and RHA280 × RHA801 (Figs. 2, 6, 7). F9 and R24 are complementary to conserved DNA sequences in the first and second exons, respectively, of both γ-TMT paralogs and flank 1, 8, 9, 10, and 134 bp INDELs in the first intron (Table 4; Supplemental Figure S1). The F9/R24 INDEL marker was screened for length polymorphisms among several mutant and wildtype inbred lines and amplified codominant DNA fragments (1,037–1,164 bp in length) from 12 inbred lines and a dominant DNA fragment (786 bp) from 11 of the 12 inbred lines; the 786 bp DNA fragment was not amplified from RHA801 (Fig. 6). Genomic DNA fragments amplified from R112 and LG24 were sequenced to identify the origin of each. The 786 bp DNA fragment was shorter than any predicted from the reference sequence and was discovered to be a randomly amplified polymorphic DNA (RAPD) (Fig. 6). When the F9/R24 INDEL marker was genotyped in RHA280 × RHA801, the RAPD marker (786 bp DNA fragment) segregated and mapped to linkage group 16 (data not shown). The 1,164 bp DNA fragment isolated from LG24 harbored alleles from both γ-TMT paralogs, whereas the 1,037 bp DNA fragment isolated from R112 harbored the allele from γ-TMT-1 only. γ-TMT-1 and 2 are either very tightly linked or tandemly duplicated (recombinants were not observed). The γ-TMT-1 and γ-TMT-2 loci were genotyped in R112 × LG24 and RHA280 × RHA801 using the F9/R24 INDEL, mapped to LG 8, and cosegregated with the g locus (Fig. 2).
The association between MT-2 and d discovered by Tang et al. (2006) was substantiated in the present study by genotyping a codominant INDEL marker (F24/R25) for MT-2 in the R112 × LG24 F2 (n = 94) and RHA280 × RHA801 RIL (n = 94) mapping populations (Figs. 2, 7). The MT-2 locus mapped to LG 4 and cosegregated with the d locus.
γ-TMT and MT-2 produce novel tocopherol profiles through intragenic and intergenic interactions
The development of codominant INDEL markers diagnostic for wildtype and mutant γ-TMT and MT-2 alleles facilitated the estimation of intragenic and intergenic effects and degree of dominance of the two loci among R112 × LG24 F2 progeny (Tables 5, 6). Of the 24 intragenic and intergenic effects, 21 were significant, and the two methyltransferase loci were associated with 94–97% of the variability for α-, β-/γ-, and δ-T percentages (Table 6). The additive effects of γ-TMT were twofold greater than the additive effects of MT-2, and of opposite sign because the mutant alleles were transmitted by different parents (d by R112 and g by LG24). The dominance effects of γ-TMT were several-fold greater than the dominance effects of MT-2 (the latter were non-significant for β-/γ- and δ-T). The degree of dominance for the γ-TMT locus was nearly complete (0.86) to complete (1.00). The wildtype γ-TMT allele (g +) was dominant to the mutant γ-TMT allele (g), as predicted for a null or knockout mutation. The degree of dominance for the MT-2 locus was nearly additive (0.12) to incompletely dominant (0.49), as predicted for a partial loss- or gain-of-function mutation.
Isolation and genetic mapping of tocopherol cyclase (TC)
GenBank (http://www.ncbi.nlm.nih.gov/) and the Compositae Genome Database (CGPdb, http://cgpdb.ucdavis.edu/) were screened for sunflower cDNA and genomic DNA sequences homologous to Arabidopsis TC (GeneBank acc. nos. AL022537 and AT4g32770; Porfirova et al. 2002; Sattler et al. 2003). BLAST analyses identified two ESTs in the CGPdb, QHB20A01 (720 bp) and QHI13M21 (502 bp), with significant homology to Arabidopsis TC (9e-28 and e−20, respectively). The cDNA clone for QHB20A01 was isolated, sequenced, and supplied 1,540 bp of the TC coding sequence. The complete 5′-UTR was isolated by nested 5′-RACE PCR using R16 as the inner and R27 as the outer primer (Table 4). The 3′-UTR in the initial contig was discovered to be complete by sequencing cDNA fragments amplified by 3′-RACE PCR using F42 as the inner and F39 as the outer primer.
Complete TC cDNA sequences were isolated from NMS373, R112, LG24, RHA280, and RHA801 by RT-PCR using developing seed RNAs. Full-length TC genomic DNA sequences were subsequently isolated by sequencing and assembling overlapping genomic DNA fragments amplified by PCR; nine exons were identified and encode 383 amino acids (GenBank acc. no. DQ229845-49; Supplemental Figures S4, S5). The amino acid similarity between sunflower and Arabidopsis tocopherol cyclases was 65%. We identified two INDELs and one SNP in non-coding sequences, one synonymous and two non-synonymous SNPs in coding sequences, and two haplotypes: NMS373, LG24, RHA280, and RHA801 (haplotype 1; 5,354 bp) and R112 (haplotype 2; 5,389 bp). Neither of the non-synonymous SNPs—CTT (Leu-32) to CAT (His-32) and AAT (Asn-279) to GAT (Asp-279)—were associated with g or d mutant phenotypes (Supplemental Figure S5).
TC transcription was not significantly different in leaves of NMS373, R112, LG24, and SRA16, but was twofold lower in developing seeds (25-DAF) of SRA16 than NMS373, R112, and LG24 (data not shown); SRA16 is wildtype for g and d (Table 1). We developed an INDEL marker for TC by positioning a pair of primers (F34/R38) upstream and downstream of a 38 bp INDEL found in intron 5 (Fig. 6; Supplemental Figure S4). Two F34/R38 INDEL marker alleles (195 and 233 bp) were observed among 12 mutant and wildtype inbred lines. The TC locus was genotyped in the R112 × LG24 F2 mapping population using the F34/R38 INDEL marker, mapped to linkage group 8, and segregated independently of g and d (Figs. 2, 7).
Discussion
The development of seed oils with dramatically different tocopherol profiles has been accomplished in sunflower without genetic engineering by identifying spontaneous and induced loss-of-function mutations that disrupt the synthesis of α-T and enhance the synthesis of β-, γ-, and δ-T (Demurin 1993; Demurin et al. 1996; Velasco et al. 2004a, b). The diversity of tocopherol profiles found in sunflower is unparalleled because wildtypes normally accumulate > 90% α-T, the ultimate product of the tocopherol biosynthetic pathway, and because other tocopherols accumulate when one or more upstream methyltransferase activities are disrupted by mutations (Table 1; Tang et al. 2006). The three non-allelic methyltransferase mutations identified in sunflower (m, g, and d), when coupled with the wildtype, produce an unprecedented spectrum of tocopherol profiles.
Our analyses shed light on the genetic mechanism underlying the g = Tph 2 mutation of Demurin (1993), substantiated the association between d and MT-2 (Tang et al. 2006), and traced the origin of the mutant MT-2 allele (d) segregating in B109 × LG24 and R112 × LG24 to B109 and R112. LG24 carries a wildtype d allele (d +) not found in any of the wildtype inbred lines we screened (Tang et al. 2006). The prevalence of the d + allele (or of quantitatively different d alleles) in wildtype germplasm is not known.
The cDNA polymorphisms discovered in γ-TMT-1 and γ-TMT-2 were diagnostic for the g = Tph 2 mutation; however, none of the DNA polymorphisms found between wildtype and mutant γ-TMT-1 and γ-TMT-2 alleles were associated with the mutant phenotype—the γ-TMT-1 and γ-TMT-2 alleles found in the mutant (LG24) and one of the wildtype (RHA801) inbred lines were identical (haplotypes 4 and 5, respectively; Figs. 3, 4, 5; Supplemental Figures S1, S2). Greatly diminished transcription of one paralog (γ-TMT-1; Fig. 4), complete loss of transcription of the other paralog (γ-TMT-2; Fig. 5), loss of a transcription initiation site (T1; Fig. 5), and alternative splicing (LG24-1-AS; Supplemental Figure S3) are characteristic of transposable element-induced mutations, particularly transposon insertions in introns and upstream regulatory sequences (Johns et al. 1985; Varagona et al. 1992; Weil et al. 1992; Brown 1996; Marillonnet and Wessler 1997; Lal et al. 1999, 2003; Selinger and Chandler 2001; Cui et al. 2003; Greco et al. 2005). Transposon-induced mutations often diminish or completely disrupt transcription, cause alternate splicing, and suppress or activate transcription initiation sites. We identified several DNA fragments homologous to class I and II transposable elements (Bennetzen 2000; Feschotte et al. 2002) in the introns of γ-TMT (Tang et al. 2006); however, none were associated with the mutant phenotype. Because g, γ-TMT-1, and γ-TMT-2 cosegregated, the mutation must be very tightly linked to the γ-TMT locus and may disrupt regulatory sequences.
The γ-TMT paralogs found in RHA801 and LG24 are either very tightly linked or tandemly duplicated (Figs. 2, 3, 7). The two paralogs cosegregated in the R112 × LG24 F2 and RHA280 × RHA801 RIL mapping populations (Figs. 2, 7). Moreover, the duplication (transmitted by LG24) was present in g g inbred lines developed by phenotypic selection and SSD among R112 × LG24 F2, F3, and F4 progeny (Figs. 3, 4, 6; Tang et al. 2006). We speculated that the duplication might be associated with the apparently strong γ-TMT activity found in developing seeds of sunflower wildtypes; however, paralogs were not identified in the other three wildtype inbred lines we screened (NMS373, R112, and RHA280). The latter either carry one homolog or identical paralogs of γ-TMT. The second possibility cannot be ruled out, but seems remote because several SNPs and INDEL distinguish the paralogs discovered in RHA801 and LG24—the nucleotide identity between γ-TMT-1 (haplotype 4) and γ-TMT-2 (haplotype 5) was 91.7% (Supplemental Figure S1). Unless NMS373, RHA280, and other wildtypes carry identical paralogs, the duplication and α-T percentage are uncorrelated and the activity of a single γ-TMT locus is sufficient to produce wildtype tocopherol profiles. Physical mapping of the γ-TMT locus is underway using clones isolated from a bacterial artificial chromosome (BAC) library developed for a wildtype inbred line (HA383; unpublished data; http://www.genome.clemson.edu), should shed light on the structure of the locus and presence or absence of duplications in other wildtypes, and supply DNA sequences for identifying the g mutation, if present, in upstream regulatory sequences.
References
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Bergmüller E, Porfirova S, Dörmann P (2003) Characterization of an Arabidopsis mutant deficient in γ-tocopherol methyltransferase. Plant Mol Biol 52:1181–1190
Bramley PM, Elmadfa I, Kafatos A, Kelly FJ, Manios Y, Roxborough HE, Schuch W, Sheehy PJA, Wagner K-H (2000) Vitamin E. J Sci Food Agric 80:913–938
Brigelius-Flohe R, Traber MG (1999) Vitamin E: function and metabolism. FASEB J 13:1145–1155
Brown JWS (1996) Arabidopsis intron mutations and pre-mRNA splicing. Plant J 10:771–780
Burton GW, Webb A, Ingold KU (1985) A mild, rapid, and efficient method of lipid extraction for use in determining vitamin E/lipid ratios. Lipids 20:29–39
Cahoon EB, Hall SE, Ripp KG, Ganzke TS, Hitz WD, Coughlan SJ (2003) Metabolic redesign of vitamin E biosynthesis in plants for tocotrienol production and increased antioxidant content. Nat Biotech 21:1082–1087
Cheng Z, Sattler S, Maeda H, Sakuragi Y, Bryant DA, DellaPenna D (2003) Highly divergent methyltransferases catalyze a conserved reaction in tocopherol and plastoquinone synthesis in cyanobacteria and photosynthetic eukaryotes. Plant Cell 15:2343–2356
Christen S, Woodall AA, Shigenaga MK, Southwell-Keely PT, Duncan MW, Ames BN (1997) Gamma-tocopherol traps mutagenic electrophiles such as NO(X) and complements alpha-tocopherol: physiological implications. Proc Natl Acad Sci USA 94:3217–3222
Cook W, Miles D (1992) Nuclear mutations affecting plastoquinone accumulation in maize. Photosynth Res 31:99–111
Cui X, Hsiac AP, Liu F, Ashlockd DA, Wise RP, Schnable PS (2003) Alternative transcription initiation sites and polyadenylation sites are recruited during Mu suppression at the rf2a locus of maize. Genetics 163:685–698
Demurin Y (1993) Genetic variability of tocopherol composition in sunflower seeds. Helia 16:59–62
Demurin Y, Skoric D, Karlovic D (1996) Genetic variability of tocopherol composition in sunflower seeds as a basis of breeding for improved oil quality. Plant Breed 115:33–36
Devaraj S, Traber MG (2003) Gamma-tocopherol, the new vitamin E? Am J Clin Nutr 77:530–531
Dolde D, Vlahakis C, Hazebroek J (1999) Tocopherols in breeding lines and effects of plant location, fatty acid composition, and temperature during development. J Am Oil Chem Soc 76:349–355
Eitenmiller RR (1997) Vitamin E content of fats and oils: nutritional implications. Food Technol 51:78–81
Falconer DS, Mackay TF (1996) Introduction to quantitative genetics. Longman, Harlow
Feschotte C, Jiang N, Wessler SR (2002) Plant transposable elements: where genetics meets genomics. Nat Rev Genet 3:329–341
Greco R, Ouwerkerk PBF, Pereira A (2005) Supperession of an atypically spliced rice CACTA transposon transcript in transgenic plants. Genetics 169:2383–2387
Grusak MA, DellaPenna D (1999) Improving the nutrient composition of plants to enhance human nutrition and health. Annu Rev Plant Physiol Plant Mol Biol 50:133–161
Hass CG, Leonard SW, Miller JF, Slabaugh MB, Traber MG, Knapp SJ (2003) Genetics of tocopherol (Vitamin E) composition mutants in sunflower. In: Abstract of plant and animal genome conference XI, San Diego, CA, USA,11–15 January 2003. http://www.intl-pagorg/11/abstracts/P7b_P821_XI.html
Havaux M, Eymery F, Porfirova S, Rey P, Dormann P (2005) Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17:3451–3469
Hensley K, Benaksas EJ, Bolli R, Comp P, Grammas P, Hamdheydari L, Mou S, Pye QN, Stoddard MF, Wallis G, Williamson KS, West M, Wechter WJ, Floyd RA (2004) New perspectives on vitamin E: gamma-tocopherol and carboxyelthylhydroxychroman metabolites in biology and medicine. Free Radic Biol Med 36:1–15
Jiang Q, Elson-Schwab I, Courtemanche C, Ames BN (2000) Gamma-tocopherol and its major metabolite, in contrast to alpha-tocopherol, inhibit cyclooxygenase activity in macrophages and epithelial cells. Proc Natl Acad Sci USA 97:11494–11499
Johns MA, Mottinger J, Freeling M (1985) A low copy number, copia-like transposon in maize. EMBO J 4:1093–1102
Kamal-Eldin A, Appelqvist LA (1996) The chemistry and antioxidant properties of tocopherols and tocotrienols. Lipids 31:671–701
Kanwischer M, Porfirova S, Bergmüller E, Dormann P (2005) Alterations in tocopherol cyclase activity in transgenic and mutant plants of Arabidopsis affect tocopherol content, tocopherol composition, and oxidative stress. Plant Physiol 137:713–723
Kolkman JM, Slabaugh MB, Bruniard JM, Berry S, Bushman BS, Olungu C, Maes N, Abratti G, Zambelli A, Miller JF, Leon A, Knapp SJ (2004) Acetohydroxyacid synthase mutations conferring resistance to imidazolinone or sulfonylurea herbicides in sunflower. Theor Appl Genet 109:1147–1159
Lal SK, Choi J-H, Shaw JR, Hannah LC (1999) A splice site mutant of maize activates cryptic splice sites, elicits intron inclusion and exon exclusion, and permits branch point elucidation. Plant Physiol 121:411–418
Lal SK, Giroux MJ, Brendel V, Vallejos CE, Hannah LC (2003) The maize genome contains a helitron insertion. Plant Cell 15:381–391
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lynch M, Walsh B (1998) Genetics and analysis of quantitative traits. Sinauer, Sunderland
Marillonnet S, Wessler SR (1997) Retrotransposon insertion into the maize waxy gene results in tissue-specific RNA processing. Plant Cell 9:967–978
Michelmore RW, Paran I, Kesseli V (1991) Identification of markers linked to disease resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Miller JF, Vick BA (2001) Registration of four high linoleic sunflower germplasms. Crop Sci 41:602
Miller JF, Zimmerman DC, Vick BA, Roath WW (1987) Registration of sixteen high oleic sunflower germplasm lines and bulk populations. Crop Sci 27:1323
Murray MG, Thompson WR (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Ohrvall M, Sundlof G, Vessby B (1996) Gamma, but not alpha, tocopherol levels in serum are reduced in coronary heart disease patients. J Intern Med 239:111–117
Pfluger P, Kluth D, Landes N, Bumke-Vogt C, Brigelius-Flohe R (2004) Vitamin E: underestimated as an antioxidant. Redox Rep 9:249–254
Podda M, Weber C, Traber MG, Packer L (1996) Simultaneous determination of tissue tocopherols, tocotrienols, ubiquinols, and ubiquinones. J Lipid Res 37:893–901
Porfirova S, Bergmüller E, Tropf S, Lemke R, Dormann P (2002) Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc Natl Acad Sci USA 99:12495–12500
Roath WW, Miller JF, Gulya T (1986) Registration of sunflower parental lines HA 821 and HA 822. Crop Sci 26:217
Rocheford TR, Wong JC, Egesel CO, Lambert RJ (2002) Enhancement of vitamin E levels in corn. J Am Coll Nutr 21:191S–198S
Sattler SE, Cahoon EB, Coughlan SJ, DellaPenna D (2003) Characterization of tocopherol cyclases from higher plants and cyanobacteria: evolutionary implications for tocopherol synthesis and function. Plant Physiol 132:2184–2195
Sattler SE, Gillilanda LU, Magallanes-Lundbacka M, Pollard M, DellaPenna D (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16:1419–1432
Selinger DA, Chandler VL (2001) B-Bolivin, an allele of the maize b1 gene with variable expression, contains a high copy retrotransposon-related sequence immediately upstream. Plant Physiol 125:1363–1379
Sheppard AJ, Pennington JAT, Weihrauch JL (1993) Analysis and distribution of vitamin E in vegetable oils and foods. In: Packer L, Fuch J (eds) Vitamin E in health and disease. Marcel Dekker, New York, pp 9–31
Shintani DK, DellaPenna D (1998) Elevating the vitamin E content of plants through metabolic engineering. Science 282:2098–2100
Shintani DK, Cheng Z, DellaPenna D (2002) The role of 2-methyl-6-phytylbenzoquinone methyltransferase in determining tocopherol composition in Synechocystis sp. PCC6803. FEBS Lett 511:1–5
Sokal RR, Rohlf FJ (1981) Biometry: the principles and practice of statistics in biological research. 2nd edn. WH Freeman, San Francisco, pp 1–859
Tang S, Yu JK, Slabaugh MB, Shintani DK, Knapp SJ (2002) Simple sequence repeat map of the sunflower genome. Theor Appl Genet 105:1124–1136
Tang S, Kishore VK, Knapp SJ (2003) PCR-multiplexes for a genome-wide framework of simple sequence repeat marker loci in cultivated sunflower. Theor Appl Genet 107:6–19
Tang S, Hass CG, Knapp SJ (2005) Candidate genes for mutations causing changes in tocopherol composition in sunflower. In: Abstract of plant and animal genome conference XIII, San Diego, CA, USA, 15–19 January 2005. http://www.intl-pag.org/13/abstracts/PAG13_W071.html
Tang S, Hass CG, Knapp SJ (2006) Ty3/gypsy-like retrotransposon knockout of a 2-methyl-6-phytyl-1,4-benzoquinone methyltransferase is non-lethal, uncovers a cryptic paraologus mutation, and produces novel tocopherol (vitamin E) profiles in sunflower. Theor Appl Genet (in press)
Traber MG (2004a) The ABCs of vitamin E and beta-carotene absorption. Am J Clin Nutr 80:3–4
Traber MG (2004b) Vitamin E, nuclear receptors and xenobiotic metabolism. Arch Biochem Biophys 423:6–11
Traber MG, Sies H (1996) Vitamin E in humans: demand and delivery. Annu Rev Nutr 16:321–347
Van Eenennaam AL, Lincoln K, Durrett TP, Valentin HE, Shewmaker CK, Thorne GM, Jiang J, Baszis SR, Levering CK, Aasen ED, Hao M, Stein JC, Norris SR, Last RL (2003) Engineering vitamin E content: from Arabidopsis mutant to soy oil. Plant Cell 15:3007–3019
Varagona MJ, Purugganan M, Wessler SR (1992) Alternative splicing induced by insertion of retrotransposons into the maize waxy gene. Plant Cell 4:811–820
Velasco L, Domínguez J, Fernández-Martínez JM (2004a) Registration of T589 and T2100 sunflower germplasms with modified tocopherol profiles. Crop Sci 44:362–363
Velasco L, Pérez-Vich B, Fernández-Martínez JM (2004b) Novel variation for the tocopherol profile in a sunflower created by mutagenesis and recombination. Plant Breed 123:490–492
Wagner KH, Kamal-Eldin A, Elmadfa I (2004) Gamma-tocopherol—an underestimated vitamin? Ann Nutr Metab 48:169–188
Weil CF, Marillonnet S, Burr B, Wessler SR (1992) Changes in state of the Wx-m5 allele of maize are due to intragenic transposition of Ds. Genetics 130:75–85
Wong JC, Lambert RT, Tadmor Y, Rocheford TR (2003) QTL associated with accumulation of tocopherols in maize. Crop Sci 43:2257–2266
Yu JK, Tang S, Slabaugh MB, Heesacker A, Cole G, Herring M, Soper J, Han F, Chu WC, Webb DM, Thompson L, Edwards KJ, Berry S, Leon AJ, Olungu C, Maes N, Knapp SJ (2003) Towards a saturated molecular genetic linkage map for cultivated sunflower. Crop Sci 43:367–387
Acknowledgements
This work was supported by funding to S.J.K. from the National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education, and Extension Service Plant Genome Program (Grant No. 2003-35300-15184), the Paul C. Berger Endowment at Oregon State University, the Georgia Research Alliance, and the University of Georgia Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gebhardt
Catherine G. Hass and Shunxue Tang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Hass, C.G., Tang, S., Leonard, S. et al. Three non-allelic epistatically interacting methyltransferase mutations produce novel tocopherol (vitamin E) profiles in sunflower. Theor Appl Genet 113, 767–782 (2006). https://doi.org/10.1007/s00122-006-0320-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0320-4