Abstract
RT-qPCR is a widely used method in gene expression and transcriptome studies. Normalization based on reference genes is necessary to accurately analyze RT-qPCR data. Thus, an accurate and systematic evaluation of these reference genes before experiments are conducted is necessary. In this study, 18 candidate reference genes were evaluated under various experimental conditions covering a range of tissue types and cultivars, NaCl, CaCl2 and temperature treatments, hormones (6-BA, ABA and NAA) and a set of osmotic stress (mannitol and PEG6000) treatments. Gene expression across 48 pear samples was evaluated using geNorm, NormFinder and BestKeeper statistical algorithms. Actin2/7 (ACT2/7), ubiquitin extension protein (UBI) and Yellow-leaf-specific gene 8 (YLS8) exhibited the most stable expression across all the pear samples tested. While in the other experimental groups, different sets of samples had their own best reference genes. In addition, the gene expression of PbCBL7, a member of the calcineurin B-like protein, was measured across all the 48 samples using the best three reference genes, it displayed variation in gene expression across different tissues and cultivars, and exhibited diverse up- or down-regulated expression patterns under various treatments, which indicate that PbCBL7 may play a role in response to specific abiotic stress in pear. These results are valuable for future research on gene expression and abiotic stress tolerance in pear.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gene expression analysis could improve our understanding of various biological processes and has become much more prevalent. RT-qPCR is a powerful tool widely used on gene expression analysis due to its high specificity, reproducibility, sensitivity, broad dynamic range and no post-PCR processing. These advantages allow a straightforward comparison and make it become the most appropriate choice for gene expression analysis (Garson et al. 2005; Li et al. 2010; Martin et al. 2008). Despite its numerous advantages, several serious flaws have been reported, including the lack of reliable reference genes for normalization (Obrero et al. 2011). To obtain reliable results, normalization is crucial for RT-qPCR studies to control for sample differences (Tong et al. 2009). Since the reference genes are exposed to the same preparation steps as the gene of interest, they are most frequently used among a set of methods proposed to normalize RT-qPCR data and to avoid the possible errors caused by the minor variations of the experimental conditions (Gutierrez et al. 2008; Han et al. 2013; Huggett et al. 2005; Schmidt and Delaney 2010; Zhu et al. 2013).
Ideal reference genes such as actin (ACT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), α- or β-tubulin (TUA, TUB) and ubiquitin (UBQ) are considered to be constitutively expressed and unaffected by experimental conditions. Thus, these genes are commonly used for normalization without any validation (Brunner et al. 2004; Chi et al. 2012; Marum et al. 2012). However, a few recent studies have shown that the widely used reference genes vary considerably in response to some specific experimental conditions (Marino et al. 2008; Morgante et al. 2011; Nicot et al. 2005), that if not chosen properly, the reference genes may be unsuitable for normalization purposes owing to large measurement errors (Dekkers et al. 2012; Obrero et al. 2011). Therefore, to ensure proper normalization, it must be demonstrated that the reference genes of choice have high expression stability independent of the varying experimental conditions.
In the recent decades, several studies on validating the importance of the reference genes for each particular experimental condition have been performed mainly on plants species including Arabidopsis thaliana, tobacco, tomato, chicory, peanut, potato, peach, soybean, cucumber, litchi, flax and zucchini (Chi et al. 2012; Czechowski et al. 2005; Expósito-Rodríguez et al. 2008; Huis et al. 2010; Jian et al. 2008; Maroufi et al. 2010; Nicot et al. 2005; Obrero et al. 2011; Remans et al. 2008; Schmidt and Delaney 2010; Tong et al. 2009; Wan et al. 2010; Zhong et al. 2011). To identify the suitable reference genes in a range of samples, statistical analysis procedures including geNorm, NormFinder and BestKeeper have been developed and applied (Andersen et al. 2004; Pfaffl et al. 2004; Vandesompele et al. 2002).
The pear (Pyrus L.), which belongs to the subfamily Pomoideae in the Rosaceae family, is a popular and commercial crop. It is also the third most important fresh fruit species which has been widely cultivated in the world (Wu et al. 2013). The birch-leaf pear (Pyrus betulifolia Bunge) originated in China and is one of the most widely used rootstock in pear production. It has a strong adaptability and is tolerant to cold, drought, waterlogging and infertility. Increasing the stress resistance of pear will improve its production and quality (Chang et al. 2012b). However, studies on plant stress resistance have focused on crops including Arabidopsis, tomato and barley (Jakab et al. 2005; Jiang et al. 2012; Pandey et al. 2011; Visioni et al. 2013). Understanding the expression patterns of some key genes in the stress tolerance pathway of the pear would give insight into the mechanisms that underlie the processes from germination to harvest, and may help to reveal the changes in gene expression under various conditions. In addition, with the release of the pear genome sequence, functional genomics research on the pear based heavily on gene expression analysis is becoming more popular and will facilitate the understanding of pear development (Wu et al. 2013). Recently, RT-qPCR has always been reported in gene expression analyses in pear (Zhang et al. 2011, 2013), but up to now, no comprehensive or systematic evaluation of candidate housekeeping genes has been undertaken in pear.
In the present study, the expression profiles of 18 candidate reference genes, including Actin2/7(ACT2/7), α-tubulin (TUA), translation elongation factor 2 (TEF2), 18S ribosomal RNA (18SrRNA), ubiquitin extension protein (UBI), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), elongation factor 1 alpha (EF-1α), Glyceraldehyde-3-phosphate dehydrogenase C subunit 1 (GAPC1), Actin 11 (ACT11), ubiquitin-conjugating enzyme 8 (UBC8), Yellow-leaf-specific gene 8 (YLS8), TIP41-like family (TIP41), Protein phosphatase 2A subunit A2 (PP2A), F-box family protein (F-box), Pentatricopeptide repeat superfamily protein (PPR2), Sand family protein (SAND), ubiquitin-conjugating enzyme 21 (UBC21) and ubiquitin 10 (UBQ10) were evaluated using RT-qPCR analysis to determine the optimal reference genes across 48 pear cDNA samples. The expression levels of these internal control genes were tested in different pear tissues and cultivars, and within a range of abiotic stress and hormone treatments. To evaluate the expression stability of reference genes, three common statistical methods, geNorm, NormFinder and BestKeeper, were used.
In addition, our previous work on PbCBL7 gene, a member of calcineurin B-like protein, has revealed that this gene can enhance E. coli tolerance towards a variety of stresses (data not shown). Given that the newly identified PbCBL7 gene may play an important role in anti-adversity stress in pear, the newly identified reference gene combination in this study was further used to examine and normalize its expression profiles. The results obtained will provide guidelines for reference gene selection for gene expression analysis under different experimental conditions in pear and also provide reference for future analysis on suitable RT-qPCR normalization conditions in other crops.
Materials and methods
Plant materials
We obtained the birch-leaf pear (Pyrus betulifolia Bunge), an important rootstock that is highly resistant to salt, from the national germplasm orchard of Institute of Horticulture of Jiangsu Academy of Agricultural Sciences, Nanjing, Jiangsu, China. Pear samples of varying genotypes and tissues and from various conditions (hormonal treatments (ABA, 6-BA and NAA), stress treatments (low and high temperature, NaCl, PEG6000, CaCl2 and mannitol)) were obtained.
For the experimental group comprising different cultivars, the flesh of ‘Yuanhuang (YH)’, ‘Sucui1 (SC1)’and ‘Cuiguan (CG)’ pears was collected at a stage equivalent to commercial ripeness, which is at about from 11.9 to 14.4 % soluble solids concentration (SSC) for the selected fruits from 8-year-old trees and free from visual symptoms of any disease or blemishes. These samples were directly stored at −80 °C for further use. The peel and leaves were collected from the fruits and 8-year-old trees, respectively, of ‘YH’, ‘SC1’, ‘CG’ ‘Xizilu (XZL)’ and ‘Cuilu (CL)’. For the experimental group comprising different tissue types, young leaf, old leaf, petiole, peel and flesh samples were taken from 8-year-old ‘Sucui2 (SC2)’ trees growing under natural conditions. For the experimental group comprising samples subjected to a range of abiotic stress including NaCl, Mannitol, CaCl2, PEG6000 and temperature treatments, seedlings of P. betulifolia Bunge were grown in 1/2 MS nutrient solution containing NaCl solution at varying concentrations (0, 100, 200, 300 and 400 mM at room temperature for 6 h), mannitol solution at varying concentrations (0, 100 and 200 mM at room temperature for 6 h), 100 mM CaCl2, or 10 % (w/v) PEG6000 solution at varying time points (2 h, 6 h, 12 h, 24 h). Seedlings were also exposed to high (42 °C) and low temperatures (4 °C) for 6 h, and seedlings which were exposed to room temperature (24 °C) are used as the control. The leaves were collected following these experimental treatments. For the experimental group comprising samples subjected to different hormone treatments, seedlings of P. betulifolia Bunge were grown in 1/2 MS nutrient solution containing different concentrations of hormones (6-BA (20 µM), ABA (50 µM), NAA (10 µM)) at various duration (2 h, 6 h, 12 h and 24 h). Leaves harvested at 0 h for each treatment were treated as controls. Plant materials were collected in triplicate, frozen in liquid nitrogen, and stored at −80 °C until total RNA extraction.
Total RNA extraction and cDNA synthesis
Total RNA was isolated from 100 mg of samples using an EASYspin Plus Plant RNA mini kit (Aidlab Biotech, China). DNase I (TaKaRa, Japan) was added to eliminate any trace of genomic DNA contamination, according to the manufacturer’s procedures. The integrity of the RNA samples was evaluated by 1.2 % (w/v) agarose gel electrophoresis, and the A260/280 and A260/230 nm absorption ratios were determined using a BioPhotometer Plus (Eppendorf, Germany) to estimate RNA concentration and quality. Clear bands corresponding to 28S and 18S rRNA and the absence of a smear indicate minimal degradation of total RNA. For cDNA synthesis, the RNA has to have an optical density absorption ratio A260/280 of more than 1.8. First-strand cDNA was synthesized using a cDNA amplification kit (TaKaRa) according to the user manual. Briefly, 1 µg RNA, 50 µM oligodT (18) and 10 mM dNTP mix were added together to incubate at 65 °C for 5 min, then placed on ice for 5 min. After that, 4 µl 5 × RT buffer, 20 U RNase Inhibitor and 200 U Prime Script RTase were added and then incubated at 42 °C for 60 min. Finally, the RT reaction was terminated by incubating at 70 °C for 15 min. Each RNA sample was controlled for genomic DNA contamination without reverse transcriptase addition into cDNA synthesis mixture. All samples were diluted 1:10 with RNase-free water, prior to its use as a template in RT-qPCR. Samples were stored at −20 °C.
Primer design and RT-qPCR amplified product verification
18 candidate reference genes were chosen for further investigation to be able to find the most stably expressed reference genes to be used in RT-qPCR analysis (Table S1). This group of genes comprised several classical internal control genes which are most commonly used as reference for expression analysis and the others based on previous reports (Han et al. 2013; Tong et al. 2009). The nucleotide sequences of pear genes were found in the pear genome database (http://peargenome.njau.edu.cn:8004/), and the chosen pear genes were then used to query against the Arabidopsis protein database using BLASTX to obtain the homologous genes. The functional annotations of these pear reference genes were also based on Arabidopsis thaliana. Primers were designed using Beacon Designer 7.0 (Premier Biosoft International, USA) with the following criteria to ensure optimal polymerization efficiency and to reduce the impact of RNA integrity on gene expression in RT-qPCR: primers were selected with a melting temperature of 56–60 °C, optimal primer length of 17–24 bp, and GC content of 45–55 %. Primers were designed to amplify products within the range of 76–199 bp (Table S2). Specificity of the amplification product for each primer pair was verified by the presence of a single band at the expected size with agarose gel electrophoresis, and by the presence of a single peak in RT-qPCR melting curve products.
Quantitative RT-PCR
RT-qPCR was carried out in 96-well plates with an ABI7500 Real-Time PCR Detection System (Applied Biosystems, USA) using SYBR Green Master ROX (Roche, Japan), following the manufacturer’s instructions. Reaction mixtures and conditions used in this study according to the method reported by Xu et al. (2012). At the end of the reactions, a melting curve analysis was performed to evaluate primer specificity. Controls without template and a reverse transcription negative were included for each primer pair to detect possible reagent contamination and to ensure that there was no potential genomic DNA contamination. Each real-time PCR reaction was performed in triplicate (technical replicates) on three individual plants (biological replicates).
Data analysis
The expression level of each selected reference gene was determined by the number of amplification cycles (C t) needed to reach a specific threshold level of the detection. The PCR efficiency showed in Table S2 was calculated for each gene with LinRegPCR program from raw fluorescence data taken from the ABI7500 detection system (Ramakers et al. 2003; Tong et al. 2009). Various analysis tools including geNorm, NormFinder and BestKeeper were used to estimate gene expression stability. The expression stability as well as the optimal combination of candidate genes to be used for normalization was determined by geNorm software. It provides a stability measure (M), excludes the least stable gene, and creates a line graph by a stepwise method to show stability of the selected genes (Vandesompele et al. 2002). NormFinder uses an ANOVA-based model to estimate intra- and inter-group variations and provides a direct measure for integrating all consistent results (Andersen et al. 2004). BestKeeper estimates the expression stability of reference genes by calculating the C t value, and provides a comparative analysis based on pairwise correlations of all candidate reference genes (Pfaffl et al. 2004).
Expression of PbCBL7 gene under different abiotic stresses
To evaluate the expression profiles of PbCBL7 gene under various experimental conditions, its relative transcript levels were normalized by the geometric average of the three most reliable reference genes selected in all the sample pools in this study through the equation of relative ratio target gene/control = \((E_{{{\text{Target}}\,{\text{gene}}}}^{{ - {\text{C}}_{\text{t}} \,{\text{Target}}\,{\text{gene}}}} )/(E_{\text{Control}}^{{ - {\text{C}}_{\text{t}} \,{\text{Control}}}})\) (Pfaffl 2001; Shi et al. 2012). Calculations were carried out with Microsoft Excel software. The data were statistically analyzed with DPS Version 7.55 software (DPS Institute, Hangzhou, China) using Duncan’s multiple range test at the P < 0.05 level of significance.
Results
Verification of the amplified product
To select the best reference genes for pear gene expression profiling, the primers of the 18 common reference genes (Table S2) were designed for RT-qPCR analysis. Melting curves and agarose gel electrophoresis results revealed that there were no primer dimers or other non-specific amplification (Fig. 1a, b). In addition, no signals were detected from the controls of template lacking reverse transcriptase, indicating no genomic DNA contamination.
C t value analysis
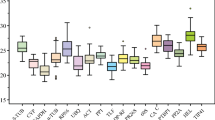
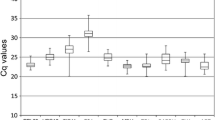
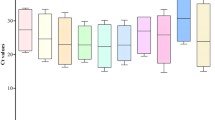
In an RT-qPCR assay, a positive reaction is measured by accumulation of a fluorescent signal. The number of cycles required for the fluorescent signal to cross the threshold is called the C t value, which is used to evaluate the transcriptional levels of the reference genes. The results would provide an overview of the abundance of all the selected genes in the samples. In this study, the C t values of the 18 reference genes showed a relative wide range, with the lowest (14.30) in 18SrRNA to the highest (34.14) in PPR2, while the majority of these values ranged from 18.2 to 27.4 in all tested pear samples (Fig. 2). It took only 14.30 amplification cycles to reach the threshold fluorescence for 18SrRNA, indicating that it was highly expressed compared with the other genes. The individual reference genes showed different expression levels across all samples used in this study. 18SrRNA showed the smallest expression variation (below 7 cycles), while PPR2 and TUA showed the highest gene expression variation (about 12 cycles) among all the selected genes. The wide expression ranges of the 18 tested reference genes indicated that no single reference gene had a constant expression under all experimental conditions in pear samples. Therefore, it is necessary to select a relatively reliable reference gene to normalize gene expression profiling under certain conditions.
GeNorm analysis
Figure 3 shows the 18 candidate reference genes with M values that are less than 1.5. This is usually regarded as acceptable in terms of expression variability. Interestingly, the most stable reference gene was not identical among different sample pools. For all the 48 samples, ACT2/7 and UBI were the most stably expressed genes, and TUA was the least stable gene (Fig. 3a). The least stable gene in the group under different CaCl2 and hormonal treatments was also TUA, which was consistent with the findings above. The most stably expressed genes were ACT11/UBQ10 and ACT2/7/EF-1α under different CaCl2 and hormonal treatments, respectively (Fig. 3e, g). In the experimental group comprising different tissues, geNorm identified TIP41 as the unstable gene, whereas PP2A and GAPDH were regarded as the best reference genes (Fig. 3b). In the experimental group comprising different cultivars, UBI and UBC8 were considered as the best genes, while PPR2 was thought to be the least stable (Fig. 3c). In the experimental group comprised of samples subjected to different concentrations of NaCl, ACT2/7 and UBQ10 were the most stable genes, and GAPC1 was the least stable gene (Fig. 3d). In the experimental group comprising of samples subjected to different temperatures, TEF2 and UBC21 were the most stably expressed genes, and 18SrRNA was the least stable gene (Fig. 3f). In the experimental group comprising different degrees of osmotic stress, YLS8 and PP2A were the most stably expressed genes, and UBQ10 was the least stable gene (Fig. 3h).
We considered the average expression stability (M) and the pairwise variation (V) when using geNorm to evaluate gene expression stability. To determine the necessity of adding the next internal control gene and the optimal number required for effective normalization, the pairwise variations Vn/Vn+1 between two sequential normalization factors NFs (NFn and NFn+1) were introduced. According to the cut-off value of 0.15 reported by Vandesompele et al. (2002), in the groups for all the samples analyzed together and different tissues and cultivars comparison, three reference genes should be selected (V3/4 < 0.15), while in the rest groups, two reference genes were sufficient (V2/3 ≤ 0.15) (Fig. 4).
Determination of the optimal number of reference genes. Every bar represents change in normalization accuracy upon stepwise addition of more reference genes according to the ranking in Fig. 3. The pairwise variation (Vn/Vn+1) was calculated from the normalization factors NFn and NFn+1, with a recommended cut-off threshold of 0.150
NormFinder analysis
A more stable gene expression is indicated by lower average expression stability values. Table 1 shows the results calculated by NormFinder. The output of NormFinder with different sample pools was highly variable. When all 48 samples taken together, ACT2/7, UBI and UBC8 showed remarkable stability in their expression levels and were always highly ranked; while 18SrRNA, TUA and TIP41 exhibited unstable expression profiles and were always included among the unstable genes.
When analyzed for the established groups with NormFinder separately, YLS8 was calculated to be the most stable single gene in the group of different cultivars, whereas PPR2 was the most variable one. NormFinder revealed ACT2/7 to be the most stable in the experimental group of different tissues, which was consistent with the results obtained from the groups under different NaCl, CaCl2 and osmotic stress treatment. In this context, the least stable genes were TUA in NaCl and CaCl2 treatment groups and UBQ10 in osmotic stress treatments groups. In groups involving exposure to various hormones and temperatures, TIP41 was the most stable gene with the stability value of 0.174 and 0.034, while TUA and 18SrRNA were the most unstable with the stability values of 0.709 and 1.399, respectively (Table 1).
BestKeeper analysis
Stable reference genes always showed a strong correlation with the BestKeeper Index. The results analyzed by this method are presented in Table 2. High coefficients of correlation always indicate that these gene pairs exhibit similar expression patterns. When all samples were taken together for analysis, ACT2/7, UBI, YLS8 and SAND showed strong correlation with the BestKeeper Index (r > 0.96), and ranked among the top four positions. These results were consistent with that derived from geNorm and NormFinder. ACT2/7 was consistently stably expressed, except in the groups involving different cultivars and temperature treatments wherein YLS8 and TEF2 were the most stable reference genes, respectively. 18SrRNA consistently ranked poorly in almost all series, becoming the least reliable reference gene.
Expression profile analysis of PbCBL7
To investigate the effect of different cultivars and tissues as well as abiotic stress on gene expression in pear, the transcript level of PbCBL7 in 48 pear samples was assessed by RT-qPCR. The expression level of PbCBL7 showed varied according to context after being normalized to ACT2/7, UBI and YLS8, which were the best reference gene combination selected by geNorm, NormFinder and BestKeeper algorithm across all tested samples. The results showed that PbCBL7 was expressed abundantly in petiole, leaf and peel and weakly in flesh (Fig. 5a). In the leaves of different cultivars, PbCBL7 exhibited the highest expression level in ‘CL’, but in peel and flesh, PbCBL7 was expressed the highest in ‘YH’ (Fig. 5b–d). PbCBL7 level was down-regulated under salt and mannitol stress, exhibiting the lowest expression at 200 mM of NaCl (Fig. 5f, g). We speculated that the 100 or 200 mM NaCl stress treatment may not initiate the defense system in pear plant and at this time the expression of PbCBL7 is negatively regulated by salt stress, but with the increase of concentration, the defense system began to engage and this resulted in enhanced transcription of PbCBL7 gene. The transcript level was higher under cold- or heat stress than at room temperature which indicates that PbCBL7 gene was regulated by high or low temperatures (Fig. 5e). Since the CBL protein family is mediated by calcium ions, we measured the transcript level of PbCBL7 upon exposure to CaCl2. PbCBL7 was up-regulated in response to CaCl2, reached maximum at 12 h then decreased, similar to what was observed under PEG6000 treatment. Furthermore, different hormone treatments were also carried out, and interestingly, PbCBL7 was consistently up-regulated under 6-BA treatment, while under ABA treatment, PbCBL7 was induced in a transient way and reached a maximum at 2 h, then consistently decreased. With NAA treatment, PbCBL7 level reached the maximum at 2 h, thereafter reached the minimum at 6 h then consistently increased after treatment for 12 h and 24 h (Fig. 6).
Expression analysis of PbCBL7 by RT-qPCR. Different experimental conditions including different tissues (a) and cultivars (b–d) and varying treatments [temperature (e), NaCl (f) and Mannitol (g)]. The data show the relative transcript levels of the target gene as normalized to the best reference gene combination, ACT2/7, UBI and YLS8. Mean values and standard errors (bars) were obtained from three real-time quantitative PCR reactions. Different letters indicate significant differences at P < 0.05 according to Duncan’s multiple range tests
Expression analysis of PbCBL7 using qPCR in response to Ca2+ signal, PEG6000 and hormones stresses. The results were shown in relative transcript levels of the target genes normalized with the best reference gene combination, ACT2/7, UBI and YLS8. CK control, h hour. Significant up-regulation with respect to control was shown by *P<0.05 or ** P<0.01. Error bars indicate standard errors
Discussion
RT-qPCR is verified to be a sensitive and accurate technique for gene expression analysis (Guénin et al. 2009; Li et al. 2010). Ideal reference genes whose transcription levels remained invariable under varying experimental conditions will greatly improve the reliability of RT-qPCR data when evaluating the expression levels of genes of interest (Klie and Debener 2011; Schmidt and Delaney 2010; Zhu et al. 2013). Several common candidate reference genes are currently being used as internal controls regardless of their instability and variance. If not chosen properly, the internal control genes would not fulfill its role as a standard in gene expression analysis, leading to a wrong conclusion (Dekkers et al. 2012; Obrero et al. 2011). The present study is the first systematic survey on the expression stability of 18 genes used as internal controls for RT-qPCR analysis under different experimental conditions in 48 different pear samples. The expression stabilities of the 18 selected reference genes showed fluctuations and discrepancies across the different sample groups, indicating that none of the internal controls have invariable expression levels across varying conditions.
The expression levels of the reference genes for RT-qPCR are determined by C t values (Klie and Debener 2011; Martin et al. 2008; Zhu et al. 2013). C t values that are too high (C t > 30) or too low (C t < 15) are generally considered to be unsuitable as reference genes (Wan et al. 2011). Evaluation of Ct values, however, lacks sensitivity and reliability. Thus, to choose the best reference genes for RT-qPCR analysis, we used various statistical approaches including geNorm, NormFinder and BestKeeper to minimize bias in the quantification of gene expression (Tong et al. 2009; Wan et al. 2010; Zhu et al. 2013).
ACT2/7, UBI and YLS8 consistently ranked as most stable by the three algorithms when all the samples were considered. With the different experimental conditions, different sets of samples had their own best reference genes. For example, ACT2/7/UBQ10 is the best reference gene combination when studying samples exposed to NaCl, whereas PP2A/GAPDH did better than ACT2/7/UBQ10 when studying samples of different tissues. UBI/UBC8 was the best combination for different cultivars.
ACT2/7, the most frequently used reference gene, is one of the major components of cytoplasmic microfilaments in eukaryotic cells (Hu et al. 2009). ACT2/7 expression is rather variable in peach and A. thaliana, but mostly stable in soybean and radish (Jian et al. 2008; McDowell et al. 1996; Tong et al. 2009; Xu et al. 2012). In this study, ACT2/7 consistently ranked as most stable, and thus considered as a suitable reference gene. However, compared with ACT2/7, another candidate reference gene ACT11 in the actin gene family showed most stability under CaCl2 treatment, as determined by geNorm. UBI was evaluated as the most stable reference gene in the group of different cultivars by geNorm. These findings are consistent with the results obtained from cucumber (Wan et al. 2010). UBQ10 was determined as an unsatisfactory reference gene in soybean and rice (Jain et al. 2006; Jian et al. 2008), but displayed stable expression in A. thaliana and tomato (Czechowski et al. 2005; Løvdal and Lillo 2009). However, in this study, UBQ10 ranked well under both NaCl and temperature treatment, but ranked poorly under different osmotic treatments. TEF2 is needed for elongation and mRNA transcription in eukaryotes (Xu et al. 2012). In the group exposed to varying temperatures, TEF2 exhibited stable expression patterns and was abundantly and constantly transcribed, consistent with previous studies in peach samples (Tong et al. 2009). In previous studies, TIP41 showed stable expression level across tissues and during development in tomato, but performed poorly during development of grape berry and in the leaves and roots of A. thaliana under copper or cadmium exposure (Czechowski et al. 2005; Expósito-Rodríguez et al. 2008; Reid et al. 2006). In the present study, TIP41 was the most unstable in groups involving different tissues. Among the members of the ubiquitin-conjugating enzyme gene family, it was reported in Ammopiptanthus mongolicus that UBC5 and UBC1 represent the best combination for normalization of differentially expressed genes under cold stress, UBC2 under salt stress, and UBC4 and UBC5 under heat stress (Shi et al. 2012). In this study, UBC8 showed the most stable expression in groups of different cultivars and UBC21 was the most suitable gene in samples exposed to different temperatures, as analyzed by geNorm. PP2A, another novel reference gene, was reported to be stably expressed in rabbiteye blueberry (Vashisth et al. 2011), while in this study, PP2A showed the most stable expression in groups involving different tissues and samples exposed to osmotic stress, as assessed by geNorm. On the other hand, EF-1α was the least reliable control in Arabidopsis, Salvia miltiorrhiza and barley (Czechowski et al. 2005; Jarošová and Kundu 2010; Yang et al. 2010), but in this study, this gene was the most stable in groups exposed to different hormones, as assessed by geNorm. TUA is stably expressed during development in soybean and in different tissues in poplar (Brunner et al. 2004; Jian et al. 2008). In this study, TUA level was highly unstable under conditions involving different cultivars and exposure to NaCl, CaCl2, temperature and hormones. GAPDH, involved in basic cellular functions, is one of the most stable genes in barley and grapevine (Jarošová and Kundu 2010; Reid et al. 2006), but unstable in tobacco (Schmidt and Delaney 2010), as evaluated by geNorm, this gene showed poor expression stability except for the group involving different tissues. 18SrRNA has failed to be selected as an ideal reference gene because it displayed poor stability in most cases in this research, consistent with findings in broomrape tissues (Gonzalez-Verdejo et al. 2008).
There is some discrepancy among the results obtained by geNorm, NormFinder and BestKeeper, which may be due to differences in algorithms and analytical procedures (Andersen et al. 2004; Pfaffl et al. 2004; Vandesompele et al. 2002). Taken together, these results indicate that a reference gene with consistent expression under certain conditions may not be suitable for normalization of gene expression under other experimental conditions. Variability in the expression of reference genes may partly be explained by the fact they may not only involve in basal cell metabolism but also join in other cellular functions (Mafra et al. 2012; Wan et al. 2011).
The combination of two or more stable genes represents the ideal internal control for RT-qPCR analysis, which would improve the accuracy significantly (Silveira et al. 2009). Therefore, to get an accurate expression profiling among multiple samples, more than one reference genes should be used (Brunner et al. 2004; Coito et al. 2012; Nicot et al. 2005; Xu et al. 2012; Zhong et al. 2011).
The results obtained from the three algorithms were comprehensive in determining suitable reference genes for qPCR normalization (Chang et al. 2012a; Klie and Debener 2011). Thus, we can conclude that expression levels of all the 18 genes were variable to some extent, but that of ACT2/7, UBI and UBC8 were in general the most stable. Thus, a combination of ACT2/7, YLS8 and PP2A would be appropriate as a reference panel for normalizing gene expression data among different tissues, whereas the combination of YLS8, UBC8 and UBC21 would be most suitable for different cultivars. Compared with others, ACT2/7 and UBQ10 represented the most suitable reference gene combination when the samples were treated with NaCl, and ACT2/7 and YLS8 represented the most suitable combination when samples were treated with CaCl2. TEF2 and YLS8 represented the most suitable combination when the temperature was altered. ACT2/7 and EF-1α were the most favorable genes in samples exposed to different hormones, and ACT2/7 and GAPDH were stably expressed under osmotic stress. In contrast, TUA, 18SrRNA and TIP41 were the most unstable under several experimental conditions and should generally not be used as reference genes.
In addition, the three best reference genes, ACT2/7, UBI and YLS8, were used to normalize the expression level of PbCBL7 in the 48 different pear samples. Calcineurin B-like proteins (CBLs) is a class of calcium-binding proteins which play an important role in the regulation of responses of plant to environmental stress (Gu et al. 2008; Kudla et al. 1999; Luan 2009; Zhang et al. 2008). Conditions that could adversely impact pear production include cold stress, heat stress, drought stress, flooding and salt stress. The expression profiles of CBLs in different cultivars and tissues as well as in samples exposed to various abiotic stresses would provide a preliminary understanding of the complicated stress tolerance mechanisms in pear.
In Arabidopsis, 10 CBL genes have been characterized, and several CBLs have been shown to function in an abiotic stress-specific signaling cascade (Pandey et al. 2004; Shi et al. 1999). While in Populus, PeCBL1, 9, 2, 5 and 3 may play a role in response to specific abiotic stress (Zhang et al. 2008). Our previous study on PbCBL7 has revealed that this gene can enhance E. coli tolerance towards a variety of stresses.
In this study, PbCBL7 was expressed in different cultivars and tissues of pear as well as in samples exposed to various stress conditions. Interestingly, expression level varied in different tissues and cultivars, and also exhibited diverse up- or down-regulated expression patterns under various treatments, which further indicate that PbCBL7 may also play a role in response to specific abiotic stress in pear.
In summary, this present study provides new evidence for the importance of appropriate reference genes in RT-qPCR analysis under given experimental conditions in pear. All 18 candidate reference genes selected showed different expression stabilities in each group, and among them, ACT2/7, UBI and YLS8 exhibited the most stable expression across all the tested pear samples, while in the other experimental groups, different sets of samples had their own best reference genes. The results of the present study provide useful guidelines for further studies on gene expression analysis using RT-qPCR in pear.
Author contribution statement
XY prepared the plant materials, carried out the RT-qPCR experiments and drafted the manuscript. LH, LX, WZ and YQ participated in the data analysis by geNorm, NormFinder and BestKeeper. CY and LJ conceived of the study, and participated in its design and coordination and revise the manuscript. The final manuscript was read and approved by all the authors.
Abbreviations
- ABA:
-
Abscisic acid
- bp:
-
Base pair
- NAA:
-
1-Naphthaleneacetic acid
- PEG6000:
-
Polyethylene Glycol 6000
- RT-qPCR:
-
Quantitative real-time polymerase chain reaction
- 6-BA:
-
6-Benzylaminopurine
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4:14
Chang E, Shi S, Liu J, Cheng T, Xue L, Yang X, Yang W, Lan Q, Jiang Z (2012a) Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. PLoS One 7:e33278
Chang YH, Li H, Cong Y, Lin J, Sheng BL (2012b) Characterization and expression of a phytochelatin synthase gene in birch-leaf pear (Pyrus betulaefolia Bunge). Plant Mol Biol Rep 30:1329–1337
Chi XY, Hu RB, Yang QL, Zhang XW, Pan LJ, Chen N, Chen MN (2012) Validation of reference genes for gene expression studies in peanut by quantitative real-time RT-PCR. Mol Genet Genomics 287:167–176
Coito JL, Rocheta M, Carvalho L, Amâncio S (2012) Microarray-based uncovering reference genes for quantitative real time PCR in grapevine under abiotic stress. BMC Res Notes 5:220
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HWM, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53:28–37
Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131
Garson JA, Grant PR, Ayliffe U, Ferns RB, Tedder RS (2005) Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J Virol Methods 126:207–213
Gonzalez-Verdejo CL, Die JV, Nadal S, Jimenez-Marin A, Moreno MT, Roman B (2008) Selection of housekeeping genes for normalization by real-time RT-PCR: analysis of Or-MYB1 gene expression in Orobanche ramosa development. Anal Biochem 379:176–181
Gu Z, Ma B, Jiang Y, Chen Z, Su X, Zhang H (2008) Expression analysis of the calcineurin B-like gene family in rice (Oryza sativa L.) under environmental stresses. Gene 415:1–12
Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, Gutierrez L (2009) Normalization of qRT-PCR data: the necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60:487–493
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J 6:609–618
Han B, Yang Z, Samma MK, Wang R, Shen W (2013) Systematic validation of candidate reference genes for qRT-PCR normalization under iron deficiency in Arabidopsis. Biometals 26:403–413
Hu RB, Fan CM, Li HY, Zhang QZ, Fu YF (2009) Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. BMC Mol Biol 10:93
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6:279–284
Huis R, Hawkins S, Neutelings G (2010) Selection of reference genes for quantitative gene expression normalization in flax (Linum usitatissimum L.). BMC Plant Biol 10:71
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651
Jakab G, Ton J, Flors V, Zimmerli L, Métraux JP, Mauch-Mani B (2005) Enhancing Arabidopsis salt and drought stress tolerance by Chemical priming for its abscisic acid responses. Plant Physiol 139:267–274
Jarošová J, Kundu J (2010) Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol 10:146
Jian B, Liu B, Bi YR, Hou WS, Wu CX, Han TF (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Jiang Y, Liang G, Yu D (2012) Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol Plant 5:1375–1388
Klie M, Debener T (2011) Identification of superior reference genes for data normalisation of expression studies via quantitative PCR in hybrid roses (Rosa hybrida). BMC Res Notes 4:518
Kudla J, Xu Q, Harter K, Gruissem W, Luan S (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA 96:4718–4723
Li QF, Sun SSM, Yuan DY, Yu HX, Gu MH, Liu QQ (2010) Validation of candidate reference genes for the accurate normalization of real-time quantitative RT-PCR data in rice during seed development. Plant Mol Biol Rep 28:49–57
Løvdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization intomato subjected to nitrogen, cold, and light stress. Anal Biochem 387:238–242
Luan S (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14:37–42
Mafra V, Kubo KS, Alves-Ferreira M, Ribeiro-Alves M, Stuart RM, Boava LP, Rodrigues CM, Machado MA (2012) Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. PLoS One 7:e31263
Marino ER, Borges AA, Perez AB, Perez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131
Maroufi A, Bockstaele EV, Loose MD (2010) Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol 11:15
Martin RC, Hollenbeck VG, Dombrowski JE (2008) Evaluation of reference genes for quantitative RT-PCR in Lolium perenne. Crop Sci 48:1881–1887
Marum L, Miguel A, Ricardo CP, Miguel C (2012) Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS One 7:e35113
McDowell JM, Huang S, McKinney EC, An YQ, Meagher RB (1996) Structure and evolution of the actin gene family in Arabidopsis thaliana. Genet 142:587–602
Morgante CV, Guimarães PM, Martins ACQ, Araújo ACG, Leal-Bertioli SCM, Bertioli DJ, Brasileiro ACM (2011) Reference genes for quantitative reverse transcription-polymerase chain reaction expression studies in wild and cultivated peanut. BMC Res Notes 4:339
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914
Obrero Á, Die JV, Romén B, Gómez P, Nadal S, Gonzalez-Verdejo CL (2011) Selection of reference genes for gene expression studies in zucchini (Cucurbita pepo) using qPCR. J Agric Food Chem 59:5402–5411
Pandey GK, Cheong YH, Kim KN, Luan S (2004) The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16:1912–1924
Pandey S, Nookaraju A, Upadhyaya CP, Gururani MA, Venkatesh J, Kim DH, Park SW (2011) An update on biotechnological approaches for improving abiotic stress tolerance in tomato. Crop Sci 51:2303–2324
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes, and sample integrity: bestKeeper-Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515
Ramakers C, Ruijter JM, Deprez RH, Moorman AF (2003) Assumption free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66
Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27
Remans T, Smeets K, Opdenakker K, Mathijsen D, Vangronsveld J, Cuypers A (2008) Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227:1343–1349
Schmidt GW, Delaney SK (2010) Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol Genet Genomics 283:233–241
Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11:2393–2406
Shi J, Liu M, Shi J, Zheng G, Wang Y, Wang J, Chen Y, Lu C, Yin W (2012) Reference gene selection for qPCR in Ammopiptanthus mongolicus under abiotic stresses and expression analysis of seven ROS-scavenging enzyme genes. Plant Cell Rep 31:1245–1254
Silveira ÉD, Alves-Ferreira M, Guimarães LA, da Silva FR, Carneiro VT (2009) Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol 9:84
Tong ZG, Gao ZH, Wang F, Zhou J, Zhang Z (2009) Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol Biol 10:71
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Vashisth T, Johnson LK, Malladi A (2011) An efficient RNA isolation procedure and identification of reference genes for normalization of gene expression in blueberry. Plant Cell Rep 30:2167–2176
Visioni A, Tondelli A, Francia E, Pswarayi A, Malosetti M, Russell J, Thomas W, Waugh R, Pecchioni N, Romagosa I, Comadran J (2013) Genome-wide association mapping of frost tolerance in barley (Hordeum vulgare L.). Plant. Genomics 14:424
Wan HJ, Zhao ZG, Qian CT, Sui YH, Malik AA, Chen JF (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261
Wan H, Yuan W, Ruan M, Ye Q, Wang R, Li Z, Zhou G, Yao Z, Zhao J, Liu S, Yang Y (2011) Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem Biophys Res Commun 416:24–30
Wu J, Wang Z, Shi Z (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23:396–408
Xu Y, Zhu X, Gong Y, Xu L, Wang Y, Liu L (2012) Evaluation of reference genes for gene expression studies in radish (Raphanus sativus L.) using quantitative real-time PCR. Biochem Biophys Res Commun 424:398–403
Yang Y, Hou S, Cui G, Chen S, Wei J, Huang L (2010) Characterization of reference genes for quantitative real-time PCR analysis in various tissues of Salvia miltiorrhiza. Mol Biol Rep 37:507–513
Zhang H, Yin W, Xia X (2008) Calcineurin B-Like family in Populus: comparative genome analysis and expression pattern under cold, drought and salt stress treatment. Plant Growth Regul 56:129–140
Zhang X, Allan AC, Yi Q, Chen L, Li K, Shu Q, Su J (2011) Differential Gene expression analysis of Yunnan red pear, Pyrus Pyrifolia, during fruit skin coloration. Plant Mol Biol Rep 29:305–314
Zhang H, Zhang S, Qin G, Wang L, Wu T, Qi K, Zhang S (2013) Molecular cloning and expression analysis of a gene for sucrose transporter from pear (Pyrus bretschneideri Rehd.) fruit. Plant Physiol Biochem 73:63–69
Zhong HY, Chen JW, Li CQ, Chen L, Wu JY, Chen JY, Lu WJ, Li JG (2011) Selection of reliable reference genes for expression studies by reverse transcription quantitative realtime PCR in litchi under different experimental conditions. Plant Cell Rep 30:641–653
Zhu J, Zhang L, Li W, Han S, Yang W, Qi L (2013) Reference gene selection for quantitative real-time PCR normalization in caragana intermedia under different abiotic stress conditions. PLoS One 8:e53196
Acknowledgments
This work was supported by grants from the Program for the National Natural Science Foundation of China (Grant No. 31372051) and Post-doctoral Foundation of Jiangsu, China (Grant No. 1302006B).
Conflict of interest
All authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by S. Abe.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, Y., Li, H., Li, X. et al. Systematic selection and validation of appropriate reference genes for gene expression studies by quantitative real-time PCR in pear. Acta Physiol Plant 37, 40 (2015). https://doi.org/10.1007/s11738-015-1784-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-015-1784-0