Abstract
It is known that somatic mutations arising during animal growth and ageing contribute to the development of neurodegenerative and other animal diseases. For plants, several studies showed that small-scale somatic DNA mutations accumulated during Arabidopsis life cycle. However, there is a lack of data on the influence of environmental stresses on somatic DNA mutagenesis in plants. In this study, we analyzed the effects of ultraviolet C (UV-C) irradiation, high soil salinity, and cadmium (CdI3) stresses on the level of small-scale somatic DNA mutations in Arabidopsis thaliana. The number of DNA mutations was examined in the Actin2 3′UTR (Actin-U1), ITS1-5.8rRNA-ITS2 (ITS), and ribulose-1,5-biphosphate carboxylase/oxygenase (rbcL) DNA regions. We found that somatic mutation levels considerably increased in CdI3-treated Arabidopsis plants, while the mutation levels declined in the UV-C- and NaCl-treated A. thaliana. Cadmium is a mutagen that is known to inhibit DNA repair processes. The detected stress-induced alterations in somatic DNA mutation levels were accompanied by markedly increased expression of base excision repair genes (AtARP, AtDME, AtDML2, AtDML3, AtMBD4, AtROS, AtUNG, and AtZDP), nucleotide excision repair genes (AtDDB1a, AtRad4, and AtRad23a), mismatch repair genes (AtMSH2, AtMSH3, and AtMSH7), and photoreactivation genes (AtUVR2, AtUVR3). Thus, the results demonstrated that UV-C, high soil salinity, and cadmium stresses influence both the level of DNA mutations and expression of DNA repair genes. Salt- and UV-induced activation of DNA repair genes could contribute to the stress-induced decrease in somatic mutation level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Somatic mutagenesis is the genomic instability of somatic cells, e.g., DNA damage and mutations randomly arising during the growth and development of an organism. The stochastic process of somatic mutagenesis is believed to be an important mechanism of animal ageing (Vijg 2004; Lombard et al. 2005). The so-called somatic mutation theory proposes that the accumulating somatic mutations and DNA damage cause abnormalities in functioning and regulation of various genes that leads to cell mosaicism and gradual organism degeneration, cell neoplastic transformation, and eventual death. A number of investigations confirm the somatic mutation theory of ageing, demonstrating that the number of somatic mutations increased with age or observing time in both animal and plant DNA (Moskalev et al. 2013; Vijg and Suh 2013; Dubrovina and Kiselev 2016).
Somatic mutations can arise spontaneously and are also induced by a variety of environmental and anthropogenic factors. Spontaneous mutations include errors in DNA replication, spontaneous lesions, and transposable genetic elements (Griffiths et al. 2000), and are found at a low frequency in plants (Dubrovina and Kiselev 2016). For plants, several studies showed that small-scale somatic mutations, e.g., nucleotide substitutions, insertions, and deletions, accumulate in the nuclear, mitochondrial, and chloroplast DNA regions of Arabidopsis thaliana during its life cycle (Caetano-Anolles 1999; Boyko et al. 2006; Pla et al. 2000; Golubov et al. 2010; Kiselev et al. 2015a). However, there is a lack of data on the influence of environmental stresses on the somatic mutation accumulation and expression of the DNA repair genes during plant life cycle. Although common environmental stress factors were not generally supposed to result in direct DNA damage and substantial mutation accumulation in plants, three studies demonstrated that heat, cold, UV, drought, flood, and salt stresses can induce microsatellite instability, point mutation accumulation, or strand breaks in Arabidopsis (Boyko et al. 2010; Yao and Kovalchuk 2011; Willing et al. 2016). Boyko et al. (2010) and Yao and Kovalchuk (2011) analyzed recombination, mutation, and microsatellite instability frequencies in the unfunctional β-glucuronidase transgene, while mutagenesis processes in transgenes and intact DNA sequences could be different. It has been shown the UV-B radiation naturally found in the environmental solar radiation provokes DNA damage (Ulm et al. 2004; Willing et al. 2016). Using whole-genome sequencing, Willing et al. (2016) found that the photolyase-induced direct reversal of pyrimidine dimers made a considerable contribution to the genetic stability across Arabidopsis generations exposed to UV-B.
The present study aimed to analyze whether somatic mutation frequency changes in response to such stresses, as high soil salinity (high NaCl concentrations), UV-C irradiation, and heavy metal stress (high cadmium concentrations). This study also aimed to analyze transcription levels of DNA repair genes and the whole plant viability (survival rates, time for seed formation, and life span) in response to these abiotic stresses. For this purpose, we have chosen three DNA regions of A. thaliana accumulating relatively high number of small-scale point mutations during the Arabidopsis life cycle as shown earlier (Kiselev et al. 2015a). Somatic mutation accumulation in the chosen DNA regions was analyzed for 2-month-old A. thaliana that has been treated with UV-C, salt stress, or cadmium. The obtained data were discussed and compared with the known information on stress-induced somatic DNA mutagenesis in plants.
Materials and methods
Plant material and growth conditions
Seven-day-old seedlings (A. thaliana ecotype Columbia L., stored by our lab) were pre-cultured on 1/2 Murashige and Skoog (MS) medium for 1 week and then planted in pots filled with commercially available rich soil in a controlled environmental chamber at + 22 °C (Sanyo MLR-352, Panasonic, Japan) kept on a 16/8 h day/night cycle at a light intensity of ~ 100 μmol m−2 s−1. For ultraviolet (UV)-induced stress, A. thaliana plants were exposed to UV-C after cultivation for 8 weeks under normal conditions. The plants were irradiated for 10 min at a distance of 15 cm above pots and peak output being 254 nm as described (Tyunin and Kiselev 2016). For cadmium stress, A. thaliana was pre-cultured on 1/2 MS medium for 7 days and then grown in pots with 40 and 100 mg CdI3 per kg of soil for additional 7 weeks. For salt stress, A. thaliana was pre-cultured on 1/2 MS medium for 7 days and grown in pots pre-soaked in the 50 or 100 mM NaCl solutions for additional 7 weeks. We used these concentrations of CdI3 and NaCl, since most plants died under higher concentrations of CdI3 and NaCl in the first 2 weeks (we tested 200 and 300 mM NaCl, 150 and 200 mg CdI3 per kg of soil). We scored survival rates of the stressed A. thaliana in percentages by calculating the number of still green survived plants and the number of pale yellow wilted plants.
DNA extraction and DNA sequencing
Total DNA was purified from A. thaliana leaves using the 8-week-old NaCl and CdI3-treated A. thaliana and 9-week-old UV-C-treated A. thaliana (1 week after the UV-C irradiation) as described (Kiselev et al. 2015b). We isolated DNA from 20 mg of dried and mixed tissues (all types of tissues were mixed) for each individual plant. For PCR amplification, partial sequences of the nuclear Actin2 gene (Actin-U1), the nuclear internal transcribed spacer sequence ITS1 and ITS2 of ribosomal DNA with 5.8S rRNA (ITS), and a chloroplast gene encoding ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) were chosen. The primer sequences and the PCR conditions were given in Kiselev et al. (2015a) and in Table S1. For PCR amplification reactions, we used a mix (1:6) of Pfu and Taq polymerases (Silex M, Russia). This blend of polymerases has been shown to exhibit an error rate of 0.08–0.09 nt per 1000 nt (Kiselev et al. 2009). The total level of small-scale mutations, including single-nucleotide substitutions, insertions, and deletions, per 1000 nt was determined as described earlier (Kiselev et al. 2015a). The PCR products were purified from agarose gels using the Cleanup Mini Kit (Evrogen, Moscow, Russia). Then, the purified products were subcloned and sequenced as described (Kiselev et al. 2011, 2013). Twenty-four clones of the Actin-U1, ITS, and rbcL of A. thaliana were sequenced using pJet forward sequencing primer and pJet reverse sequencing primer. The 24 clones for each DNA region were obtained from three biological replicates (8 clones per each individual plant). Thus, we used three independent PCR products for the Actin-U1, ITS, and rbcL DNA regions.
RNA isolation and real-time qRT-PCRs
Total RNA was isolated as described (Dubrovina et al. 2016) from 2-month-old plants exposed to UV-C, high soil salinity, and cadmium stresses. For UV-C stress, RNA was extracted 20 min, 1 day, and 1 week after the exposure. Concentration of the total RNA was measured using NanoPhotometer P-300 (Implen, Germany). Complementary DNAs were synthesized using the cDNA synthesis Kit with Oligo(dT)15 primer (Silex, Moscow, Russia) as described (Aleynova et al. 2016). Then, we used real-time quantitative PCR (qRT-PCR) to amplify the following cDNAs: endonuclease AtARP (NM_129709); DNA damage-binding protein AtDDB1a (AY074257); DNA demethylases AtDME (NM_120538), AtDML2 (NM_111836), AtDML3 (NM_119567), and AtROS (NM_129207); DNA glycosylases AtMBD4 (BT028919) and AtUNG (BT029175); mismatch repair proteins AtMSH2 (NM_113607), AtMSH3 (NM_118686), and AtMSH7 (NM_180299); DNA polymerase AtPol (DQ446242); DNA repair proteins AtRAD4 (BT010359), and AtRAD23 (NM_101486); photolyases AtUVR2 (NM_101109) and AtUVR3 (NM_001035626); and DNA 3′-phosphatase AtZDP (NM_180255). The qRT-PCRs were performed using a Real-Time PCR Kit (Syntol, Moscow, Russia), the EvaGreen Real-time PCR dye (Biotium, Hayward, USA), and primers that are listed in Table S2 in a thermocycler supplied with Multicolor Real-Time PCR Detection System (DNA Technology, Moscow, Russia). The analysis of qRT-PCR data was performed using the \(2^{{ - \Delta \Delta C_{\text{T}} }}\) method (Livak and Schmittgen 2001). AtActin2 (GenBank NM_112764) and AtGAPDH (GenBank NM_111283) were used as endogenous controls to normalize variance in the quality and amount of A. thaliana cDNA used in each real-time RT-PCR experiment (Kiselev et al. 2017).
Results
Plant survival, seed formation, and life span after UV-C irradiation, under high soil salinity, and under cadmium stress
In the present study, we treated A. thaliana plants by UV-C, high soil salinity, and cadmium stresses. While most A. thaliana plants were still alive 1 week after the UV-C treatment (Table 1), half of the leaves in the rosettes withered and new young leaves were produced. Under cadmium stress, only small rosettes (6–8 yellow–green leaves) that did not give seeds were present after 7 weeks of cadmium stress (Fig. S1). For salt stress, the plants produced seeds after cultivation for 4–5 months, despite growth of the stressed plants (50 mM NaCl) was considerably inhibited (Fig. S1; Table 1). There were no viable 5-month-old plants under high soil salinity stress (100 mM NaCl). It should be also noted that the life span of the survived Arabidopsis plants was prolonged to 6 months under 50 mM NaCl stress, while the plants lived for 4 months under the control conditions (Table 1). Under the control conditions, the 4-month-old plants were adult rosettes with yellow dry leaves and seedpods. However, the NaCl-stressed plants (50 mM NaCl) showed this phenotype only after 6 months of cultivation.
The effect of UV-C, salt, and cadmium stresses on somatic DNA mutation frequency and mutation types
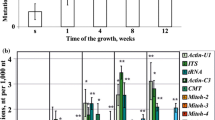
We analyzed nucleotide substitution, insertion, and deletion frequencies in the three chosen DNA region of A. thaliana plants after UV-C irradiation, under high salinity stress, and under cadmium application. We sequenced a portion of the nuclear Actin2 3′UTR region, the nuclear ITS1-5.8 rRNA-ITS2 intergenic region, and the chloroplast ribulose-1,5-bisphosphate carboxylase/oxygenase gene. For nucleic acid extraction, we used only those Arabidopsis leaves that were directly exposed to UV-C (still present after UV irradiation) and other stresses. Earlier, we analyzed somatic mutation levels in nine randomly selected coding and non-coding DNA sequences of A. thaliana depending on the time of Arabidopsis growth (Kiselev et al. 2015a). The Actin-U1, ITS and rbcL regions showed the highest number of mutations accumulating with the plant age in the 8-week-old A. thaliana plants. Therefore, in this study, we extracted DNA and RNA from 8-week-old A. thaliana exposed to 7 weeks of continuous cadmium and salt stresses, and analyzed the frequency of small-scale mutation events per 1000 nt of the Actin-U1, ITS, and rbcL nucleotide sequences (Fig. 1). We found that the somatic mutation frequency elevated after CdI3 treatment from 1.4 per 1000 nt in control plants to 3 per 1000 nt in the CdI3-treated plants (Fig. 1a). The somatic mutation frequency was the highest in the ITS region (3.6 per 1000 nt Fig. 1b) in the CdI3-treated plants. These results correlated with data published earlier that the highest mutation frequency was in the non-coding DNA regions ITS and Actin-U1 regions (Kiselev et al. 2015a). The data indicate that somatic mutation rate is higher in non-coding DNA sequences than in protein-coding regions in plants.
Nucleotide substitution, deletion, and insertion (indels) frequencies in the partial Actin-U1, ITS, and rbcL DNA regions of A. thaliana. The data are presented as the average level of mutations (a), the level of mutations for each analyzed DNA region (b), and the level of nucleotide substitutions and indels. The DNA was purified from A. thaliana plants collected 8 weeks (for control, NaCl, and CdI3) or 9 weeks (for UV-C) after germination. A. thaliana was pre-cultured on 1/2 MS medium for 7 days and then planted in pots. For salt stress, 7-day-old seedlings were grown in pots pre-soaked in the 50 or 100 mM NaCl solutions (NaCl 50 and 100). For cadmium stress, 7-day-old seedlings were planted in pots containing 40 and 100 mg CdI3 per kg of soil (CdI3 40 and 100). For ultraviolet (UV)-induced stress, 8-week-old after germination plants were exposed UV-C as described (Tyunin and Kiselev 2016) and the DNA was extracted 1 week after the UV-C irradiation. The data are presented as mean ± standard error. The 24 clones for each DNA region were obtained from three biological replicates (8 clones per each plant). Means on each graph followed by the same letter were not different using one-way analysis of variance (ANOVA) with Tukey’s pairwise comparisons. p < 0.05 was considered to be statistically significant
Single-nucleotide substitutions were the major type of the detected somatic mutations in the analyzed DNA sequences of A. thaliana (Fig. 1c). All detected nucleotide substitutions were unique, and, therefore, they did not result from PCR amplification of one mutated allele (Fig. S2). A significant decrease in small-scale mutation levels has been detected in the DNA of the UV-C- and NaCl-treated plants in contrast to that in the DNA of CdI3-treated plants (Fig. 1a). This decrease in both the substitution and indel levels was detected in all analyzed DNA regions (Fig. 1b, c). A:T → G:C transitions were the prevalent type of the detected single-nucleotide substitutions accumulating during A. thaliana growth under the control and stress conditions (~ 74%, Table 2). The number of G:C → A:T transitions and A → T and A → C transversions increased in 1.4, 2.1, and 1.9 times, respectively, after UV-C irradiation (Table 2). It is possible that the UV-induced accumulation of G:C → A:T transitions is a result of deamination of methylated cytosines under UV-C irradiation as proposed Ossowski et al. (2010). The number of T → G and A → C transversions increased under the NaCl treatment in 2.8 and 3 times, while the number of G → A and C → T transitions decreased in 1.2–1.7 and 1.5–5 times, respectively (Table 2). CdI3 treatment increased the number of A → T transversions in 1.7–2.9 times and induced C → A transversions up to 3.5–4% (Table 2).
The effect of UV-C, salt, and cadmium stresses on transcription of DNA repair genes in A. thaliana
To elucidate the molecular mechanisms contributing to somatic mutagenesis observed after UV-C irradiation, under high soil salinity, and under cadmium stress, the expression of 17 DNA repair genes encoding DNA glycosylases (AtMBD4 and AtUNG1), mismatch repair proteins (AtMSH2, AtMSH3, AtMSH7), DNA damage-binding protein (AtDDB1a), endonuclease (AtARP), DNA polymerase (AtPol), DNA demethylases (AtDML2, AtDML3, AtROS, and AtDME), photolyase (AtUVR2, AtUVR3), DNA repair proteins (AtRad4, AtRad23a), and DNA 3′-phosphatase (AtZDP) was analyzed using real-time qRT-PCR (Figs. 2, 3). The genes selected for the expression analyses belong to base excision repair or BER (AtARP, AtDME, AtDML2, AtDML3, AtMBD4, AtROS, AtUNG, and AtZDP), nucleotide excision repair or NER (AtDDB1a, AtRad4, and AtRad23a), mismatch repair or MMR (AtMSH2, AtMSH3, and AtMSH7), and photoreactivation (AtUVR2, AtUVR3) pathways (Spampinato 2017).
Expression of 8 genes from the base excision repair (BER) reparation pathway in response to ultraviolet C (UV-C) irradiation, high soil salinity, and cadmium stress. Transcription levels of AtARP (a), AtDME (b), AtDML2 (c), AtDML3 (d), AtMBD4 (e), AtROS (f), AtUNG (g), and AtZDP (h) were analyzed in 2-month-old A. thaliana. A. thaliana was pre-cultured on 1/2 MS medium for 7 days and then planted in pots. Control—total RNA was extracted from 8-week-old Arabidopsis rosettes growing under the standard conditions. UV-C: 20 m, 1 day, 7 days—the total RNA was extracted after 20 min, 1 day, and 7 days of UV-C exposure of the 8-week-old Arabidopsis rosettes. NaCl: 50, 100—the total RNA was extracted from 8-week-old Arabidopsis rosettes growing in soil pre-soaked with 50 and 100 mM NaCl. CdI3: 40, 100—the total RNA was extracted from 8-week-old Arabidopsis rosettes growing in the presence of 40 and 100 mg CdI3 per kg of soil. Data are shown as averages (n = 24) ± standard error calculated from two independent experiments. cDNA for each plant for each gene was amplified six times for Arabidopsis actin and six times for GAPDH. Means on each graph followed by the same letter were not different using one-way analysis of variance (ANOVA) with Tukey’s pairwise comparisons. p < 0.05 was considered to be statistically significant
Expression of nine genes from the nucleotide excision repair (NER), mismatch repair (MMR), and photoreactivation pathways was analyzed in response to ultraviolet C (UV-C) irradiation, high soil salinity, and cadmium stresses. Transcription levels of NER genes AtDDB1a (a), AtRad4 (b), and AtRad23a (c); MMR genes AtMSH2 (d), AtMSH3 (e), AtMSH7 (f); photoreactivation genes AtUVR2 (g), AtUVR3 (h); and a DNA polymerase gene involved in all reparation systems AtPol were analyzed in 2-month-old A. thaliana. A. thaliana was pre-cultured on 1/2 MS medium for 7 days and then planted in pots. Control—total RNA was extracted from 8-week-old Arabidopsis rosettes growing under the standard conditions. UV-C: 20 m, 1 day, 7 days—the total RNA was extracted after 20 min, 1 day, and 7 days of UV-C exposure of the 8-week-old Arabidopsis rosettes. NaCl: 50, 100—the total RNA was extracted from 8-week-old Arabidopsis rosettes growing in soil pre-soaked with 50 and 100 mM NaCl. CdI3: 40, 100—the total RNA was extracted from 8-week-old Arabidopsis rosettes growing in the presence of 40 and 100 mg CdI3 per kg of soil. Data are shown as averages (n = 24) ± standard error calculated from two independent experiments. cDNA for each plant for each gene was amplified six times for Arabidopsis actin and six times for GAPDH. Means on each graph followed by the same letter were not different using one-way analysis of variance (ANOVA) with Tukey’s pairwise comparisons. p < 0.05 was considered to be statistically significant
We analyzed transcription levels of the DNA repair genes 20 min, 1 day, and 1 week after the UV-exposure. The UV-C treatment increased transcription levels of 14 DNA repair genes out of 17 analyzed genes: BER (tARP, AtDME, AtDML2, AtDML3, AtMBD4, AtUNG, and AtZDP), NER (AtRad4, AtRad23a), MMR (AtMSH3, AtMSH7), and photoreactivation (AtUVR2, AtUVR3) genes (Figs. 2, 3). Expression of the DNA polymerase AtPol, which is involved in all these systems, was also elevated after UV-C irradiation (Fig. 3i). Expression of AtROS, AtDDB1a, and AtMSH2 did not respond to the UV-C treatment (Figs. 2f, 3a, d). One week after the UV-C treatment, transcription of the DNA repair genes, except for AtMBD4 and AtRad4, returned to that in the control plants. Continuous CdI3 stress significantly increased expression of six BER genes (AtARP, AtDME, AtDML3, AtROS, AtUNG, and AtZDP), three MMR genes (AtMSH2, AtMSH3, and AtMSH7), and a gene encoding the DNA polymerase involved DNA repair (AtPol) (Figs. 2, 3). We did not detect activated expression of the NER or photoreactivation genes. High soil salinity stress considerably increased expression of AtDME, AtUNG, AtDDB1a, AtRad4, and AtPol genes from BER and NER repair systems (Figs. 2b, g, 3a, b, i). Under salinity stress, expression of the MMR and photoreactivation genes was not changed. Therefore, salinity stress had the least effect on expression of the DNA repair genes under the analyzed conditions.
Discussion
Plants as sessile organisms had to develop an efficient system to cope with a variety of environmental and anthropogenic factors influencing plant genetic integrity and viability. Such natural environmental factors as UV and ionizing radiation, transposons and viruses, or oxygen-free radicals, in combination with mutation induction by various anthropogenic factors are well known to provoke DNA damage and induce mutation accumulation (Müller et al. 2000; Tuteja et al. 2001; Lombard et al. 2005; Jiang et al. 2014; Willing et al. 2016). In addition to the inducible mutations, DNA mutations can spontaneously arise in the plant DNA due to a variety of factors, such as occasional DNA replication and recombination errors, spontaneous DNA repair defects, or generation of natural mutagenic byproducts in cell metabolism (Smith 1992). Plant DNA repair system serves to limit the inducible and spontaneous mutations to at an optimal level that does not interfere with plant survival and stress resistance (Britt 1996; Tuteja et al. 2001; Hu et al. 2016). However, there is a lack of information on the responses of the plant DNA repair machinery to common environmental stresses and on the support of plant genetic integrity in response to stresses.
The commonly known data for animals report that UV light or oxygen-free radicals generated after a number of stresses are naturally occurring environmental mutagens (Smith 1992; Müller et al. 2000; Lombard et al. 2005; Tuteja et al. 2001). There is still a lack of studies analyzing the effects of different environmental stresses on somatic DNA mutation accumulation in plants. We analyzed the influence of UV-induced stress, high soil salinity, and cadmium stress conditions on the level of small-scale somatic mutations detected in three selected DNA regions of the A. thaliana. Two studies (Boyko et al. 2010; Yao and Kovalchuk 2011) demonstrated that UV-C and chlorine ions increased the point mutation numbers in Arabidopsis carrying unfunctional β-glucuronidase transgene. Jiang et al. (2014) analyzed mutations arising in the genomes of several independent lineages of A. thaliana growing under high soil salinity conditions and showed that stressed lineages accumulated ~ 100% more mutations than the control lineages. Using whole-genome sequencing, Willing et al. (2016) demonstrated that treatment with UV-B induced a higher number of mutations per haploid genome per generation in A. thaliana than cultivation without this treatment (1.2–2.2-fold increase).
In our study, we analyzed accumulation of somatic mutations during Arabidopsis life cycle and did not study accumulation of transgenerational mutations. UV-C and NaCl treatments resulted in a decreased level of somatic DNA mutations accumulated in the analyzed DNA regions in our study (Fig. 1). The lowered rate of somatic DNA mutations after the UV-C treatment and under the high soil salinity stress can be explained by improved reparation of the DNA mutations due to increased expression of DNA repair genes detected in this study (Fig. 2). The activation DNA repair genes belonged to different reparation systems, namely BER, NER, MMR, and photoreactivation (Spampinato 2017). Thus, we showed activation of all those systems under tested stress conditions.
In the mentioned papers, mutations were calculated in several independent Arabidopsis lineages (Jiang et al. 2014) and in 21-day-old plants (Boyko et al. 2010; Yao and Kovalchuk 2011). Mechanisms of transgenerational DNA mutagenesis observed in lineages can differ from somatic DNA mutagenesis occurring during plant development and ageing. Furthermore, Yao and Kovalchuk (2011) used young 14-day-old Arabidopsis plants for the UV-C exposure, while 8-week-old Arabidopsis rosettes were used in our study. It is possible that younger Arabidopsis plants were more sensitive to UV-C treatment compared to the 8-week-old plants. Transgenerational mutations are passed through plant reproductive structures and gametes. However, plant gametes are known to form late during development through differentiation of somatic meristematic cells (Buss 1983), and, therefore, genetic changes emerged in these somatic cells are likely to be propagate across further plant generations. In our work, we collected A. thaliana tissues from the whole rosettes (all above-ground types of tissues were mixed and grinded). Thus, we obtained PCR products from the DNA of both differentiated and undifferentiated (meristematic cells). However, differentiated cells predominate over undifferentiated meristematic cells in plants, and, thus, the obtained data primarily showed the processes of somatic mutagenesis in differentiated tissues. It is possible that NaCl and UV-C stresses had a different effect (e.g., down-regulation or no effect) on expression of the DNA repair genes in meristematic cells as compared with differentiated ones. Thus, this question needs additional experiments.
Ossowski et al. (2010) analyzed accumulation of occasional de novo mutations in the complete nuclear genomes of five A. thaliana lines cultivated for 30 generations. According to the results obtained, most single-nucleotide substitutions were G:C → A:T transitions. Willing et al. (2016) supported this finding, demonstrating that the majority of nucleotide substitutions were G:C → A:T transitions in A. thaliana. Ossowski et al. (2010) hypothesized that two main factors, namely deamination of methylated cytosines and ultraviolet light irradiation, resulted in this biased spectrum of Arabidopsis transgenerational mutagenesis. This study revealed that most of the detected single-nucleotide substitutions arising during A. thaliana growth both under normal and abiotic stress conditions were A:T → G:C transitions (~ 74%, Table 2) with the A → G and T → C transitions being the prevalent type of substitution, which is in accordance with our previous mutation accumulation data (Kiselev et al. 2015b). Similarly, A:T → G:C transitions predominated over other types of single-nucleotide substitutions accumulating during long-term cultivation of the ginseng and rice callus cultures (Noro et al. 2007; Kiselev et al. 2009, 2011, 2013). This difference may be explained by the possible differences in the transgenerational and somatic DNA mutagenesis. However, since plant viability was lowered after the UV-C and the NaCl treatments, the stress treatments could select for more healthy plants and thus enable detection of decreased somatic mutation levels. The cultivation under NaCl at a concentration of 50 mM could also select for more healthy plants which possessed prolonged life span (Table 1). It should be also noted that Boyko et al. (2010) and Yao and Kovalchuk (2011) analyzed recombination, mutation, and microsatellite instability frequencies in the unfunctional β-glucuronidase transgene, while mutagenesis processes in transgenes and intact DNA sequences could be different.
Cadmium is a mutagen that is known to interfere with and inhibit DNA repair processes (Jin et al. 2003). The results of the present study showed an activation of DNA repair genes under CdI3 stress. Therefore, cadmium inhibited the DNA repair machinery after the transcriptional activation of DNA repair genes. It is possible that the increased level of somatic DNA mutations accumulating under cadmium stress conditions contributed to the premature senescence and death without seed formation of A. thaliana.
In conclusion, the data showed that abiotic stress conditions, such as UV-C irradiation and high soil salinity stress, up-regulated expression of the DNA repair genes, and therefore, the number of the small-scale mutations decreased. A. thaliana growing under constant salt stress conditions exhibited prolonged life span compared with that of plants growing under standard conditions. It is possible that treatment with some stress conditions, which do not inhibit activities of DNA repair enzymes, may be used for DNA refinement from the accumulated somatic mutations.
Author contribution statement
KVK contributed to all the experiments, data analysis, interpretation of the results, and paper preparation. ZVO performed the experiments and analyzed the effects of UV-C, salt, and cadmium stresses on Arabidopsis plants. ASD was involved in the stress experiments, data analysis, and paper preparation. ARS performed RNA extraction and qRT-PCRs. APT performed DNA sequencing and was involved in the stress experiments.
References
Aleynova OA, Grigorchuk VP, Dubrovina AS, Rybin VG, Kiselev KV (2016) Stilbene accumulation in cell cultures of Vitis amurensis Rupr. overexpressing VaSTS1, VaSTS2, and VaSTS7 genes. Plant Cell Tissue Organ Cult 125:329–339
Boyko A, Zemp F, Filkowski J, Kovalchuk I (2006) Double-strand break repair in plants is developmentally regulated. Plant Physiol 141:488–497
Boyko A, Golubov A, Bilichak A, Kovalchuk I (2010) Chlorine ions but not sodium ions alter genome stability of Arabidopsis thaliana. Plant Cell Physiol 51:1066–1078
Britt AB (1996) DNA damage and repair in plants. Ann Rev Plant Physiol Plant Mol Biol 47:75–100
Buss LW (1983) Evolution, development, and the units of selection. Proc Natl Acad Sci USA 80:1387–1391
Caetano-Anolles G (1999) High genome-wide mutation rates in vegetatively propagated bermudagrass. Mol Ecol 8:1211–1221
Dubrovina AS, Kiselev KV (2016) Age-associated alterations in the somatic mutation and DNA methylation levels in plants. Plant Biol (Stuttg) 18:185–196
Dubrovina AS, Aleynova OA, Kiselev KV (2016) Influence of overexpression of the true and false alternative transcripts of calcium-dependent protein kinase CPK9 and CPK3a genes on the growth, stress tolerance, and resveratrol content in Vitis amurensis cell cultures. Acta Physiol Plant 38:78
Golubov A, Yao Y, Maheshwari P, Bilichak A, Boyko A, Belzile F, Kovalchuk I (2010) Microsatellite instability in Arabidopsis increases with plant development. Plant Physiol 154:1415–1427
Griffiths AJF, Miller JH, Suzuki DT et al (2000) An introduction to genetic analysis, 7th edn. W.H. Freeman, New York
Hu ZB, Cools T, De Veylder L (2016) Mechanisms used by plants to cope with DNA damage. Annu Rev Plant Biol 67:439–462
Jiang CF, Mithani A, Belfield EJ, Mott R, Hurst LD, Harberd NP (2014) Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res 24:1821–1829
Jin YH, Clark AB, Slebos RJC, Al-Refai H, Taylor JA, Kunkel TA, Resnick MA, Gordenin DA (2003) Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat Genet 34:326–329
Kiselev KV, Turlenko AV, Tchernoded GK, Zhuravlev YN (2009) Nucleotide substitutions in rolC and nptII gene sequences during long-term cultivation of Panax ginseng cell cultures. Plant Cell Rep 28:1273–1278
Kiselev KV, Shumakova OA, Tchernoded GK (2011) Mutation of Panax ginseng genes during long-term cultivation of ginseng cell cultures. J Plant Physiol 168:1280–1285
Kiselev KV, Dubrovina AS, Shumakova OA (2013) DNA mutagenesis in 2- and 20-year-old Panax ginseng cell cultures. In Vitro Cell Dev Biol Plant 35:1525–1532
Kiselev KV, Tyunin AP, Ogneva ZV, Dubrovina AS (2015a) Age-associated alterations in somatic mutation level in Arabidopsis thaliana. Plant Growth Regul 75:493–501
Kiselev KV, Dubrovina AS, Tyunin AP (2015b) The methylation status of plant genomic DNA influences PCR efficiency. J Plant Physiol 175:59–67
Kiselev KV, Aleynova OA, Grigorchuk VP, Dubrovina AS (2017) Stilbene accumulation and expression of stilbene biosynthesis pathway genes in wild grapevine Vitis amurensis Rupr. Planta 245:151–159
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408
Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW (2005) DNA repair, genome stability, and aging. Cell 120:497–512
Moskalev AA, Shaposhnikov MV, Plyusnina EN, Zhavoronkov A, Budovsky A, Yanai H, Fraifeld VE (2013) The role of DNA damage and repair in aging through the prism of Koch-like criteria. Ageing Res Rev 12:661–684
Müller J, Sigel RK, Lippert B (2000) Heavy metal mutagenicity: insights from bioinorganic model chemistry. J Inorg Biochem 79:261–265
Noro Y, Takano-Shimizu T, Syono K, Kishima Y, Sano Y (2007) Genetic variations in rice in vitro cultures at the EPSPs–RPS20 region. Theor Appl Genet 114:705–711
Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94
Pla M, Jofré A, Martell M, Molinas M, Gómez J (2000) Large accumulation of mRNA and DNA point modifications in a plant senescent tissue. FEBS Lett 472:14–16
Smith KC (1992) Spontaneous mutagenesis: experimental, genetic and other factors. Mutat Res 277:139–162
Spampinato CP (2017) Protecting DNA from errors and damage: an overview of DNA repair mechanisms in plants compared to mammals. Cell Mol Life Sci 74:1693–1709
Tyunin AP, Kiselev KV (2016) Alternations in VaSTS gene cytosine methylation and t-resveratrol production in response to UV-C irradiation in Vitis amurensis Rupr. cells. Plant Cell Tissue Organ Cult 124:33–45
Tuteja N, Singh MB, Misra MK, Bhalla PL, Tuteja R (2001) Molecular mechanisms of DNA damage and repair: progress in plants. Crit Rev Biochem Mol Biol 36:337–397
Ulm R, Baumann A, Oravecz A, Máté Z, Adám E, Oakeley EJ, Schäfer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA 101:1397–1402
Vijg J (2004) Impact of genome instability on transcription regulation of aging and senescence. Mech Ageing Dev 125:747–753
Vijg J, Suh Y (2013) Genome instability and aging. Annu Rev Physiol 75:645–668
Willing EM, Piofczyk T, Albert A, Winkler JB, Schneeberger K, Pecinka A (2016) UVR2 ensures transgenerational genome stability under simulated natural UV-B in Arabidopsis thaliana. Nat Commun 7:13522
Yao Y, Kovalchuk I (2011) Abiotic stress leads to somatic and heritable changes in homologous recombination frequency, point mutation frequency and microsatellite stability in Arabidopsis plants. Mutat Res Fundam Mol Mech Mutagen 707:61–66
Acknowledgements
This work was supported by a Grant from the Russian Foundation for Basic Research (16-04-00839a).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Horbowicz.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kiselev, K.V., Ogneva, Z.V., Dubrovina, A.S. et al. Altered somatic mutation level and DNA repair gene expression in Arabidopsis thaliana exposed to ultraviolet C, salt, and cadmium stresses. Acta Physiol Plant 40, 21 (2018). https://doi.org/10.1007/s11738-017-2600-9
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-017-2600-9