Abstract
Stilbenes (resveratrol, piceid, viniferins, etc.) are acting as phytoalexins and possess valuable biologically active properties. To improve stilbene production in grapevine, the effect of VaSTS1, VaSTS2, and VaSTS7 gene overexpression on stilbene production in callus cell cultures of wild grapevine Vitis amurensis Rupr. were accessed. Overexpression of VaSTS1 increased the total content of stilbenes by 1.2–2.1 times up to 0.36 mg per g of the dry weight (mg g−1 dry wt) or 0.036 % of dry wt, while overexpression of VaSTS2 and VaSTS7 increased the content of stilbenes by 1.5–3.5 and 2.5–2.8 times (up to 0.48 and 0.60 mg g−1 dry wt), respectively. P-coumaric acid (CA), a precursor of stilbenes, induced stilbene content and production in all cell lines with the highest positive effect on stilbene accumulation being observed in the STS2- and STS7-transgenic cell lines. After the CA feeding, the stilbene content increased up to 1.09–1.16 mg g−1 dry wt in the STS2- and one STS7-transgenic cell lines and only up to 0.35 and 0.40 mg g−1 dry wt in the control and STS1-transgenic cell lines, respectively. Thus, overexpression of VaSTS1, VaSTS2, and VaSTS7 genes had a positive effect on stilbene accumulation in the cells of V. amurensis cultivated in vitro. T-resveratrol and t-resveratrol glycosides were the prevalent stilbenes synthesized in the STS-transgenic calli before and after CA feeding. The results obtained are innovative for plant biotechnology, since stilbene production in plant cell cultures overexpressing a STS gene has not been studied until now.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stilbenes are natural compounds occurring in a number of unrelated plant families, including Fabaceae (peanut), Pinaceae (pine), or Vitaceae (grape). A considerable number of stilbene structures are based on the structure of resveratrol, which is the key precursor in the stilbene biosynthesis (Chong et al. 2009). Trans-resveratrol or t-resveratrol may be metabolized to form other stilbenes, such as viniferins (oxidation), pterostilbene (methylation), or piceid (glycosylation). T-resveratrol is widely known as a molecule that is beneficial to human health (Shankar et al. 2011; Suwalsky et al. 2015), and this explains the high level of interest in t-resveratrol among different research groups.
Resveratrol is synthesized via the phenylpropanoid pathway (Langcake and Pryce 1977) where stilbene synthase (STS, EC 2.3.1.95) directly catalyzes the reaction of resveratrol formation condensing three molecules of malonyl-CoA and one molecule of cumaryl-CoA to form the stilbene backbone. STS genes usually form small families, but in Vitis vinifera STSs exist as a multigene family yielding 48 STS members where at least 32 members were shown to be functional (Parage et al. 2012). STSs have differences in amino acid structures and exhibit various gene expression patterns, suggesting functional diversification and possible variations in enzymatic activities among different members (Parage et al. 2012; Vannozzi et al. 2012). We detected expression of at least 10 STS genes that differ in their deduced amino acid sequences and expression patterns in callus cell cultures of wild grapevine V. amurensis, which is a close species to V. vinifera (Dubrovina et al. 2010; Kiselev et al. 2012). The level of VaSTS1 expression was the highest among the expression levels of the 10 analyzed STS genes, but VaSTS1 poorly responded at the transcriptional level to various treatments (Dubrovina et al. 2010). Expression of other STS genes, e.g. VaSTS2 and VaSTS7, was differentially up-regulated in response to salicylic acid, calcium ionophore, or transformation with agrobacterial rol genes (Kiselev et al. 2010, 2012).
Plant cell cultures are currently thought to be promising producers of plant secondary metabolites for the pharmaceutical industry (Kawiak et al. 2011). While a number of STS gene transfer experiments in the genomes of whole plants have been performed to induce production of stilbenes (reviewed in Delaunois et al. 2009; Kiselev 2011), there are no reports on STS gene overexpression into plant cell cultures. Analyses of stilbene production in untreated plant cell cultures revealed low levels of stilbene accumulation, less than 0.01 % dry wt (Tassoni et al. 2005), while in elicited cells the stilbene levels can reach up to 0.1 % dry wt (Lima et al. 2012). The aim of the present study was to establish STS-transgenic grapevine cell cultures and to analyze stilbene production in the transformed cells. The results obtained show that overexpression of different STS genes of V. amurensis, namely STS1, STS2, and STS7, improved stilbene production by callus cell cultures of V. amurensis.
Materials and methods
Plant materials and growth conditions, Vitis amurensis cell culture
V2 callus culture of the wild-growing grape Vitis amurensis Rupr. (Vitaceae) was established in 2002 (Kiselev et al. 2013). The KA0 empty vector-transformed cell line was obtained in 2012 by co-cultivation of the V2 cell suspension with Agrobacterium tumefaciens GV3101::pMP90 strain containing pZP-RCS2-nptII vector (Tzfira et al. 2005), which contained only the kanamycin (Km) resistance gene, nptII, as described previously (Aleynova-Shumakova et al. 2014). Then, for transgenic cell selection and stilbene production analysis the grape cell cultures were cultivated at 35-day subculture intervals in the dark at 24–25 °C in test tubes (height 150 mm, internal diameter 146 mm) with 7–8 ml of the solid Murashige and Skoog (MS) modified WB/A medium supplemented with 0.5 mg 6-benzylaminopurine l−1, 2 mg α-naphthaleneacetic acid l−1, and 8 g l−1 agar (Dubrovina et al. 2010). Reagents for cell cultures were purchased from Sigma Chemical Co. (MO, USA) and Serva Feinbiochemica GmbH & Co. (Heidelberg, Germany). P-coumaric acid (CA) was obtained from Sigma (St. Louis, USA). Sterile solutions of CA (as ethanol solutions), were added to the autoclaved media aseptically in the desired concentrations (0.1 and 0.5 mM). The callus tissue samples were harvested from the 35 days old cultures during their linear growth phase and when the highest resveratrol content was observed. These samples were weighed and used for quantitative real-time PCR, RNA isolation, and stilbene analysis. The calli were dried using hot air flow (50 °C for 2 h), and then weighed and used for stilbene detection.
Isolation, cloning, and sequencing of VaSTS1, VaSTS2, and VaSTS7 transcript variants
Total RNA isolation was performed using hexadecyltrimethylammonium bromide (CTAB)-based extraction protocol (Kiselev et al. 2013). Complementary DNAs were synthesized at +37 °C using moloney murine leukemia virus (MMLV) RT (the RT-PCR Kit, Silex M, Moscow, Russia) as described previously (Kiselev et al. 2007). The PCR products were isolated from agarose gels using a Cleanup Mini Kit (Evrogen, Moscow, Russia).
Full length cDNAs of VaSTS1, VaSTS2, and VaSTS7 transcripts were amplified by RT-PCR from cDNA from grape V2 cell culture using primers 5′ATG GCK TCW GTK GAG GAA and 5′ATT TGT AAY TGT AGG AAT G designed to the known sequence of the VaSTS gene of V. amurensis (GenBank acc. no GQ167204, EU659863, EU659868). cDNA was cloned to pJet (Fermentas, Vilnius, Lithuania) and sequenced using ABI 3130 Genetic Analyzers as described (Kiselev et al. 2013).
Overexpression of VaSTS1, VaSTS2, and VaSTS7 genes in cell cultures of V. amurensis
To generate the constructions for plant cell transformation, the full-length cDNAs of VaSTS1, VaSTS2, and VaSTS7 transcript variants were amplified by PCR using the forward primer 5′GCT CGA GCT CAT GGC KTC WGT KGA GGA A and the reverse primer 5′TCG AGG ATC CAT TTG TAA YTG TAG GAA TGA from pTZ57-VaSTSs. The forward primer contained SacI restriction site and reverse primers contained BamHI restriction site, which are underlined. The full-length and cDNAs of VaSTS1, VaSTS2, and VaSTS7 genes were cloned into the pSAT1 vector (Tzfira et al. 2005) by the SacI and BamHI sites under the control of the double cauliflower mosaic virus (CaMV) 35S promoter. Then, the expression cassettes from pSAT1 with the VaSTS1, VaSTS2, and VaSTS7 variants were cloned into the pZP-RCS2-nptII vector by the PalAI (AscI) sites (Tzfira et al. 2005). The pZP-RCS2-nptII construction also carried the nptII gene under the control of the double CaMV 35S promoter. Plasmid DNA samples (pSAT1 and pZP-RCS2-nptII) were kindly provided by Professor Alexander Krichevsky (State University of New York, Stony Brook, USA). The used restriction enzymes were obtained from SibEnzyme (Novosibirsk, Russia).
The overexpression constructs with the VaSTS1, VaSTS2, and VaSTS7 genes (pZP-RCS2-VaSTS1-nptII, pZP-RCS2-VaSTS2-nptII, and pZP-RCS2-VaSTS7-nptII) and empty vector (pZP-RCS2-nptII) were introduced into the A. tumefaciens strain GV3101::pMP90 and transformed into the V. amurensis suspension culture V2 by co-cultivation with the bacterial cells as described (Kiselev et al. 2013).
After transformation, the calli were cultivated for a 3-month period in the presence of 10–15 mg of Km l−1 to select transgenic cells and for a 4-month period in the presence of 250 mg of cefotaxime (Cf) l−1 to suppress the bacteria. The absence of A. tumefaciens was confirmed by PCR of the VirB2 gene using primers 5′ATG CGA TGC TTT GAA AGA TAC CG and 5′TTA GCC ACC TCC AGT CAG CG as described (Aleynova-Shumakova et al. 2014).
For analyzing mRNA levels of the exogenous VaSTS1 exogenous transgene, we used primers STS1 5′CTG TTG TGC TGC ATA GCA TTC designed to the 3′ end of the protein coding region of VaSTS1 and primer pSAT1a 5′GAG AGA CTG GTG ATT TTT GCG designed to the CaMV 35S terminator in the pSAT1 vector (Tzfira et al. 2005). For analyzing mRNA levels of the exogenous VaSTS2 exogenous transgene, we used primers STS2 5′GGT GAA GGA TTG GAT TGG G designed to the 3′ end of the protein coding region of VaSTS2 and primer pSAT1a. For analyzing mRNA levels of the exogenous VaSTS7 exogenous transgene, we used primer STS7 5′GGA TTG GGG AGT ATT ATT TGG designed to the 3′ end of the protein coding region of VaSTS7 and primer pSAT1a.
Chemicals
All solvents were of high performance liquid chromatography (HPLC) grade. Analytical standards: t-resveratrol, t-piceid and p-coumaric acid were obtained from Sigma-Aldrich (St. Louis, MO, USA), ε-viniferin was obtained from Panreac AppliChem (GmbH, Germany).
Analytical chromatography
Liquid chromatography with a high-resolution mass spectrometry
Transgenic lines harboring the pZP-RCS2-nptII, and pZP-RCS2-VaSTS1-nptII, pZP-RCS2-VaSTS2-nptII, and pZP-RCS2-VaSTS7-nptII constructs cultivated on solid WB/A medium (35 days) were analyzed for the presence of stilbenes.
HPLC with UV and high-resolution mass spectrometry (HPLC–UV–HRMS) for identification (components 1–5, Fig. 1) was performed by using LCMS-IT-TOF tandem mass-spectrometer (Shimadzu, Japan) including with LC-20AD Prominence liquid chromatograph (Shimadzu, Japan), an SPD-M20A photodiode array detector, and an ion-trap/time-of-flight mass spectrometer. An analytical Ascentis C18 column (100 mm, 2.1-mm i.d., 3-μm part size, Supelco, Bellefonte, Pennsylvania, USA) for separation was applied. The mass spectral (MS) parameters included electrospray ionization (ESI) with simultaneous negative and positive ion detection with a resolution of 12,000. The following settings were used: the range of m/z detection was 100–1000, the drying gas (N2) pressure was 195 kPa, the nebulizer gas flow rate was 1.5 l/min, the ion source potential changed from −3.8 to 4.5 kV, and the interface temperature was 200 °C. All MS2 experiments were run by adjusting the collision energy to the point where the precursor ion had an abundance of 50 % relative to the precollision intensity. The pressure of the collision gas (Ar) was 0.003 Pa. The range of m/z detection for the precursor ion fragments was 50–600. Separation was carried out at following conditions: the column temperature was 40 °C, the mobile phase consisted of 0.1 % aqueous acetic acid (A) and acetonitrile (B). The following elution gradient with a flow rate of 0.25 mL/min was used: 0 min 0 % B; 5 min 0 % B; 40 min 100 % B and then eluent B until 50 min.
A representative HPLC–UV profile for the extracts obtained from the KA0 cell culture of V. amurensis recorded at 310 nm. The callus tissue samples were harvested from the 35-day-old cultures. Line A—KA0 cells cultivated in standard conditions; line B—KA0 cells treated with 0.5 mM p-coumaric acid (CA); line C—ST1-IV cells cultivated in standard conditions; line D—ST1-IV cells treated with 0.5 mM CA; line E—ST2-I cells cultivated in standard conditions; line F—ST2-I cells treated with 0.5 mM CA; line G—ST7-I cells cultivated in standard conditions; line H—ST7-I cells treated with 0.5 mM CA. t-resveratrol diglucoside (1), t-piceid or t-resveratrol glucoside (2), t-resveratrol (3), ε-viniferin (4), δ-viniferin (5), coumaroyldihexoside (6), coumaric acid derivative A hexoside (7), caffeoyldihexoside (8), coumaroylhexoside (9), caffeoylhexoside (10), and coumaric acid derivative A (11)
Liquid chromatography with a low-resolution mass spectrometry
HPLC with MS and UV detection (HPLC–MS–UV) for identification (components 6–11, Fig. 1) and quantification of all components was performed using an 1260 Infinity analytical HPLC system (Agilent Technologies, Santa Clara, California, USA), equipped with a G1315D photodiode array detector, G1311C quaternary pump, G1316A column oven and G1329B auto sampler. The HPLC system was connected with a Bruker HCT ultra PTM Discovery System (Bruker Daltonik GmbH, Bremen, Germany), equipped with ion trap mass spectrometer. The extracts were separated on Zorbax C18 column (150 mm, 2.1-mm i.d., 3.5-μm part size, Agilent Technologies, USA); the column temperature was 40 °C. The mobile phase consisted of a gradient elution of 0.1 % aqueous acetic acid (A) and acetonitrile (B). The gradient profile with a flow rate of 0.2 mL/min was: 0 min 0 % B; 35 min 40 % B; 40 min 50 % B. 50 min 100 % B and then eluent B until 60 min. The injected volume was 1–5 μL. The MS analysis was carried out with electrospray ionization (ESI) and negative ions detection. The following settings were used: the range of m/z detection was 100–1000, the drying gas (N2) flow rate was 8 l/min, the nebulizer gas (N2) pressure was 175 kPa, the ion source potential was −4.0 kV and the drying gas temperature was 325 °C. The all MS2 signals were recording with the fragmentation amplitude 1 V. UV spectra were recorded in the 200–400 nm range, and chromatograms for quantification were acquired at 310 nm. All determined components of the extracts were calculated on the base of four-point regression curves built with the available standards. All solutions were filtered through a 0.45-μm membrane (Millipore, Bedford, MA, USA) prior to inject.
Statistical analysis
Statistical analysis was performed using the Statistica 10.0 program (StatSoft Inc, Boston, USA). The data are presented as mean ± standard error (SE) and were tested by paired Student’s t test. The 0.05 level was selected as the point of minimal statistical significance in all analyses.
Results
Stilbene identification
Compound 1 (Supplementary Table 1) of extract from grape KA0 cell line showed the spectral data corresponded to described earlier (Jerkovic et al. 2007). On the negative MS2 spectrum of [M − H]− precursor ions with m/z 551 only two ions were presented. These ions with m/z 389 and 227 corresponded to the fragments formed by consistent elimination of one and two dehydrated molecules of hexose. On the positive MS2 spectrum of [M + H]+ precursor ions the fourteens fragment ions was presented. More intensive ions with m/z 391 and 229 corresponded to the fragments formed by elimination of one and two dehydrated molecules of hexose. Other fragment ions with m/z 373, 355 and 337 formed by losses two molecules of hexose coupled with one, two and tree molecules of water, respectively. Also the ions with m/z 535 (loss H2O), 517 (loss 2H2O) and 499 (loss 3H2O) were presented on the MS2 spectrum. High-resolution ESI mass-spectrum of compound 1 showed the positive [M + H]+ quasi-molecular ions with m/z 553.1935 (calculated 553.1916) and negative [M − H]− quasi-molecular ions with m/z 551.1781 (calculated 551.1770). These values corresponded to molecular formula of resveratrol dihexoside (C26H32O13). MS2 pattern of the compound corresponded to the described earlier trans-resveratrol di-glucoside. Also, UV spectrum of the compound was similar with the spectrum of trans-isomer of resveratrol. All chromatographic and MS characteristics of compounds 2–4 (Supplementary Table 1) strongly corresponded to the characteristics of standard compounds and suggested with literature data (Jerkovic et al. 2007; Mulinacci et al. 2010).
Compound 5 (Supplementary Table 1) of the extract was determined as trans-δ-viniferin. High-resolution ESI mass-spectrum of compound 5 showed the positive [M + H]+ quasi-molecular ions with m/z 455.1491 (calculated 455.1489) and negative [M − H]− quasi-molecular ions with m/z 453.1376 (calculated 453.1343). Results of calculation of molecular formula (C28H22O6) showed proper for viniferin elemental composition. All MS and UV spectral characteristics of the compound were similar with the data described earlier for trans-δ-isomer of viniferin (Jerkovic et al. 2007; Mulinacci et al. 2010). Next compounds (6, 8, 9, 10, Supplementary Table 2) of extract had the similar spectral data with the data described in literature for hydroxycinnamic acids derivatives (Ieri et al. 2012; Zhao et al. 2014). Compounds 6 and 9 were assigned with the structures of coumaroyldihexoside and coumaroylhexoside, respectively. Their negative ESI–MS spectra contained quasi-molecular ions with composition [M − H]− and cluster ions with compositions [M + CH3COO]−. Their MS2 spectra of [M − H]− precursor ions contained the similar fragment ions with m/z 163 corresponded to coumaric acid anions. These ions formed by elimination two molecules of dehydrated hexose from compound 6 and a molecule of dehydrated hexose from compound 9. Also in MS2 spectra of the negative quasi-molecular ions of both compounds the fragment ions with m/z 119 was presented. These ions corresponded to decarboxylation product of coumaric acid anions and formed by simultaneously elimination molecules of dehydrated hexose with CO2 molecules from the quasi-molecular ions. Quasi-molecular ions of compound 8 and 10 had MS2 fragmentation pattern similar with the MS2 pattern of compounds 6 and 9. The fragment ions shown in Supplementary Table 2 were formed by elimination of dehydrated hexose molecules and CO2 from quasi-molecular ions of the compounds. Values of m/z of these ions differed from the respective m/z values of ions of coumaric acid derivatives on 16 a.m.u. This number corresponded to an oxygen atom. Therefore, we assigned the structures of caffeoyldihexoside and caffeoylhexoside to the compounds 8 and 10, respectively.
Our analysis revealed the presence of two unknown compounds—number 7 and 11 (Supplementary Table 2). On the base of MS2 spectral data of their quasi-molecular ions, we assumed these compounds to be coumaric acid derivatives. Their MS2 spectra contained fragment ions with m/z 163 and 119, typical for coumaric acid derivative ions fragmentation (see above). Also UV spectral characteristics of the compounds 7 and 11 were similar to those of coumaric acid. The m/z values of quasi-molecular ions of the compound 7 differed from the m/z values of respective ions of the compound 11 on 162 a.m.u. This number corresponded to a dehydrated molecule of hexose. Moreover, according to MS2 data (Supplementary Table 2), fragmentation of the quasi-molecular ions of the compound 7 showed the presence of the fragment ions with m/z 228 corresponding to the m/z values of quasi-molecular ions of the compound 11. Therefore, we suggested the compound 7 to be compound 11 hexoside derivative.
Genetic transformation of V. amurensis with the VaSTS1, VaSTS2, and VaSTS7 genes and selection of the transgenic cell lines
To establish grape cell cultures overexpressing the full-length VaSTS1, VaSTS2, and VaSTS7 genes, the V2 suspension culture of V. amurensis was incubated with A. tumefaciens strains bearing constructs pZP-RCS2-nptII (empty vector, KA0 cell line), pZP-RCS2-VaSTS1-nptII (ST1 cell line), pZP-RCS2-VaSTS2-nptII (ST2 cell line), or pZP-RCS2-VaSTS7-nptII (ST7 cell line), where the transcripts of the VaSTS1, VaSTS2, and VaSTS7 genes were cloned under the control of the double CaMV 35S promoter. We selected transgenic cell aggregates in the presence of 10–15 mg Km l−1 for 3–4 months and established several Km-resistant lines. Previously, Km sensitivity of the parent V2 culture was tested (Kiselev et al. 2013), and it was shown that these calli completely ceased to grow during the second month of cultivation at such low Km concentration as 10 mg per liter (mg l−1). During the first month after transformation, we selected the fast-growing calli from separate primary small aggregates, which appeared in the presence of Km and established several Km-resistant independent clonal lines ST1-I, II, and IV transformed with the VaSTS1 gene; ST2-I and II transformed with the VaSTS2 gene; ST7-I, II, and III transformed with the VaSTS7 gene. The formation of stable callus phenotypes of the used cell lines was observed 3 months after the removal of Km from the nutrient medium.
The independently transformed ST1-I, II, IV; ST2-I, II; ST7-I, II, and III cell lines were used for further investigation. These calli represented friable vigorously growing homogenous tissues, which did not appear to have undergone differentiation. We did not observe the formation of any differentiated structures, such as root- or shoot-like structures in the transformed cell lines on the WB/A medium with standard hormone composition and on the media with decreased levels of 6-benzylaminopurine and α-naphthaleneacetic acid in the dark. The KA0 calli reproduced morphological, growth, and biosynthetic characteristics of the parent V2 culture. The KA-0 cell line was used as a control in all further experiments, including the present investigation. The semiquantitative RT-PCR has shown that the nptII gene was transcribed in all obtained transgenic cell lines (data not shown). The absence of A. tumefaciens was confirmed using PCR to control the presence or absence of the VirB2 gene (data not shown).
We analyzed expression of the exogenous VaSTS1, VaSTS2, and VaSTS7 genes (transgenes) in the obtained cell lines (Fig. 2). All STS-transgenic cell lines actively expressed the exogenous VaSTS1, VaSTS2, or VaSTS7 genes, and their expression levels were approximately at the same level in all the analyzed cell lines.
Quantification of the exogenous VaSTS1, VaSTS2, and VaSTS7 expression performed by real-time RT-PCR in the obtained transgenic cell lines of V. amurensis. The callus tissue samples were harvested from the 35-day-old cultures. qRT-PCR data were obtained from at least two independent experiments and are averages of eight technical replicates for each independent experiment (four qPCR reactions normalized to VaActin1 and four qPCR reactions normalized to VaGAPDH expression) and presented as mean ± standard error. *p < 0.05; **p < 0.01 versus values of VaSTSs expression in the empty vector-transformed KA0 cell line
Production of stilbenes by the VaSTS1, VaSTS2, and VaSTS7-transformed cell lines of V. amurensis and their growth parameters
Using HPLC, we identified five stilbenes in the control cell line KA0: t-resveratrol diglucoside (1), t-piceid or t-resveratrol glucoside (2), t-resveratrol (3), ε-viniferin (4), and δ-viniferin (5) (Fig. 1A). Also, there were other peaks on the HPLC–UV profile which could correspond to other compounds, but their areas were much less than the areas of above mentioned stilbenes. Recently, t-resveratrol diglucoside, t-piceid, t-resveratrol, and viniferins were isolated from grape cell suspension cultures and from Arabidapsis plants overexpressing STS genes (Lo et al. 2007; Lima et al. 2012).
Overexpression of the used VaSTS genes increased the area peaks of the identified five stilbenes (Fig. 1c, e, g). Treatment of the KA0 and STS-transgenic V. amurensis cell lines with 0.5 mM of the CA increased area of the peaks for the identified five stilbenes and led to appearance of the 6 new peaks: coumaroyldihexoside (6), coumaric acid derivative A hexoside (7), caffeoyldihexoside (8), coumaroylhexoside (9), caffeoylhexoside (10), and coumaric acid derivative A (11) (Fig. 1b, d, f, h).
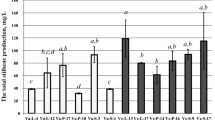
In the Table 1, we presented fresh and dry biomass accumulation (in g l−1), total stilbene content (in mg per g of dry wt), and stilbene production (in mg l−1, formula: dry wt production in g l−1× stilbene content in mg per g of dry wt) by the KA0 (empty vector-transformed cell line); ST1-I, II, IV (VaSTS1 gene), ST2-I, II (VaSTS2); and ST7-I, II, III (VaSTS7) cell lines overexpressing different VaSTS genes. The ST1, ST2, and ST7 calli reproduced growth characteristics of the empty vector control KA0 calli (Table 1).
Overexpression of the VaSTS1, STS2, and STS7 genes increased total stilbene content up to 3.5 times (up to 0.6 mg g−1 dry wt) and stilbene production up to 3.4 times (up to 6.12 mg l−1), compared to the KA0 calli, but only in the ST2-I and ST7-I cell lines this increase in stilbene accumulation was significant (Table 1). Then, we analyzed the content of individual stilbenes in the used KA0, ST1, ST2, and ST7 cell lines. The analysis revealed that overexpression of the VaSTS1 gene increased t-resveratrol content in 1.3–5 times, overexpression of the VaSTS2 gene increased t-resveratrol content in 2.3–7 times, and overexpression of the VaSTS7 gene increased t-resveratrol content in 2.7–7.3 times in comparison with the KA0 calli, but only in the ST1-IV, ST2-I, ST7-I, and ST2-II cell lines this increase was significant (Table 2). We detected only a minor increase in the content of other stilbenes in the ST1, ST2, and ST7 cell lines, but it was significant only for the ST2-I cell line (Table 2). Overall, the influence of the VaSTS overexpression on the content of other resveratrol derivatives (t-resveratrol diglucoside, t-piceid, ε-viniferin, δ-viniferin) was not as significant as the influence on t-resveratrol content.
It is possible that we did not find a stronger activation effect of VaSTS overexpression on stilbene biosynthesis due to a limitation in the precursor pool for t-resveratrol precursors, e.g. coumaroyl-CoA and malonyl-CoA. P-coumaric acid (CA) is a low-price available precursor of stilbenes and isoflavonoids, and it is known that that CA increased resveratrol accumulation in V. amurensis calli via selective enhancement of expression of individual STS genes (Shumakova et al. 2011). Therefore, we treated the control and VaSTS-overexpressing cell lines with CA in order to improve stilbene accumulation. For the CA treatments, we selected VaSTS-overexpressing cell lines with the highest stilbene content (ST1-VI, ST2-I, and ST7-I).
Incubation with CA increased total stilbene content and production in the KA0 calli in 1.5–1.9 times in comparison with the untreated KA0 (Table 3). Thus, the total stilbene accumulation in the KA0 calli after CA feeding became approximately the same as it was in the ST1 cell lines growing in standard conditions. In ST1-IV, CA increased the total stilbene content and production only in 1.1 times. The highest CA-induced increase in the content and production of stilbenes was detected in the ST2-I and ST7-I cell lines (Table 3). We also analyzed the content of individual stilbenes in the KA0, ST1, ST2, and ST7 cell lines treated with CA (Table 4). CA treatment increased the content of primarily t-resveratrol and t-resveratrol diglucoside in the used cell lines. The highest t-resveratrol content was 0.42 mg g−1 dry wt in the ST2-I cell line after feeding with 0.5 mM CA. The highest t-resveratrol diglucoside content was 0.65 % of dry wt in the ST7-I cell line after feeding with 0.5 mM CA.
Discussion
The five stilbenes, t-resveratrol, t-piceid, ε-viniferin, δ-viniferin, and t-resveratrol diglucoside, produced in the cell cultures of V. amurensis were also described in other grapevine species and other plants (Jerkovic et al. 2007; Mulinacci et al. 2010). Overexpression of the STS1, STS2, and STS7 genes in V. amurensis cell cultures increased the content of these five stilbenes up to 0.06 % dry wt and did not lead to formation of new stilbenes. Only after the CA treatment, we detected six new vigorous peaks and we defined them as p-coumaric acid derivatives. Transformation of the V. amurensis calli with three different STS genes and CA feeding increased primarily the content of t-resveratrol and t-resveratrol glycosides, while the content of other stilbenes was affected at a lower level.
Many researchers used overexpression of STS genes to increase t-resveratrol content in plant cells (Giorcelli et al. 2004; Schwekendiek et al. 2007). Plants were usually genetically modified to produce t-resveratrol derivates to assess their roles in human health and plant disease control. To the best of our knowledge, there is no information about stilbene accumulation in STS-transgenic plant cell cultures in the literature. To date, Vst1 and Stsy genes of V. vinifera have been the most common genes used for plant transformation (Delaunois et al. 2009; Kiselev 2011). Transformation of various Vitaceae species with the Vst1 gene resulted in an increase in the levels of t-resveratrol and its derivatives in transgenic plants (Kiselev 2011). The highest t-resveratrol levels (up to 0.35 mg g−1 of dry wt or 0.035 % of dry wt) were registered after the transformation of grape cells with the Vst1 gene under the control of the PR10 promoter (Coutos-Thevenot et al. 2001). In this case, the levels of t-resveratrol increased in the transgenic plants only after elicitor treatment (UV light or Botrytis infection). Notably, the t-resveratrol levels were seen in one out of 50 transgenic plants strains. This increase might be caused by mutations in the genes involved in t-resveratrol biosynthesis, which could be provoked by the integration of exogenous sequences. STS-transgenic plants usually produced varying t-resveratrol and t-piceid levels, but the average stilbene content did not exceed 0.1 % of dry wt (Giorcelli et al. 2004; Schwekendiek et al. 2007). Thus, the creation of STS-transgenic plants has been attempted in a number of studies, but none of the attempts resulted in a high t-resveratrol level similar to that detected after rol gene transformation (Kiselev et al. 2007; Dubrovina et al. 2010). Transformation of V. amurensis cells cultivated in vitro with the rol genes of A. rhizogenes has been shown to greatly enhance resveratrol production in the transformed calli (Kiselev et al., 2007; Dubrovina et al. 2010). In these transgenic cultures, t-resveratrol production was increased by more than 100-fold (up to 3 % or 30 mg g−1 of dry wt) compared with the controls or the resveratrol levels that were observed in other studies. This enhancement in resveratrol production in the rol-transformed cell cultures correlated with the activation of expression of different VaSTS genes. Cultured cells from a variety of plant species (Arachis spp., Gossypium spp., Vitis spp.) contained low resveratrol levels (up to 0.03 % or 0.3 mg g−1 of dry wt), and treatments with UV irradiation, ultrasound treatment, elicitors, precursors, or other agents resulted in slight improvements in resveratrol production (Tassoni et al. 2005; Shumakova et al. 2011; Lima et al. 2012; Santamaria et al. 2011, 2012).
We proposed that there could be a limitation in the precursor pool for the t-resveratrol biosynthesis in the V. amurensis cells cultivated in vitro. Therefore, we applied feeding the used grape cell lines with CA, which is a low-price available precursor of stilbenes and isoflavonoids. Overexpression of the VaSTS2 and VaSTS7 genes in callus cell cultures of V. amurensis combined with cultivation in the presence of high CA concentrations have led to increased stilbene levels reaching 1.1–1.2 mg g−1 dry wt of the calli. The stilbene accumulation levels were close to the most promising results that were obtained after STS gene transfers in the genome of whole plants (Schwekendiek et al. 2007). T-resveratrol and t-resveratrol diglucoside contents were increased in the VaSTS2- and VaSTS7-overexpressing V. amurensis cell lines after CA treatment at a higher degree than in the VaSTS1-overexpressing cells and KA0 control. Notably, a significant amount of the added CA was converted to coumaric acid derivates, probably through activity of 4-coumarate 3-hydroxylases and glucosyltransferases (Rodrigues et al. 2015) or an inefficient activity of 4-coumarate CoA ligase, which is known to catalyze the formation of CoA thiol esters of 4-coumarate, substrates for the STS enzymes. It is possible that we did not obtain cell lines producing stilbenes at a higher level than 1.1–1.2 mg g−1 dry wt, because there was a limitation in the stilbene precursor pool and additional enzyme activities in the grape cells preventing effective using of precursors added externally. STSs were proposed to have undergone functional diversification and might possess varying enzymatic activities (Parage et al. 2012; Vannozzi et al. 2012). This hypothesis was partially confirmed by the data obtained in the present work indicating that t-resveratrol and t-resveratrol diglucoside contents were increased in the VaSTS2- and VaSTS7-overexpressing V. amurensis cells at a higher degree compared with that in the VaSTS1-overexpressing cells.
In conclusion, the data obtained indicate that overexpression of VaSTS1, VaSTS2, and VaSTS7 genes had a positive effect on stilbene levels in grapevine cell cultures. The results presented are important for plant biotechnology, since stilbene production in plant cell cultures overexpressing a STS gene has not been studied until now.
Abbreviations
- CA:
-
P-coumaric acid
- HPLC:
-
High-performance liquid chromatography
- Km:
-
Kanamycin
- STS:
-
Stilbene synthase
- Wt:
-
Weight
References
Aleynova-Shumakova OA, Dubrovina AS, Manyakhin AY, Karetin YA, Kiselev KV (2014) VaCPK20 gene overexpression significantly increased resveratrol content and expression of stilbene synthase genes in cell cultures of Vitis amurensis Rupr. Appl Microbiol Biotechnol 98:5541–5549
Chong J, Poutaraud A, Hugueney P (2009) Metabolism and roles of stilbenes in plants. Plant Sci 177:143–155
Coutos-Thevenot P, Poinssot B, Bonomelli A, Yean H, Breda C, Buffard D, Esnault R, Hain R, Boulay M (2001) In vitro tolerance to Botrytis cinerea of grapevine 41B rootstock in transgenic plants expressing the stilbene synthase Vst1 gene under the control of a pathogen-inducible PR 10 promoter. J Exp Bot 52:901–910
Delaunois B, Cordelier S, Conreux A, Clément C, Jeandet P (2009) Molecular engineering of resveratrol in plants. Plant Biotechnol J 7(1):2–12
Dubrovina AS, Manyakhin AY, Zhuravlev YN, Kiselev KV (2010) Resveratrol content and expression of phenylalanine ammonia-lyase and stilbene synthase genes in rolC transgenic cell cultures of Vitis amurensis. Appl Microbiol Biotechnol 88:727–736
Giorcelli A, Sparvoli F, Mattivi F, Tava A, Balestrazzi A, Vrhovsek U, Calligari P, Bollini R, Confalonieri M (2004) Expression of the stilbene synthase (StSy) gene from grapevine in transgenic white poplar results in high accumulation of the antioxidant resveratrol glucosides. Transgenic Res 13:203–214
Ieri F, Pinelli P, Romani A (2012) Simultaneous determination of anthocyanins, coumarins and phenolic acids in fruits, kernels and liqueur of Prunus mahaleb L. Food Chem 135:2157–2162
Jerkovic V, Nguyen F, Nizet S, Collin S (2007) Combinatorial synthesis, reversed-phase and normalphase high-performance liquid chromatography elution data and liquid chromatography/positive atmospheric pressure chemical ionization tandem mass spectra of methoxylated and glycosylated resveratrol analogues. Rapid Commun Mass Spectrom 21:2456–2466
Kawiak A, Krolicka A, Lojkowska E (2011) In vitro cultures of Drosera aliciae as a source of a cytotoxic naphthoquinone: ramentaceone. Biotechnol Lett 33:2309–2316
Kiselev KV (2011) Perspectives for production and application of resveratrol. Appl Microbiol Biotechnol 90:417–425
Kiselev KV, Dubrovina AS, Veselova MV, Bulgakov VP, Fedoreyev SA, Zhuravlev YN (2007) The rolB gene-induced overproduction of resveratrol in Vitis amurensis transformed cells. J Biotechnol 128:681–692
Kiselev KV, Dubrovina AS, Isaeva GA, Zhuravlev YN (2010) The effect of salicylic acid on phenylalanine ammonia-lyase and stilbene synthase gene expression in Vitis amurensis cell culture. Russ J Plant Physiol 57:415–421
Kiselev KV, Shumakova OA, Manyakhin AY, Mazeika AN (2012) Influence of calcium influx induced by the calcium ionophore, A23187, on resveratrol content and the expression of CDPK and STS genes in the cell cultures of Vitis amurensis. Plant Growth Regul 68:371–381
Kiselev KV, Dubrovina AS, Shumakova OA, Karetin YA, Manyakhin AY (2013) Structure and expression profiling of a novel calcium-dependent protein kinase gene, CDPK3a, in leaves, stems, grapes, and cell cultures of wild-growing grapevine Vitis amurensis Rupr. Plant Cell Rep 32:431–442
Langcake P, Pryce RJ (1977) A new class of phytoalexins from grapevines. Experientia 33:151–152
Lima MRM, Ferreres F, Dias ACP (2012) Response of Vitis vinifera cell cultures to Phaeomoniella chlamydospora: changes in phenolic production, oxidative state and expression of defence-related genes. Eur J Plant Pathol 132:133–146
Lo C, Le Blanc JCY, Yu CKY, Sze KH, Ng DCM, Chu IK (2007) Detection, characterization, and quantification of resveratrol glycosides in transgenic arabidopsis over-expressing a sorghum stilbene synthase gene by liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom 21:4101–4108
Mulinacci N, Innocenti M, Santamaria AR, la Marca G, Pasqua G (2010) High-performance liquid chromatography/electrospray ionization tandem mass spectrometric investigation of stilbenoids in cell cultures of Vitis vinifera L., cv. Malvasia. Rapid Commun Mass Spectrom 24:2065–2073
Parage C, Tavares R, Rety S, Baltenweck-Guyot R, Poutaraud A, Renault L, Heintz D, Lugan R, Marais GAB, Aubourg S, Hugueney P (2012) Structural, functional, and evolutionary analysis of the unusually large stilbene synthase gene family in grapevine. Plant Physiol 160:1407–1419
Rodrigues JL, Araújo RG, Prather KL, Kluskens LD, Rodrigues LR (2015) Heterologous production of caffeic acid from tyrosine in Escherichia coli. Enzyme Microb Technol 71:36–44
Santamaria AR, Mulinacci N, Valletta A, Innocenti M, Pasqua G (2011) Effects of elicitors on the production of resveratrol and viniferins in cell cultures of Vitis vinifera L. cv Italia. J Agric Food Chem 59:9094–9101
Santamaria AR, Innocenti M, Mulinacci N, Melani F, Valletta A, Sciandra I, Pasqua G (2012) Enhancement of viniferin production in Vitis vinifera L. cv. Alphonse Lavallée cell suspensions by low-energy ultrasound alone and in combination with methyl jasmonate. J Agric Food Chem 60:11135–11142
Schwekendiek A, Spring O, Heyerick A, Pickel B, Pitsch NT, Peschke F, Keukeleire DD, Weber G (2007) Constitutive expression of a grapevine stilbene synthase gene in transgenic hop (Humulus lupulus L.) yields resveratrol and its derivatives in substantial quantities. J Agric Food Chem 55:7002–7009
Shankar S, Nall D, Tang SN, Meeker D, Passarini J, Sharma J, Srivastava RK (2011) Resveratrol inhibits pancreatic cancer stem cell characteristics in human and kras(G12D) transgenic mice by inhibiting pluripotency maintaining factors and epithelial-mesenchymal transition. PLoS ONE 6:e16530
Shumakova OA, Manyakhin AY, Kiselev KV (2011) Resveratrol content and expression of phenylalanine ammonia-lyase and stilbene synthase genes in cell cultures of Vitis amurensis treated with coumaric acid. Appl Biochem Biotechnol 165:1427–1436
Suwalsky M, Villena F, Gallardo MJ (2015) In vitro protective effects of resveratrol against oxidative damage in human erythrocytes. Biochim Biophys Acta Biomembr 1848:76–82
Tassoni A, Fornale S, Franceschetti M, Musiani F, Michael AJ, Perry B, Bagni N (2005) Jasmonates and Na-orthovanadate promote resveratrol production in Vitis vinifera cv. Barbera cell cultures. New Phytol 166:895–905
Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57:503–516
Vannozzi A, Dry IB, Fasoli M, Zenoni S, Lucchin M (2012) Genome-wide analysis of the grapevine stilbene synthase multigenic family: genomic organization and expression profiles upon biotic and abiotic stresses. BMC Plant Biol 12:130
Zhao Y, Li X, Zeng X, Huang S, Houa SZ, Lai XP (2014) Characterization of phenolic constituents in Lithocarpus polystachyus. Anal Methods 6:1359–1363
Acknowledgments
This work was supported by grants from the following institutions and foundations: Russian Foundation for Basic Research (14-04-31113-mol_a) and the Russian Science Foundation (14-14-00366). The Russian Science Foundation supported binary vector construction, growth analysis of the calli, and stilbene quantifications, while Russian Foundation for Basic Research supported grape cell culture transformations, selection of the transgenic calli.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Aleynova, O.A., Grigorchuk, V.P., Dubrovina, A.S. et al. Stilbene accumulation in cell cultures of Vitis amurensis Rupr. overexpressing VaSTS1, VaSTS2, and VaSTS7 genes. Plant Cell Tiss Organ Cult 125, 329–339 (2016). https://doi.org/10.1007/s11240-016-0954-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-016-0954-0