Abstract
Although previous studies reported structural changes associated with electroconvulsive therapy (ECT) in major depressive disorder (MDD), the underlying molecular basis of ECT remains largely unknown. Here, we combined two independent structural MRI datasets of MDD patients receiving ECT and transcriptomic gene expression data from Allen Human Brain Atlas to reveal the molecular basis of ECT for MDD. We performed partial least square regression to explore whether/how gray matter volume (GMV) alterations were associated with gene expression level. Functional enrichment analysis was conducted using Metascape to explore ontological pathways of the associated genes. Finally, these genes were further assigned to seven cell types to determine which cell types contribute most to the structural changes in MDD patients after ECT. We found significantly increased GMV in bilateral hippocampus in MDD patients after ECT. Transcriptome-neuroimaging association analyses showed that expression levels of 726 genes were positively correlated with the increased GMV in MDD after ECT. These genes were mainly involved in synaptic signaling, calcium ion binding and cell-cell signaling, and mostly belonged to excitatory and inhibitory neurons. Moreover, we found that the MDD risk genes of CNR1, HTR1A, MAOA, PDE1A, and SST as well as ECT related genes of BDNF, DRD2, APOE, P2RX7, and TBC1D14 showed significantly positive associations with increased GMV. Overall, our findings provide biological and molecular mechanisms underlying structural plasticity induced by ECT in MDD and the identified genes may facilitate future therapy for MDD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Electroconvulsive therapy (ECT) has been recommended as a first choice for major depressive disorder (MDD) with psychotic features, suicidal attempts, or treatment resistant due to its effectiveness and fast response (van Diermen et al., 2018). Many previous studies (Ota et al., 2015; Joshi et al., 2016; Gryglewski et al., 2019) and meta-analyses (Gbyl & Videbech, 2018; Takamiya et al., 2018; Enneking et al., 2020) reported consistent increment of gray matter volume (GMV) in a wide range of brain regions (Wang et al., 2017; Xu et al., 2019), especially in hippocampus induced by ECT in MDD patients (Joshi et al., 2016; Cao et al., 2018; Leaver et al., 2020). A large number of previous studies have demonstrated that structural plasticity is the underlying mechanism for MDD after ECT, and structural properties could effectively distinguish or predict treatment outputs (Redlich et al., 2016; Jiang et al., 2018). At the same time, efforts were also paid to investigate the role of genetic factors in the etiology of the ECT response (Benson-Martin et al., 2016; Singh & Kar, 2017; Pinna et al., 2018). These studies reported a lot of genes associated with ECT including brain-derived neurotrophic factor (BDNF) (Polyakova et al., 2015), dopamine 2 receptor gene (DRD2) (Bousman et al., 2015), dopamine receptor 3 gene (DRD3) (Dannlowski et al., 2013), and cathechol-o-methyltransferase (COMT) (Tang et al., 2020) but with relatively small sample size. Moreover, an international consortium on genetics of ECT and severe depressive disorders (Gen-ECT-ic) has been established in 2020 with the goal to collect a large sample size of > 30,000 cases to reveal genetic mechanism of treatment response to ECT (Soda et al., 2020). However, whether/how genetic factors account for structural plasticity by ECT in MDD remains unknown.
Recently, the Allen Human Brain Atlas (AHBA, http://human.brain-map.org) microarray dataset which comprises expression level data for more than 20,000 genes covering the whole brain in six donated post-mortem brains provides an indirect way to explore how spatial variations in genes’ expression related to the changes of macroscopic neuroimaging phenotypes. Moreover, a practical pipeline has been set up and has been successfully adopted in various mental disorders for relating brain-wide transcriptomic and neuroimaging data (Arnatkeviciute et al., 2019). For example, the cortical thickness (Anderson et al., 2020), functional correlations and morphometric similarity network (Li et al., 2021, 2023; Yang et al., 2023; Chen et al., 2024; Yu et al., 2024), as well as GMV (Ji et al., 2021) and structural brain network (Liu et al., 2019) in the psychiatric disorders, have been linked to gene expression data in AHBA. Therefore, AHBA provides a good opportunity to investigate relationship between transcriptional data and ECT-induced GMV alterations, which may advance our understanding of biological mechanisms underlying structural plasticity induced by ECT in MDD.
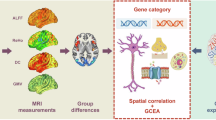
In the current study, we aimed to reveal molecular basis of GMV changes in MDD patients after ECT with longitudinal structural MRI and public AHBA transcriptomic data. First, GMV alterations induced by ECT were analyzed using two independent datasets. Then, the spatial correlations between gene expression levels and GMV alterations were performed using partial least square regression (PLS) to obtain positive associated genes. Only overlapped genes between the two independent datasets were used for further analyses. Third, functional enrichment analysis was conducted using Metascape (https://metascape.org/gp/index.html#/main/step1) to explore ontological pathways of the identified genes. Finally, these genes were further assigned to seven cell types to specify which cell types contributed most to the GMV alterations induced by ECT in the MDD patients. We also performed spatial Spearman correlations between GMV alterations and expression levels of MDD risk genes and ECT related genes. The analytical pipelines were shown in Fig. 1.
The pipeline of data analyses in this study. Individual voxel-wise gray matter volume (GMV) was calculated based on structural MRI data of MDD patients before ECT (TP1) and after ECT (TP2) from two independent datasets. Paired t-tests were used to identify voxel-wise GMV difference maps, i.e., statistical T map. Gene expression data are obtained from six donated brains in the Allen Human Brain Atlas. Gene-wise cross-samples partial least squares (PLS) regressions are performed between gene expression and t maps of GMV differences derived from the two datasets, respectively. The overlap of the genes significantly associated with GMV differences found in the two independent datasets was defined as ECT related genes in MDD patients. Finally, functional enrichment and cell subtype analyses were further performed for these overlapped genes
Materials and methods
Participants
This study included two independent datasets of MDD patients treated by ECT from University of Science and Technology of China (Dataset 1) and Anhui Medical University (Dataset 2) using the same diagnosis, inclusion and exclusion criteria. They were diagnosed as MDD according to Diagnostic and Statistical Manual of Mental Disorders-IV (DSM-IV) criteria and were assigned to ECT since they showed resistance to drug therapy or a severe suicidal tendency. Depression severity was assessed using the 17-item Hamilton Rating Scale for Depression (HAMD) at 12–24 h before the first ECT and 24–72 h after the last ECT. Patients with major neurological or other psychiatric disorders, psychoactive substances, life threatening somatic disease, head trauma followed by unconsciousness for more than five minutes, and MRI-related contraindications were excluded. Moreover, images with poor quality, such as obvious brain lesions, incomplete whole brain coverage, or imaging artifacts were also excluded. Finally, 41 and 23 patients with MDD were kept in dataset 1 and dataset 2, respectively (Table 1). All patients provided written informed consent and the study was approved by the local ethics committees of the Anhui Medical University (Approval number: 20,140,072).
ECT procedures
Modified bi-frontal ECT was performed for all patients using Thymatron System IV Integrated ECT Instruments (Somatics, Lake Bluff, IL, USA). For each patient, the first three sessions occurred on consecutive days in the first week, and the remaining was conducted every other day with a break of weekend. For each session, the initial percent energy was set based on the age of each patient (e.g., 50% for a 50-year-old patient), the stimulation strength was adjusted with an increment of 5% of the maximum charge (approximately 1,000 millicoulombs), and the percent energy was increased until seizure visually observed. The sessions were stopped when a patient’s HAMD score was less than 7. Moreover, propofol, succinylcholine and atropine were used for all patients during ECT. The details for ECT procedures could be found in our previous studies (Wang et al., 2020; Xu et al., 2020; Pang et al., 2022; Li et al., 2023).
MRI data acquisition
In our study, two independent datasets were used. For dataset 1, the structural 3D T1 images were acquired using a clinical 3.0 T Discovery GE 750 scanner. The major parameters were: repetition time (TR) = 8.16 ms, echo time (TE) = 3.18 ms, flip angle = 12°, field of view = 256 × 256 mm2, voxel size = 1 × 1 × 1 mm3 and 188 slices. For dataset 2: the structural 3D T1 images were obtained using a clinical 3.0 T GE 750 MRI scanner. The parameters were: TR = 8.676 ms, TE = 3.184 ms, flip angle = 8°, field of view = 256 × 256 mm2, matrix size = 256 × 256, voxel size = 1 × 1 × 1 mm3 and 188 slices. The structural 3D T1 MRI data were scanned twice for each subject at 12–24 h before the first ECT and 24–72 h after the last ECT.
Data preprocessing
The T1 images were preprocessed using the standard pipeline in the DPABI toolbox (http://rfmri.org/dpabi) with unified segmentation and diffeomorphic anatomical registration through the exponentiated lie algebra (DARTEL). The major steps were: (1) segment each image into gray matter, white matter, and cerebrospinal fluid; (2) normalize the segmented images using the DARTEL; (3) resample all images into a voxel size of 1.5 mm × 1.5 mm × 1.5 mm; (4) modulated by multiplying the voxel values with the Jacobian determinant (transformation matrix during registration) derived from the spatial normalization to account for individual difference; and (5) smoothed with a Gaussian kernel of 8 mm ×8 mm ×8 mm full-width at half maximum for statistical analyses.
Gene expression data processing
The gene expression data was obtained from six donated postmortem brains and only the data in the left hemisphere were used for analyzing to exclude sampling bias using a public available pipeline (https://github.com/BMHLab/AHBAprocessing) (Arnatkeviciute et al., 2019). First, the probes were resigned to genes (latest version) using the Re-annotator toolkit (https://sourceforge.net/projects/reannotator/). Then, a threshold of 50% was used as intensity-based filter. Third, RNA-seq data with highest correlation values was used to select possible probes. Finally, expression data for each participant was normalized by scaling robust sigmoid. As a result, we obtained 10,027 genes for each of 1285 samples.
GMV alterations and associated genes
Paired t-tests were performed within the gray matter mask to identify voxel-wise GMV differences in MDD patients before and after ECT to explore therapeutical effects. Then, spheres with a radius of 4.5 mm (i.e., 3 times of the voxel size) centered in MNI coordinate of each tissue sample (n = 1285) were drawn, and the mean t-value within each sphere was defined as the GMV alteration. To explore the associations with the expression levels of all genes, PLS regression was performed with gene expression data as predictor variables (Abdi & Williams, 2013). The first component of the PLS (PLS1) which was the linear combination of gene expression values most strongly correlated with regional changes in GMV was used in the current study. Then, cross-samples non-parametric Spearman rank test was performed to determine relationship between regional PLS1 weighted gene expression and regional GMV alterations. To estimate the variability of PLS1 score for each gene, bootstrapping with 1000 times was performed. Z scores were defined as the ratio of the weight of each gene to its bootstrap standard error and ranked the genes according to their contributions to PLS1 using univariate one-sample Z tests (Morgan et al., 2019). The set of genes with Z > 5 were considered as positive associated genes (Since all gene expression values and most of the t-statistic values were positive, we only focused on positive associations indicating brain regions with increased GMV showing higher gene expression). This procedure was performed separately for each dataset. The final gene sets were defined as the overlap between the two datasets (interaction).
MDD risk genes analysis
Although hundreds of MDD risk genes have been reported, only 12 highly reliable MDD risk genes defined as the overlap reported in both situ hybridization data and the 10,027 background genes of AHBA data in a previous study were analyzed (Li et al., 2021). The 12 MDD risk genes include ADRA2A, CHRM2, CNR1, HTR5A, CRH, CUX2, GAD2, HTR1A, MAOA, PDE1A, SST and TAC1. Then, we calculated a matrix of 1285 regions × 12 gene expressions. Based on this matrix, cross-sample non-parametric Spearman rank was performed to explore their relationships with GMV alterations. Moreover, the number of comparisons (n = 12) was further corrected with a significance threshold of p < 4.16 × 10− 3 (0.05/12, Bonferroni correction).
ECT related genes analysis
ECT related genes were determined based on previous studies (Nishiguchi et al., 2015; Benson-Martin et al., 2016; Minelli et al., 2016; Pinna et al., 2018; Moschny et al., 2020). The overlap genes between reported ECT genes and the identified 10,027 genes from AHBA dataset were determined and 22 ECT related genes (BDNF, DRD2, COMT, NET1, HTR2A, RGS4, APOE, ACE, VEGFA, VEGFB, VEGFC, RNF175, RNF213, TBC1D14, WSCD1, E2F1, GNB3, SIRT1, KLF4, SOX2, P2RX7, and GRIK4) were finally obtained. Then, we calculated a matrix of 1285 regions × 22 gene expressions. Based on this matrix, cross-sample non-parametric Spearman rank test was performed to explore their relationships with GMV alterations. Moreover, the number of comparisons (n = 22) was used for multiple comparisons correction with significant level at p < 0.05 (a significance threshold of p < 2.27 × 10− 3 = 0.05/22, Bonferroni correction).
Functional enrichment analyses
To understand the biological processes, molecular functions and cellular components of the identified genes, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) were performed for the positively associated genes (Zhou et al., 2019). The top 5% and top 20 items most associated with these genes were shown.
Assign genes to cell types
A recent single-cell study using postmortem human cortical samples assigned genes to different cell types and seven canonical neuron classes including microglia, endothelial cells, oligodendrocyte precursors, oligodendrocytes, astrocytes, excitatory and inhibitory neurons were defined (Seidlitz et al., 2020). Considering cellular diversity in the brain, we assigned positively associated genes to the seven canonical cell classes by overlapping the gene set of each cell type with the positively associated genes.
Results
GMV alterations induced by ECT in MDD patients
A wide range of brain regions showed increased GMV in MDD patients after ECT in both datasets (Fig. 2). GMV of bilateral hippocampus were significantly increased after correction (Gaussian random filed: GRF, a voxel level p < 0.001, and a cluster level p < 0.05) for dataset 1 and maximum statistical difference in bilateral hippocampus was also found in dataset 2.
Gray matter volume (GMV) changes in MDD patients after ECT. (A-B). Paired t-tests were performed to identify GMV changes in MDD patients before and after ECT in dataset 1 and dataset 2 and the statistical t-maps were shown, respectively. (C) The significantly increased GMV of bilateral hippocampus in dataset 1 was found in MDD patients after ECT compared to MDD patients before ECT after GRF correction
Cortical gene expressions related to GMV alterations
We found that the PLS1 weighted gene expression map was spatially correlated with the t-maps in both datasets (Fig. 3A and B). We found that 979 genes showed significantly positive association with GMV alterations in MDD patients after ECT for dataset 1 and 1592 genes for dataset 2. We identified 726 overlapped genes found in both dataset 1 and dataset 2. The top 5 and bottom 5 genes showing largest positive or negative correlations in dataset 1 and dataset 2 were shown ranked as the normalized weights of PLS1 (right panel in Fig. 3A and B).
Correlation between gene expression and gray matter volume (GMV) differences. (A-B) Scatterplots of regional PLS1 scores and regional differences in GMV for dataset 1 (A) and dataset 2 (B) were shown. The top and bottom 5 genes showing highest and lowest PLS1 loadings were shown for dataset 1 (A) and dataset 2 (B)
MDD and ECT related risk genes analysis
For the 12 selected MDD risk genes, expressions of CNR1, HTR1A, MAOA, PDE1A, and SST showed significantly positive associations with GMV alterations indicating that brain samples with higher gene expression had larger GMV increase in MDD patients after ECT (left panel in Table 2). For the 22 selected ECT related genes, expressions of BDNF, DRD2, APOE, P2RX7, and TBC1D14 showed significantly positive associations with GMV alterations (right panel in Table 2).
Enrichment analysis for GMV associated genes
The biological processes, cellular components and molecular functions for these positively associated genes were determined using GO and KEGG analyses. These genes are mainly related to synaptic signaling, cell junction organization, axon, presynapse, calcium and so on (Fig. 4).
Cell types of overlapped genes
To determine cell types of the ECT associated genes, all the identified overlapping genes we assigned to different cell types. We found that 36 of positively associated genes were mainly expressed in astrocytes, 18 in endothelial cells, 73 in excitatory neurons, 63 in inhibitory neurons, 3 in microglia, 10 in oligodendrocytes and 17 in oligodendrocyte precursors (Fig. 5).
Discussion
Although previous studies have reported genetic factors or molecular basis associated with ECT in MDD patients, the relationship between macroscopic neuroimaging changes and transcriptomic expression level remains largely unknown. In the current study, using gene expression data obtained from AHBA and two independent structural MRI datasets of MDD before and after ECT, we performed transcription-neuroimaging association analysis to explore how gene expressions accounted for GMV alterations. We found that expression levels of 726 genes showed positive correlations with GMV alterations in MDD after ECT. These genes were mainly enriched for synaptic signaling and cell junction and mostly related to excitatory and inhibitory neurons. These findings provide new evidence for the molecular basis of ECT in MDD.
We identified significantly increased GMV of bilateral hippocampus after ECT in MDD patients, which was consistent with previous findings found in human or animal models. Many previous human studies and meta-analyses reported robust structural plasticity of hippocampus following ECT (Gbyl & Videbech, 2018; Gryglewski et al., 2019; Gbyl et al., 2021). In addition, the previous animal models studies also reported increased microglia (Sepulveda-Rodriguez et al., 2019), neurogenesis and synapse formation (Chen et al., 2009), angiogenesis and proliferation of endothelial cells (Segi-Nishida et al., 2008) and gliogenesis (Jansson et al., 2009) in hippocampus following electroconvulsive shock (ECS). One previous study showed that hippocampal volume increase following ECT is an independent neurobiological effect unrelated to brain-derived neurotrophic factor (BDNF) (Bouckaert et al., 2016), and the other study showed that it is not related to BDNF but is mediated by rs699947 in the premotor region of vascular endothelial growth factor (VEGF) (Van Den Bossche et al., 2019). One possible explanation is that the expression of VEGF could lead to angiogenesis and neurogenesis, thus leading to increase in hippocampal volume.
In addition to hippocampus, a wide range of other brain regions also showed increased GMV in MDD after ECT. By assessing spatial similarity between GMV changes and gene expression levels in AHBA brains, we identified 726 positively associated genes. The enrichment analyses found that these positively associated genes are mainly involved in synaptic signaling pathway. This result is consistent with a previous study which reported that the associated genes with morphometric similarity networks (MSN) changes in MDD also primarily participate in synaptic signaling (Li et al., 2021). One possible explanation is that this pathway influences synaptic maturation and stability in depression (Duman & Aghajanian, 2012), whereas ECS (Chen et al., 2018), even a single administration (Chen et al., 2020), could significantly increase number of synapses. A previous study also demonstrated that ECS causes transient down-regulation of key molecules to stabilize synaptic structure and to prevent Ca2 + influx as well as a simultaneous increase in neurotrophic factors providing a short time window of increased structural synaptic plasticity (Nordgren et al., 2013). Our results together with previous findings offered a better understanding of biological mechanisms underlying structural plasticity induced by ECT in MDD patients and highlight synaptic signaling pathway as potential mechanism of ECT.
Moreover, we found excitatory and inhibitory neurons with the largest proportion of the positively associated genes with ECT-related GMV alterations in MDD patients. The excitatory neurons use glutamate as the main neurotransmitter to propagate excitatory (Spruston, 2008) while inhibitory neurons mainly use GABA as the main inhibitory neurotransmitter for inhibitory control (Fino et al., 2013). These two neuron types were found to be related to dysregulation of gene expression in MDD (Li et al., 2021) and were considered as potential therapy targets for MDD (Northoff & Sibille, 2014). Recently, a single-nucleus transcriptomic study also reported greatest dysregulation occurred in deep layer excitatory neurons in prefrontal cortex in MDD patients (Nagy et al., 2020). Although our results about upregulation of gene expression related to excitatory and inhibitory neurons are not direct evidence, they linked GMV alterations induced by ECT in MDD patients to cell type-specific transcriptional signatures.
As for single gene, the PEA15 was the strongest positively correlated gene in both datasets according to the ranked PLS1 loadings indicating that it might be more likely to contribute to GMV increase induced by ECT. The PEA15 is a small protein that regulates multiple cellular functions through its interaction with various signaling molecules (Danziger et al., 1995) and has been demonstrated to modulate signaling pathways for cell proliferation (Renault et al., 2003). Many previous studies reported that PEA15 could promote cancer cell survival (Tang et al., 2019) as well as neuronal survival under conditions of reduced glucose availability (Huang et al., 2018). Moreover, differential PEA15 phosphorylations may distinctly regulate proliferative and/or apoptotic signal in astrocytes following status epilepticus (Park & Kang, 2018). Thus, the higher expression level of PEA15 may lead to more cell survival and cell proliferation resulting in increased GMV in the brain.
Among MDD risk genes, HTR1A was the strongest positively correlated gene and showed opposite associations with alterations of MSN in MDD patients (Li et al., 2021). This seemingly contradictory pattern suggested that gene expressions associated with disease progression of MDD could also be modulated by ECT. Indeed, many previous genetic studies have proven that HTR1A gene was involved in the pathogenesis of MDD and behavioral response to antidepressant treatment (Wang et al., 2018; Basu et al., 2019). Moreover, HTR1A is also suggested to be a postsynaptic heteroreceptor that mediates responses to released 5-HT in several brain regions including hippocampus (Samuels et al., 2015). Thus, it is explainable that we identified positive association between gene expression level of HTR1A and GMV alterations induced by ECT.
Similarly, DRD2 was the strongest positively correlated gene among all ECT related genes. DRD2 is one of dopamine receptors and is essential for signaling in the brain for multiple brain functions (Stojanovic et al., 2017). Previous studies reported that dopaminergic transmission is diminished in MDD (Dunlop & Nemeroff, 2007; Admon et al., 2017; Pecina et al., 2017), and ECT could increase dopaminergic activity particularly through induction of dopamine release and sensitivity of dopamine receptors (Kobayashi et al., 2017; Yang et al., 2020). Two previous studies explored the effect of the DRD2 on ECT efficacy and both reported a similar heterozygotic trend for remission (Huuhka et al., 2008; Bousman et al., 2015).
Despite meaningful findings, several methodological issues should be mentioned in the current study. First of all, the AHBA data were obtained from six post-mortem individuals without psychiatric diagnoses whereas the neuroimaging data were obtained from MDD patients before and after ECT. It is impossible to avoid individual’s variation of gene expressions. Future transcription-neuroimaging association studies are needed to verify our results by using gene expression and neuroimaging data obtained from the same individuals. Second, the bias of hemisphere selection is unavoidable since only gene expressions from left hemisphere rather than the whole brain in all six brains were used. Third, no significant GMV alterations were identified after GRF corrections in dataset 2, which might be resulted from the relatively small sample size. Finally, our study is exploratory, thus, more cautions are needed to explain our results.
Conclusions
In conclusion, this exploratory study linked the structural plasticity to gene expression levels using transcription-neuroimaging association analysis and identified 726 genes positively associated with GMV alterations induced by ECT in MDD. These genes were mainly associated with synaptic signaling, cell junction and calcium signaling and were primarily belonged to excitatory and inhibitory neurons. Together, our findings offered a better understanding of biological mechanisms underlying structural plasticity induced by ECT in the MDD.
Data availability
T1 images of MDD patients were scanned at University of Science and Technology of China (Dataset 1) and Anhui Medical University (Dataset 2). All data is available upon reasonable request to the corresponding authors. GMV images were calculated using the standard pipeline at DPABI (http://rfmri.org/dpabi). The gene expression analysis was performed using AHBA processing pipeline (https://github.com/BMHLab/AHBAprocessing). The probe-to-gene annotations were obtained by the Re-annotator toolkit (https://sourceforge.net/projects/reannotator/). Functional enrichments were analyzed using Metascape (https://metascape.org/gp/index.html#/main/step1). The PLS analysis was performed using codes released by Kirstie Jane (https://github.com/KirstieJane/NSPN_WhitakerVertes_PNAS2016/blob/master/SCRIPTS/).
References
Abdi, H., & Williams, L. J. (2013). Partial least squares methods: Partial least squares correlation and partial least square regression. Methods in Molecular Biology, 930, 549–579.
Admon, R., Kaiser, R. H., Dillon, D. G., Beltzer, M., Goer, F., Olson, D. P., Vitaliano, G., & Pizzagalli, D. A. (2017). Dopaminergic Enhancement of Striatal Response to reward in Major Depression. American Journal of Psychiatry, 174, 378–386.
Anderson, K. M., Collins, M. A., Kong, R., Fang, K., Li, J., He, T., Chekroud, A. M., Yeo, B. T. T., & Holmes, A. J. (2020). Convergent molecular, cellular, and cortical neuroimaging signatures of major depressive disorder. Proceedings of the National Academy of Sciences of the United States of America, 117, 25138–25149.
Arnatkeviciute, A., Fulcher, B. D., & Fornito, A. (2019). A practical guide to linking brain-wide gene expression and neuroimaging data. Neuroimage, 189, 353–367.
Basu, A., Chadda, R. K., Sood, M., Kaur, H., & Kukreti, R. (2019). A preliminary association study between serotonin transporter (5-HTTLPR), receptor polymorphisms (5-HTR1A, 5-HTR2A) and depression symptom-clusters in a north Indian population suffering from Major Depressive Disorder (MDD). Asian J Psychiatr, 43, 184–188.
Benson-Martin, J. J., Stein, D. J., Baldwin, D. S., & Domschke, K. (2016). Genetic mechanisms of electroconvulsive therapy response in depression. Hum Psychopharmacol, 31, 247–251.
Bouckaert, F., Dols, A., Emsell, L., De Winter, F. L., Vansteelandt, K., Claes, L., Sunaert, S., Stek, M., Sienaert, P., & Vandenbulcke, M. (2016). Relationship between hippocampal volume, serum BDNF, and Depression Severity following electroconvulsive therapy in late-life depression. Neuropsychopharmacology : Official Publication of the American College of Neuropsychopharmacology, 41, 2741–2748.
Bousman, C. A., Katalinic, N., Martin, D. M., Smith, D. J., Ingram, A., Dowling, N., Ng, C., & Loo, C. K. (2015). Effects of COMT, DRD2, BDNF, and APOE Genotypic Variation on Treatment Efficacy and Cognitive Side effects of Electroconvulsive Therapy. The Journal of Ect, 31, 129–135.
Cao, B., Luo, Q., Fu, Y., Du, L., Qiu, T., Yang, X., Chen, X., Chen, Q., Soares, J. C., Cho, R. Y., Zhang, X. Y., & Qiu, H. (2018). Predicting individual responses to the electroconvulsive therapy with hippocampal subfield volumes in major depression disorder. Scientific Reports, 8, 5434.
Chen, F., Madsen, T. M., Wegener, G., & Nyengaard, J. R. (2009). Repeated electroconvulsive seizures increase the total number of synapses in adult male rat hippocampus. European Neuropsychopharmacology, 19, 329–338.
Chen, F., Ardalan, M., Elfving, B., Wegener, G., Madsen, T. M., & Nyengaard, J. R. (2018). Mitochondria are critical for BDNF-Mediated synaptic and vascular plasticity of Hippocampus following repeated Electroconvulsive seizures. International Journal of Neuropsychopharmacology, 21, 291–304.
Chen, F., Danladi, J., Wegener, G., Madsen, T. M., & Nyengaard, J. R. (2020). Sustained ultrastructural changes in rat hippocampal formation after repeated Electroconvulsive seizures. International Journal of Neuropsychopharmacology, 23, 446–458.
Chen, K., Yang, J., Li, F., Chen, J., Chen, M., Shao, H., He, C., Cai, D., Zhang, X., Wang, L., Luo, Y., Cheng, B., & Wang, J. (2024). Molecular basis underlying default mode network functional abnormalities in postpartum depression with and without anxiety. Human Brain Mapping, 45, e26657.
Dannlowski, U., Domschke, K., Birosova, E., Lawford, B., Young, R., Voisey, J., Morris, C. P., Suslow, T., Konrad, C., Kugel, H., Ohrmann, P., Bauer, J., Schoning, S., Zavorotnyy, M., Diemer, J., Arolt, V., Baune, B. T., & Zwanzger, P. (2013). Dopamine D(3) receptor gene variation: Impact on electroconvulsive therapy response and ventral striatum responsiveness in depression. International Journal of Neuropsychopharmacology, 16, 1443–1459.
Danziger, N., Yokoyama, M., Jay, T., Cordier, J., Glowinski, J., & Chneiweiss, H. (1995). Cellular expression, developmental regulation, and phylogenic conservation of PEA-15, the astrocytic major phosphoprotein and protein kinase C substrate. Journal of Neurochemistry, 64, 1016–1025.
Duman, R. S., & Aghajanian, G. K. (2012). Synaptic dysfunction in depression: Potential therapeutic targets. Science, 338, 68–72.
Dunlop, B. W., & Nemeroff, C. B. (2007). The role of dopamine in the pathophysiology of depression. Archives of General Psychiatry, 64, 327–337.
Enneking, V., Leehr, E. J., Dannlowski, U., & Redlich, R. (2020). Brain structural effects of treatments for depression and biomarkers of response: A systematic review of neuroimaging studies. Psychological Medicine, 50, 187–209.
Fino, E., Packer, A. M., & Yuste, R. (2013). The logic of inhibitory connectivity in the neocortex. The Neuroscientist : A Review Journal Bringing Neurobiology, Neurology and Psychiatry, 19, 228–237.
Gbyl, K., & Videbech, P. (2018). Electroconvulsive therapy increases brain volume in major depression: A systematic review and meta-analysis. Acta Psychiatrica Scand, 138, 180–195.
Gbyl, K., Rostrup, E., Raghava, J. M., Andersen, C., Rosenberg, R., Larsson, H. B. W., & Videbech, P. (2021). Volume of hippocampal subregions and clinical improvement following electroconvulsive therapy in patients with depression. Progress in Neuropsychopharmacology and Biological Psychiatry, 104, 110048.
Gryglewski, G., Baldinger-Melich, P., Seiger, R., Godbersen, G. M., Michenthaler, P., Klobl, M., Spurny, B., Kautzky, A., Vanicek, T., Kasper, S., Frey, R., & Lanzenberger, R. (2019). Structural changes in amygdala nuclei, hippocampal subfields and cortical thickness following electroconvulsive therapy in treatment-resistant depression: Longitudinal analysis. British Journal of Psychiatry, 214, 159–167.
Huang, Q., Voloudakis, G., Ren, Y., Yoon, Y., Zhang, E., Kajiwara, Y., Shao, Z., Xuan, Z., Lebedev, D., Georgakopoulos, A., & Robakis, N. K. (2018). Presenilin1/gamma-secretase protects neurons from glucose deprivation-induced death by regulating miR-212 and PEA15. The Faseb Journal, 32, 243–253.
Huuhka, K., Anttila, S., Huuhka, M., Hietala, J., Huhtala, H., Mononen, N., Lehtimaki, T., & Leinonen, E. (2008). Dopamine 2 receptor C957T and catechol-o-methyltransferase Val158Met polymorphisms are associated with treatment response in electroconvulsive therapy. Neuroscience Letters, 448, 79–83.
Jansson, L., Wennstrom, M., Johanson, A., & Tingstrom, A. (2009). Glial cell activation in response to electroconvulsive seizures. Progress in Neuropsychopharmacology and Biological Psychiatry, 33, 1119–1128.
Ji, Y., Zhang, X., Wang, Z., Qin, W., Liu, H., Xue, K., Tang, J., Xu, Q., Zhu, D., Liu, F., & Yu, C. (2021). Genes associated with gray matter volume alterations in schizophrenia. Neuroimage, 225, 117526.
Jiang, R., Abbott, C. C., Jiang, T., Du, Y., Espinoza, R., Narr, K. L., Wade, B., Yu, Q., Song, M., Lin, D., Chen, J., Jones, T., Argyelan, M., Petrides, G., Sui, J., & Calhoun, V. D. (2018). SMRI biomarkers predict Electroconvulsive Treatment outcomes: Accuracy with Independent Data sets. Neuropsychopharmacology : Official Publication of the American College of Neuropsychopharmacology, 43, 1078–1087.
Joshi, S. H., Espinoza, R. T., Pirnia, T., Shi, J., Wang, Y., Ayers, B., Leaver, A., Woods, R. P., & Narr, K. L. (2016). Structural plasticity of the Hippocampus and Amygdala Induced by Electroconvulsive Therapy in Major Depression. Biological Psychiatry, 79, 282–292.
Kobayashi, K., Imoto, Y., Yamamoto, F., Kawasaki, M., Ueno, M., Segi-Nishida, E., & Suzuki, H. (2017). Rapid and lasting enhancement of dopaminergic modulation at the hippocampal mossy fiber synapse by electroconvulsive treatment. Journal of Neurophysiology, 117, 284–289.
Leaver, A. M., Vasavada, M., Kubicki, A., Wade, B., Loureiro, J., Hellemann, G., Joshi, S. H., Woods, R. P., Espinoza, R., & Narr, K. L. (2020). Hippocampal subregions and networks linked with antidepressant response to electroconvulsive therapy. Molecular psychiatry.
Li, J., Seidlitz, J., Suckling, J., Fan, F., Ji, G. J., Meng, Y., Yang, S., Wang, K., Qiu, J., Chen, H., & Liao, W. (2021). Cortical structural differences in major depressive disorder correlate with cell type-specific transcriptional signatures. Nature Communications, 12, 1647.
Li, Y., Yu, X., Ma, Y., Su, J., Li, Y., Zhu, S., Bai, T., Wei, Q., Becker, B., Ding, Z., Wang, K., Tian, Y., & Wang, J. (2023). Neural signatures of default mode network in major depression disorder after electroconvulsive therapy. Cerebral Cortex, 33, 3840–3852.
Liu, F., Tian, H., Li, J., Li, S., & Zhuo, C. (2019). Altered voxel-wise gray matter structural brain networks in schizophrenia: Association with brain genetic expression pattern. Brain Imaging Behav, 13, 493–502.
Minelli, A., Congiu, C., Ventriglia, M., Bortolomasi, M., Bonvicini, C., Abate, M., Sartori, R., Gainelli, G., & Gennarelli, M. (2016). Influence of GRIK4 genetic variants on the electroconvulsive therapy response. Neuroscience Letters, 626, 94–98.
Morgan, S. E., Seidlitz, J., Whitaker, K. J., Romero-Garcia, R., Clifton, N. E., Scarpazza, C., van Amelsvoort, T., Marcelis, M., van Os, J., Donohoe, G., Mothersill, D., Corvin, A., Pocklington, A., Raznahan, A., McGuire, P., Vertes, P. E., & Bullmore, E. T. (2019). Cortical patterning of abnormal morphometric similarity in psychosis is associated with brain expression of schizophrenia-related genes. Proc Natl Acad Sci U S A, 116, 9604–9609.
Moschny, N., Zindler, T., Jahn, K., Dorda, M., Davenport, C. F., Wiehlmann, L., Maier, H. B., Eberle, F., Bleich, S., Neyazi, A., & Frieling, H. (2020). Novel candidate genes for ECT response prediction-a pilot study analyzing the DNA methylome of depressed patients receiving electroconvulsive therapy. Clin Epigenetics, 12, 114.
Nagy, C., Maitra, M., Tanti, A., Suderman, M., Theroux, J. F., Davoli, M. A., Perlman, K., Yerko, V., Wang, Y. C., Tripathy, S. J., Pavlidis, P., Mechawar, N., Ragoussis, J., & Turecki, G. (2020). Single-nucleus transcriptomics of the prefrontal cortex in major depressive disorder implicates oligodendrocyte precursor cells and excitatory neurons. Nature Neuroscience, 23, 771–781.
Nishiguchi, M., Kikuyama, H., Kanazawa, T., Tsutsumi, A., Kaneko, T., Uenishi, H., Kawabata, Y., Kawashige, S., Koh, J., & Yoneda, H. (2015). Increases in iPS transcription factor (Oct4, Sox2, c-Myc, and Klf4) gene expression after modified electroconvulsive therapy. Psychiatry Investig, 12, 532–537.
Nordgren, M., Karlsson, T., Svensson, M., Koczy, J., Josephson, A., Olson, L., Tingstrom, A., & Brene, S. (2013). Orchestrated regulation of Nogo receptors, LOTUS, AMPA receptors and BDNF in an ECT model suggests opening and closure of a window of synaptic plasticity. PLoS One, 8, e78778.
Northoff, G., & Sibille, E. (2014). Why are cortical GABA neurons relevant to internal focus in depression? A cross-level model linking cellular, biochemical and neural network findings. Molecular Psychiatry, 19, 966–977.
Ota, M., Noda, T., Sato, N., Okazaki, M., Ishikawa, M., Hattori, K., Hori, H., Sasayama, D., Teraishi, T., Sone, D., & Kunugi, H. (2015). Effect of electroconvulsive therapy on gray matter volume in major depressive disorder. Journal of Affective Disorders, 186, 186–191.
Pang, Y., Wei, Q., Zhao, S., Li, N., Li, Z., Lu, F., Pang, J., Zhang, R., Wang, K., Chu, C., Tian, Y., & Wang, J. (2022). Enhanced default mode network functional connectivity links with electroconvulsive therapy response in major depressive disorder. Journal of Affective Disorders, 306, 47–54.
Park, J. Y., & Kang, T. C. (2018). The differential roles of PEA15 phosphorylations in reactive astrogliosis and astroglial apoptosis following status epilepticus. Neuroscience Research, 137, 11–22.
Pecina, M., Sikora, M., Avery, E. T., Heffernan, J., Pecina, S., Mickey, B. J., & Zubieta, J. K. (2017). Striatal dopamine D2/3 receptor-mediated neurotransmission in major depression: Implications for anhedonia, anxiety and treatment response. European Neuropsychopharmacology, 27, 977–986.
Pinna, M., Manchia, M., Oppo, R., Scano, F., Pillai, G., Loche, A. P., Salis, P., & Minnai, G. P. (2018). Clinical and biological predictors of response to electroconvulsive therapy (ECT): A review. Neuroscience Letters, 669, 32–42.
Polyakova, M., Schroeter, M. L., Elzinga, B. M., Holiga, S., Schoenknecht, P., de Kloet, E. R., & Molendijk, M. L. (2015). Brain-derived neurotrophic factor and antidepressive effect of Electroconvulsive Therapy: Systematic review and Meta-analyses of the preclinical and clinical literature. PLoS One, 10, e0141564.
Redlich, R., Opel, N., Grotegerd, D., Dohm, K., Zaremba, D., Burger, C., Munker, S., Muhlmann, L., Wahl, P., Heindel, W., Arolt, V., Alferink, J., Zwanzger, P., Zavorotnyy, M., Kugel, H., & Dannlowski, U. (2016). Prediction of Individual Response to Electroconvulsive Therapy via Machine Learning on Structural Magnetic Resonance Imaging Data. JAMA Psychiatry, 73, 557–564.
Renault, F., Formstecher, E., Callebaut, I., Junier, M. P., & Chneiweiss, H. (2003). The multifunctional protein PEA-15 is involved in the control of apoptosis and cell cycle in astrocytes. Biochemical Pharmacology, 66, 1581–1588.
Samuels, B. A., Anacker, C., Hu, A., Levinstein, M. R., Pickenhagen, A., Tsetsenis, T., Madronal, N., Donaldson, Z. R., Drew, L. J., Dranovsky, A., Gross, C. T., Tanaka, K. F., & Hen, R. (2015). 5-HT1A receptors on mature dentate gyrus granule cells are critical for the antidepressant response. Nature Neuroscience, 18, 1606–1616.
Segi-Nishida, E., Warner-Schmidt, J. L., & Duman, R. S. (2008). Electroconvulsive seizure and VEGF increase the proliferation of neural stem-like cells in rat hippocampus. Proc Natl Acad Sci U S A, 105, 11352–11357.
Seidlitz, J., Nadig, A., Liu, S., Bethlehem, R. A. I., Vertes, P. E., Morgan, S. E., Vasa, F., Romero-Garcia, R., Lalonde, F. M., Clasen, L. S., Blumenthal, J. D., Paquola, C., Bernhardt, B., Wagstyl, K., Polioudakis, D., de la Torre-Ubieta, L., Geschwind, D. H., Han, J. C., Lee, N. R., Murphy, D. G., Bullmore, E. T., & Raznahan, A. (2020). Author correction: Transcriptomic and cellular decoding of regional brain vulnerability to neurogenetic disorders. Nature Communications, 11, 5936.
Sepulveda-Rodriguez, A., Li, P., Khan, T., Ma, J. D., Carlone, C. A., Bozzelli, P. L., Conant, K. E., Forcelli, P. A., & Vicini, S. (2019). Electroconvulsive Shock Enhances Responsive Motility and Purinergic Currents in Microglia in the Mouse Hippocampus. eNeuro 6.
Singh, A., & Kar, S. K. (2017). How electroconvulsive therapy works? Understanding the neurobiological mechanisms. Clin Psychopharmacol Neurosci, 15, 210–221.
Soda, T., McLoughlin, D. M., Clark, S. R., Oltedal, L., Kessler, U., Haavik, J., Bousman, C., Smith, D. J., Bioque, M., Clements, C. C., Loo, C., Vila-Rodriguez, F., Minelli, A., Mickey, B. J., Milev, R., Docherty, A. R., Langan Martin, J., Achtyes, E. D., Arolt, V., Redlich, R., Dannlowski, U., Cardoner, N., Clare, E., Craddock, N., Di Florio, A., Dmitrzak-Weglarz, M., Forty, L., Gordon-Smith, K., Husain, M., Ingram, W. M., Jones, L., Jones, I., Juruena, M., Kirov, G., Landen, M., Muller, D. J., Nordenskold, A., Palsson, E., Paul, M., Permoda, A., Pliszka, B., Rea, J., Schubert, K. O., Sonnen, J. A., Soria, V., Stageman, W., Takamiya, A., Urretavizacaya, M., Watson, S., Zavorotny, M., Young, A. H., Vieta, E., Rybakowski, J. K., Gennarelli, M., Zandi, P. P., Sullivan, P. F., & Baune, B. T. (2020). International Consortium on the Genetics of Electroconvulsive Therapy and severe depressive disorders (Gen-ECT-ic). Eur Arch Psychiatry Clin Neurosci, 270, 921–932.
Spruston, N. (2008). Pyramidal neurons: Dendritic structure and synaptic integration. Nature Reviews Neuroscience, 9, 206–221.
Stojanovic, T., Orlova, M., Sialana, F. J., Hoger, H., Stuchlik, S., Milenkovic, I., Aradska, J., & Lubec, G. (2017). Validation of dopamine receptor DRD1 and DRD2 antibodies using receptor deficient mice. Amino Acids, 49, 1101–1109.
Takamiya, A., Chung, J. K., Liang, K. C., Graff-Guerrero, A., Mimura, M., & Kishimoto, T. (2018). Effect of electroconvulsive therapy on hippocampal and amygdala volumes: Systematic review and meta-analysis. British Journal of Psychiatry, 212, 19–26.
Tang, B., Liang, W., Liao, Y., Li, Z., Wang, Y., & Yan, C. (2019). PEA15 promotes liver metastasis of colorectal cancer by upregulating the ERK/MAPK signaling pathway. Oncology Reports, 41, 43–56.
Tang, Z., Zhang, S., Guo, D., & Wang, H. (2020). Association between COMT gene Val108/158Met and antidepressive treatment response: A meta-analysis. Gene, 734, 144333.
Van Den Bossche, M. J. A., Emsell, L., Dols, A., Vansteelandt, K., De Winter, F. L., Van den Stock, J., Sienaert, P., Stek, M. L., Bouckaert, F., & Vandenbulcke, M. (2019). Hippocampal volume change following ECT is mediated by rs699947 in the promotor region of VEGF. Transl Psychiatry, 9, 191.
van Diermen, L., van den Ameele, S., Kamperman, A. M., Sabbe, B. C. G., Vermeulen, T., Schrijvers, D., & Birkenhager, T. K. (2018). Prediction of electroconvulsive therapy response and remission in major depression: meta-analysis. British Journal of Psychiatry, 212, 71–80.
Wang, J., Wei, Q., Bai, T., Zhou, X., Sun, H., Becker, B., Tian, Y., Wang, K., & Kendrick, K. (2017). Electroconvulsive therapy selectively enhanced feedforward connectivity from fusiform face area to amygdala in major depressive disorder. Social Cognitive and Affective Neuroscience, 12, 1983–1992.
Wang, P., Lv, Q., Mao, Y., Zhang, C., Bao, C., Sun, H., Chen, H., Yi, Z., Cai, W., & Fang, Y. (2018). HTR1A/1B DNA methylation may predict escitalopram treatment response in depressed Chinese Han patients. Journal of Affective Disorders, 228, 222–228.
Wang, L., Wei, Q., Wang, C., Xu, J., Wang, K., Tian, Y., & Wang, J. (2020). Altered functional connectivity patterns of insular subregions in major depressive disorder after electroconvulsive therapy. Brain Imaging Behav, 14, 753–761.
Xu, J., Wang, J., Bai, T., Zhang, X., Li, T., Hu, Q., Li, H., Zhang, L., Wei, Q., Tian, Y., & Wang, K. (2019). Electroconvulsive therapy induces cortical morphological alterations in major depressive disorder revealed with Surface-Based Morphometry Analysis. International Journal of Neural Systems, 29, 1950005.
Xu, J., Wei, Q., Bai, T., Wang, L., Li, X., He, Z., Wu, J., Hu, Q., Yang, X., Wang, C., Tian, Y., Wang, J., & Wang, K. (2020). Electroconvulsive therapy modulates functional interactions between submodules of the emotion regulation network in major depressive disorder. Translational Psychiatry, 10, 271.
Yang, C., Qiu, Y., Qing, Y., Xu, J., Dai, W., Hu, X., & Wu, X. (2020). Synergistic effect of electric stimulation and mesenchymal stem cells against Parkinson’s disease. Aging (Albany NY), 12, 16062–16071.
Yang, J., Chen, K., Zhang, J., Ma, Y., Chen, M., Shao, H., Zhang, X., Fan, D., Wang, Z., Sun, Z., & Wang, J. (2023). Molecular mechanisms underlying human spatial cognitive ability revealed with neurotransmitter and transcriptomic mapping. Cerebral Cortex, 33, 11320–11328.
Yu, X., Chen, K., Ma, Y., Bai, T., Zhu, S., Cai, D., Zhang, X., Wang, K., Tian, Y., & Wang, J. (2024). Molecular basis underlying changes of brain entropy and functional connectivity in major depressive disorders after electroconvulsive therapy. CNS Neuroscience & Therapeutics, 30, e14690.
Zhou, Y., Zhou, B., Pache, L., Chang, M., Khodabakhshi, A. H., Tanaseichuk, O., Benner, C., & Chanda, S. K. (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications, 10, 1523.
Funding
This work was supported by National Natural Science Foundation of China (No. 62006220, 62001462, 32071054, and 82001429), Anhui Provincial Science Fund for Distinguished Young Scholars (1808085J23) and Shenzhen Science and Technology Research Program (No. JCYJ20200109114816594, JCYJ20180507182441903).
Author information
Authors and Affiliations
Contributions
K.W, Y.T and J.X designed the study. Y.T and T.B collected the data. H.S, J.X, X.Z, X.F, K.Z and J.Z analyzed the data. H.S and J.X and wrote the manuscript with help of all authors. All the authors discussed the results and edited the manuscript. All authors approved the final version.
Corresponding authors
Ethics declarations
Ethical approval
All patients provided written informed consent and the study was approved by the local ethics committees of the Anhui Medical University (Approval number: 20140072).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, H., Bai, T., Zhang, X. et al. Molecular mechanisms underlying structural plasticity of electroconvulsive therapy in major depressive disorder. Brain Imaging and Behavior 18, 930–941 (2024). https://doi.org/10.1007/s11682-024-00884-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11682-024-00884-9