Abstract
Purpose
Nitrogen fixation by free-living diazotrophs from the atmosphere is an important pathway for nitrogen input into the soil. However, there is little information regarding soil diazotrophic community composition and diversity under long-term fertilization in rice paddy ecosystems.
Materials and methods
Using the 15N2-tracing method and nifH gene as a molecular marker, we investigated the abundance, structure, and activity of soil diazotrophic community in soil at a 30-year-old filed experimental site treated with four different fertilizer management practices: control (non-fertilization), chemical NPK fertilizers, NPK plus rice straw (NPK+RS), or NPK plus chicken manure (NPK+OM).
Results and discussion
Among all the treatments, the NPK+OM treatment significantly improved the soil nutritional status. Fertilization increased both bacteria and nifH gene abundances, with the highest values (p < 0.05) found in the NPK+OM treatment. The potential nitrogen fixation rate ranged from 14.6 to 118 μg kg−1 day−1, and the highest rates (p < 0.05) were also observed in the NPK+OM treatment. Long-term chemical NPK fertilization decreased the diversity of diazotrophic community, whereas NPK+RS and NPK+OM treatments maintained the diversity of diazotrophic community. Long-term fertilization changed diazotrophic community as compared to non-fertilization, but there were no significant differences among fertilized treatments. Most nifH sequences were closely linked to Alphaproteobacteria, which was dominated by the genera Bradyrhizobium. Relatively higher Cyanobacteria abundances were observed in the unfertilized soil as compared with fertilized soil.
Conclusions
Our results suggest that long-term fertilization increased the abundance of diazotrophs and changed their community structure, and combined use of chicken manure and chemical NPK fertilizers can significantly improve the activity of diazotrophic community.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Rice is one of the most important staple food feeding more than three billion people (Maclean 2002). Approximately 40% increase in rice yield is expected to meet the ever-expanding population in 2050s (Van Nguyen and Ferrero 2006). In this sense, more chemical or organic fertilizers will be required to satisfy the nutrient requirements for high rice productivity. In paddy soil, microbes are assumed to be adapted to the high nutrient availability, and their community composition and functional gene diversity that affect carbon (C), nitrogen(N), and phosphorus (P) cycles are closely related to rice productivity (Pan et al. 2017; Su et al. 2015). On the other hand, in contrast to upland farmlands, rice paddy soil provides a unique system to gain insights into the microbiology switches between oxic and anoxic conditions in wetland systems (Wegner and Liesack 2015).
Nitrogen is a critical nutrient for terrestrial net primary production. However, plants can not directly access N2, which makes up about 80% of the atmosphere (Santi et al. 2013). Diazotrophs, or N2-fixers, are widely distributed among the archaeal and bacterial taxa (Dixon and Kahn 2004), and contribute to plant available N thorough reducing atmospheric N2 to ammonium in the soil (Reardon et al. 2014). From paddy filed soils, a number of culturable N2-fixing microorganisms have been isolated, such as Azospirillum, Bacillus, Burkholderia, and Herbaspirillum (Boddey et al. 1995; Xie et al. 2003; Okubo et al. 2012). Additionally, many studies have observed an interesting phenomenon that paddy soil can continuously maintain its fertility for a long time under flooded condition (Ladha et al. 1997), which probably because the oxygen-limited environment has the potential to increase nitrogenase activity, therefore enhance nitrogen fixation in soil (Reed et al. 2011; Ferrando and Fernandez Scavino 2015).
The intensive use of chemical or organic fertilizers, which consists of the largest part of human interference in the global nitrogen cycle, raised environmental concerns regarding the increased greenhouse gas emissions and groundwater pollution (Dixon and Kahn 2004). Meanwhile, the abundance and diversity of diazotrophic bacteria may be suppressed by high fertilizer application in many agricultural ecosystems (Fuentes-Ramírez et al. 1999; Reed et al. 2011). Therefore, balancing the fertilizer usage and biological nitrogen fixation will be beneficial to maximizing crop yields and minimizing production costs. To address this issue, it is important to understand how diazotrophic community responds to different fertilization practices use under different agricultural systems (Yeoh et al. 2016). Chinnadurai et al. (2014) found that nifH gene abundance was closely related to soil available N concentration, and significantly higher in long-term organically managed maize soil compared to inorganic nutrient management practices; Reed et al. (2007) found that phosphorous fertilization can stimulate nitrogen fixation rate and increases soil inorganic nitrogen concentrations in a restored prairie. In addition, Berthrong et al. (2014) suggested that nitrogen fertilization rather than elevated CO2 suppressed the diazotrophic community diversity and abundance in a pine forest. So far, it is well demonstrated that different management regimes affect nifH gene abundance and community composition in upland agricultural systems (Hsu and Buckley 2009; Mirza et al. 2013; Wang et al. 2016), but less information is available for waterlogged agricultural soils. Paddy ecosystems provide an excellent case to investigate the diazotrophic community abundance and composition under anaerobic/aerobic conditions. However, few studies have attempted to explore the effects of long-term different fertilization practices on diazotrophic abundance and community structure in this unique agricultural ecosystem (Zhao et al. 2016).
In the current study, we selected a 30-year-old fertilization experimental filed with control (non-fertilization), chemical NPK, NPK plus rice straw (NPK+RS) or manure (NPK+OM). We hypothesized that diazotrophic community might be suppressed due to long-term nitrogen fertilization application in rice paddy soil. For this purpose, we have examined the abundance, structure of nitrogen-fixing communities, and their potential nitrogen fixation rate estimate by 15N2 tracing method under long-term different fertilization treatments.
2 Materials and methods
2.1 Study site and soil sampling
The long-term fertilization experimental site was established in 1986, located in Ningxiang County, Hunan Province (112°18′E, 28°07′N). This region has a continental monsoon warm and humid climate. The average elevation of the region is 36 m, with a mean annual precipitation of 1550 mm and mean temperature of 17 °C. Since 1986, the field has been under rice-rice-barley rotation. The early and late season rice was cultivated from May to August and August to October, respectively. The initial physical and chemical properties (0–20 cm depth) in 1986 was as follow: pH 6.85, organic matter 29.4 g kg−1, total N 2.0 g kg−1, total P 0.6 g kg−1, total K 20.6 g kg−1, available N 144 mg kg−1, Olsen P 12.9 mg kg−1, exchangeable K 33 mg kg−1.
Four treatments were established in this experiment: control (no fertilization; CK), chemical NPK fertilizers (NPK), and chemical NPK fertilizers plus rice straw (NPK+RS) or chicken manure (NPK+OM). The straw residue was applied at the rate of 6375 kg ha−1 year−1 with the molar N/P ratio of molar 10.5:1. Chemical NPK fertilizers were applied as urea-N at 300 kg ha−1 year−1, and as superphosphate at P2O5 100 kg ha−1 year−1, and potassium chloride at K2O 140 kg ha−1 year−1, respectively. Chicken manure containing 1.77% N, 0.80% P2O5, and 1.12% K2O at the amount of 5290 kg ha−1 year−1 with the N/P ratio of 3.1:1. Each fertilization treatment received the same levels of nitrogen, phosphorus, potassium from fertilizers. On June 2016, soil samples were collected from three replicate plots of each treatment. A composite sample from each plot was obtained by mixing five random soil cores from the plowing depth (0–20 cm). Each soil sample was divided into three parts, one was freeze-dried for DNA extractions, the second was stored at 4 °C for biological characteristics analyses, and the remaining was air-dried and passed through a 2-mm sieve for chemical analyses.
2.2 Soil analyses
Soil pH was determined using a glass combination electrode with a 1:2.5 soil to water ratio. Soil organic carbon (SOC), total nitrogen (TN), and total sulfur (TS) concentrations were determined by dry combustion using an Elemental Analyzer (Vario Macro, Elementar, Germany). Soil total phosphorus (TP) was measured by the wet acid digestion method combined with colorimetric procedures. Soil Olsen phosphorus (Olsen P) was measured by extracting with 0.5 M NaHCO3 with 1:25 (w/v) and colorimetric analysis (Olsen 1954). Exchangeable potassium was extracted with 1 M CH3COONH4 (pH 7.0) with a 1:10 (w/v) soil to solution ratio for 30 min and measured by atomic absorption spectrometry. Soil NH4 +and NO3 − contents were measured with 2 M KCl extraction, filtering through a 0.45-μm pore size ploy sulfone membrane, and then analysis with a continuous flow analyzer (SAN++; SKALAR, The Netherlands). Substrate-induced respiration (SIR) was measured after adding 0.5 mg glucose g−1 soil saturating levels of labile carbon according to Ge et al. (2016).
2.3 Potential N-fixation rate
Soil nitrogen fixation rate was measured by 15N2-based method modified from Hsu and Buckley (2009) and Keuter et al. (2014). Briefly, fresh soil (5 g dry weight) was placed into 100 mL glass jars, the soil was then flooded so that 2 mm excess water covered the soil. Then milliliters of air in the headspace of each jar was replaced with 15N2 (98% 15N), the soil samples were incubated for 15 days in the dark at 25 °C. Another set of soil samples were incubated with ambient air as controls. Each sample was replicated five times. After incubation, the soil samples were air-dried at room temperature, and then ground to pass a 200-mesh sieve for analyzing 15N enrichment using a Delta V Advantage isotope ratio mass spectrometer (Thermo Finnigan, Germany). The potential 15N fixation rate was calculated by multiplying the difference in enrichment atom percent excess between a labeled sample and the control sample and the concentration (g kg−1 soil) of total nitrogen in soil (Bei et al. 2013).
2.4 Soil DNA extraction
DNA was extracted from 0.50 g of soil using the FastDNA SPIN kit for soil (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s instructions. DNA quality and concentration were determined using a NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), then stored at − 20 °C until amplification. The DNA yields were as follows: CK, 65.0 ± 4.65 μg g−1 soil; NPK+RS, 138.8 ± 52.0 μg g−1 soil; NPK, 99.2 ± 6.73 μg g−1 soil; NPK+OM, 155 ± 10.9 μg g−1 soil.
2.5 PCR, cloning, and sequencing
Diluted DNA (1:10) was used to amplify the nifH gene (~ 360 bp) with the primers PolF (TGCGAYCCSAARGCBGACTC) and PolR (ATSGCCATCATYTCRCCGGA) (Poly et al. 2001). Each reaction mixture contained 2 μL of template DNA, 22.5 μL of 2× PCR Master Mix, 1.5 μL of each primer, 0.2 μL of bovine serum albumin (BSA) and made up to 50 μL with sterile water (ddH2O). The thermal cycle profile consisted of the following: 2 min of denaturation, followed by 35 rounds of cycles at 95 °C for 30 s, 59 °C for 30 s, and 72 °C for 45 s, and then completed by a final extension at 72 °C for 7 min. Aliquots (2 μL) of the amplified products were visualized on ethidium bromide-stained 1.2% agarose gels. The amplified products were purified according to the manufacture’s guide. The purified PCR products were cloned into Escherichia coli plasmids using the kit pGEM-T Easy Vector (Promega, Madison, WI, USA) recommended by the manufacturer. Positive colonies were selected and amplified with primers M13F (GTAAACGACGGCCAG) and M13R (CAGGAAACAGCTATGAC). Forty-two positive clones per sample were randomly selected and sequenced using the Sanger technology (MajorBio Ltd., Shanghai, China).
2.6 NifH gene sequence and analysis
NifH gene sequence analysis was carried out with BioEditor 7.0.9.0 (Hall 1999) to remove the vector sequences. Due to the low resolution of identification for the nifH gene at the DNA level (Lema et al. 2012), the nifH gene was translated into a deduced amino acid sequence using the BioEditor. The deduced amino acid sequences were defined as operation taxonomic units (OTUs) at 98 and 97% similarity using Mothur 1.38.1 (Schloss et al. 2009), the coverage was higher when OTUs defined at the 98% similarity level (Table 1). A representative sequence of each OTU was subjected to BLAST search against the GenBank database; the closest species match for query sequences were included for phylogenetic analysis. Reference sequences from the GenBank database and the respective OTUs (98% similarity) sequences were aligned using the Clustal W program (Thompson et al. 1994). A phylogenetic tree was constructed based on 98% deduced amino acid sequences of nifH gene by the Maximum likelihood method based on a Poisson correction model with the MEGA 5.0 software (Tamura et al. 2011) with 1000 bootstrap replicates.
2.7 Quantification of nifH and bacterial 16S rRNA genes
Quantitative PCR (qPCR) was performed with a real-time PCR detection system (Light Cycle 480; Roche). The nifH gene was quantified using PolF/PolR primers (Poly et al. 2001) and the bacteria quantified based on 16S rRNA gene using the primers 515F (GTGCCAGCMGCCGCGGTAA) and 907R (CCGTCAATTCCTTTGAGTTT) (Biddle et al. 2008). Each sample was prepared in three replicates in a 20 μL volume, containing 10 μL Absolute SYBER Fluorescein Mix (Thermo Scientific, Grand Island, NY), 0.4 μL forward and reverse primer, 1 μL of 1:10 diluted DNA template, and 8.2 μL double ddH2O. The template-free control reactions contained 1 μL of ddH2O instead of DNA. Thermal conditions for nifH gene were set as follows: 5 min at 95 °C, following by 40 cycles of 10 s at 95 °C, 30 s at 59 °C, and 72 °C for 40 s. For 16S rRNA gene, thermal conditions were set as follows: 5 min at 95 °C, following by 45 cycles of 10 s at 95 °C, 45 s at 53 °C, 45 s at 72 °C, and 15 s at 84 °C. Standard curves for qPCR were created using an up to 10-fold dilution series of PCR product containing a fragment with known nifH or 16S rRNA gene copy numbers.
2.8 Nucleotide sequence accession numbers
The nifH gene sequences obtained in this study were deposited in the GenBank under accession numbers KY 311079 to KY 311559.
2.9 Statistical analyses
To test the difference between the fertilization treatments, one-way analysis of variance (ANOVA), followed by Fisher’s least significant difference (LSD) post-hoc tests were performed by using SPSS (version 16.0). NifH gene sequences were subjected to Good’s coverage, rarefaction analysis, Chao1, and Shannon-Weaver diversity analysis using by Mothur 1.38.1 (Schloss et al. 2009). Redundancy (RDA) and Multivariate Regression Tree (MRT) analyses were performed to explore the diazotrophic community composition and identify the most important environmental factors (999 permutations) affecting the dizazotrophic community composition, respectively, based on diversity of nifH gene OTUs using the correlation matrix. Adonis was used to test the difference in diazotrophic community composition between treatments. RDA was performed with Vegan package, MRT was generated using Mvpart and MVPARTwrap packages in R 3.2.5 (Team RC 2016).
3 Results
3.1 Effects of long-term fertilization on soil biochemical properties
The soil properties varied significantly after 30 years under different fertilization regimes (Table 2). Among all the treatments, NPK+OM had significantly higher (p < 0.05) SOC, TN, TS, Olsen P, and exchangeable K concentrations. Specifically, compared with CK, the AP concentration increased approximately 10-fold in the NPK+OM treatment. Moreover, NPK+OM treatment significantly (p < 0.05) increased soil microbial biomass as indicated by SIR. The CK treatment had significantly higher pH than the fertilization treatments. Soil ammonium concentration was significantly higher (p < 0.05) in both NPK and NPK+OM treatments, and the lowest concentration was found in the NPK+RS treatment. No significant differences in nitrate concentrations were detected among treatments.
PCA of the soil biochemical properties showed that 91.6% of total variance was explained by the first two axes, with PC1 and PC2 accounted for 78.0 and 13.6% of the total variance, respectively (Fig. S1, Electronic Supplementary Material). Along the PC1 axis, NPK+OM clearly separated from other treatments. Therefore, the PC1 axis could be a good predictor of the soil nutritional status after different long-term fertilization regimes.
3.2 Effects of long-term fertilization on 16S rRNA gene and nifH abundances
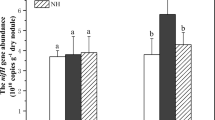
Different fertilization regimes had significant effects on soil bacterial biomass as estimated by the abundances of 16S rRNA gene (Fig. 1). The number of soil bacterial 16S rRNA gene ranged from 3.4 × 1010 to 2.0 × 1011 g−1 dry soil. Compared with CK, The NPK+OM treatment significantly increased the abundance of 16S rRNA gene, whereas no significant differences were found among CK, NPK, and NPK+RS treatments. The nifH gene copy numbers ranged from 2.8 × 108 to 1.7 × 109 g−1 dry soil. Similar to 16S rRNA gene, the highest nifH gene abundance was found in the NPK+OM treatment and the lowest was detected in the CK treatment. Regardless fertilization treatments, the increase of nifH gene copy number was positively related to the 16S rRNA gene (r = 0.84, p < 0.001). Additionally, the soil nifH gene abundance was significantly correlated (p < 0.05) with SOC, TN, AP, and SIR, and soil 16S rRNA gene abundance was significantly related (p < 0.05) with pH, SOC, TN, TS, AP, and SIR (Table 2).
3.3 Effects of long-term fertilization on potential nitrogen fixation rate
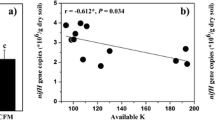
Soil potential nitrogen fixation rate estimated by the 15N2 tracing method ranged from 14.6 to 118 μg kg−1 day−1 (Fig. 2). Significantly higher (p < 0.05) rate was found in the NPK+OM treatment, whereas there were no significant differences in potential nitrogen fixation rates among CK, NPK+RS, and NPK treatments. Additionally, the potential nitrogen fixation rate was significantly (p < 0.05) correlated with 16S rRNA (r = 0.62, p = 0.0381) or nifH gene (r = 0.66, p = 0.0193) abundance.
3.4 Effect of long-term fertilization on nifH gene diversity
A total of 481 clone library sequences were identified at the 98 and 97% protein levels (Table 1), which generated 76 and 60 OTUs, respectively. The CK, NPK+RS, and NPK+OM had very similar observed OTU numbers at both OTU cutoff levels. In addition, the CK treatment had the highest Shannon and Chao1 estimator values, despite of the lowest sequencing effort (Table 1). Among all the treatments, the lowest OTU numbers and diversity estimators were detected in the NPK treatments (p < 0.05) regardless the OTU cutoff levels. Furthermore, the lowest diversity of nifH genes for NPK treatment was also proved by the rarefaction curve, which almost approached an asymptote with sampling effect at the 97% similarity level (Fig. S2, Electronic Supplementary Material).
Phylogenetic tree was generated to classify the nifH gene clusters based on the known related N-fixing genotypes. Phylogenetic analysis showed that the 481 translated nifH amino acids sequences derived from different long-term fertilization treatments were clustered in various taxonomic groups at 98% similarity level (Fig. 3). Alphaproteobacteria (14 OTUs; 271 sequences) were the most dominated nifH sequences in four treatments, followed by Deltaprobacteria (6 OTUs; 34 sequences), Betaproteobacteria (3 OTUs, 24 sequences), and Cyanobacteria (2 OTUs, 21 sequences), and the rest of the sequences belonged to the phylotypes including Firmicutes, Bacteroidia, and Actinobacteria.
Among all the sequences, OTU22, OTU1, and OTU23 were the most abundant OTUs which contained over one third of nifH sequence (Table S1, Electronic Supplementary Material). BLAST results indicated that all these three OTUs were affiliated with Bradyrhizobium species with 97–99% identity. The OTU heat map revealed that the soil in the CK treatment contained less sequences in the OTU22 than soils in other treatments, but the highest sequence numbers in OTU15 and OTU16 (Fig. 4). Moreover, OTU16 was exclusively detected in the CK treatment. OTU15 and OTU16 affiliated with the order Nostocales within the Cyanobacteria phylotypes (100 and 97% amino acid identity, respectively) (Table S1, Electronic Supplementary Material). Among the fertilized treatments, OTU7 contained sequences most abundant NPK treatment as compared with NPK+RS and NPK+OM treatments, OTU7 was closely related to the Methylobacter marinus (AAK97414) class with 93% amino acid identity. The OTU heat map showed that NPK+RS and NPK+OM had relatively less different OTU compositions. However, compared with the NPK+RS treatment, the NPK+OM treatment had contained more OTU19 and OTU25, which closely related to the Azospirillum lipoferum (ABG88868) and Alkaliflexus imshenetskii (WP_026474998), respectively (Fig. 4 and Table S1, Electronic Supplementary Material).
Maximum likelihood phylogenetic trees of translated nifH gene sequences (120 amino acids) at 98% similarity level. Bootstrap values (%) were generated from 1000 replicates, and values of > 50% are shown. The tree was rooted with a nifH protein sequence from archaeon (Methanothermococcus okinawensis). The sequences with known bacteria are indicated by species names and protein accession numbers in GenBank. OTUs are showed by OTU number and the associated with accession number, and only OTUs more than three sequences are present. The number in parentheses represents the number of clones in each OTU and the total number of clones
3.5 Diazotrophic community composition and its influencing factors
In order to explore the relationship between the soil properties and diazortrophic community composition, RDA and MRT were used in our study (Fig. 5a, b). RDA showed that long-term different fertilization treatments changed dizazotrophic community composition. The first two axes of the RDA explained 48.5% of the total diazortrophic community variation. Diazortrophic community in the CK treatment differed from other fertilization treatments (PerMANOVA; F = 2.51, p = 0.013). However, the different fertilization practices had no significantly different effects on diazotrophic community composition (PerMANOVA; F = 1.33, p = 0.17).
Redundancy analysis (a) and multivariate regression tree (b) for the nifH community composition under four different fertilization treatments. The asterisk indicates significant (p < 0.05) correlation with the nifH community composition with 999 permutations tests. The parentheses in the multivariate regression tree represent significant indicator species
The MRT analysis conformed that soil pH is an important factor affecting soil diazotrophic community composition; the whole tree explained about 40% of the variance of nifH community. Soil pH clearly separated CK from other treatments at the first split due to the high abundance of the indicator species of OTU16 (Fig. 5b). In addition, SOC concentration also affected nifH community, and its effect separated from NPK+OM treatment from other fertilized treatments.
4 Discussion
Long-term fertilizer application altered soil nutritional status in rice paddy soils. Among all the fertilization treatments, the NPK+OM showed the maximum effect in improving soil fertilities, indicating that chicken manure is an effective farming practice for maintaining soil fertility when combined with chemical fertilization application. The influence of long-term fertilization on soil fertilities can be linked to the changes in microbial community compositions, due to the importance of microbes in soil ecosystems. Soil nifH gene is one of the most important genetic markers reflecting ecological functionality or human induced disturbances (Mirza et al. 2013; Berthrong et al. 2014; Izquierdo and Klaus 2015). In rice paddy ecosystems, frequent flooding and intensive use of fertilizers affect diazotrophic communities (Wartiainen et al. 2008; Mårtensson et al. 2009; Shu et al. 2012). Most studies focus on nifH community changes as affected by fertilization managements mainly in relatively short periods (a few years or less). Conversely, very little information is available about the long-term responses under different fertilization treatments. In our study, using the molecular marker of nifH gene and the 15N2-tracing method, we reveled that long-term application of different fertilizers changed the abundance, composition, and activity of diazortrophic community.
In contrast to our hypothesis, we observed that no suppressive effects of long-term fertilization on the abundance of nifH gene. Instead, long-term fertilization resulted in increase of nifH gene and 16S rDNA gene abundances. Our results were in line with those which showed that application of straw, chemical fertilizers, or livestock had positive effects on abundances of total bacteria and diazotrophic communities (Hai et al. 2009; Sun et al. 2015a). Additionally, the NPK+OM treatment had the significant highest potential nitrogen fixation as compared to other treatments, which indicated that chicken manure application in combination with chemical NPK stimulate the abundance and activity of diazotrophic communities. The reason might be because organic manures not only improved soil nutrition and carbon availability, but also provided a better living condition for microbial communities (Sun et al. 2015b). More importantly, the organic manure had lowest molar N/P ratio (3.1:1) as compared NPK (13:1) and NPK+RS(10.5:1). The treatments with high N/P ratio may escalate soil phosphorus limitation, and therefore restrict growth of the heterotrophic diazotrophs for nitrogen fixation.
Our phylogenetic analysis showed that Alphaproteobacteria were the main source of nifH gene sequences under different fertilization treatments. More specifically, these sequences were identified as members of order Rhizobiales, and affiliated with the Bradyrhizobium-related species. In agriculture systems, Bradyrhizobium is recognized as signature land use change indicator microorganism under different edaphoclimatic conditions (Zhalnina et al. 2013). The high abundance of Bradyrhizobium species sequences may be because they are as they are oligotrophic bacteria which can survive even under nutrient-deprived and diverse conditions (Yousuf et al. 2014; Piromyou et al. 2015). Moreover, the root of rice can be colonized by Rhizobia which would results in high abundances of Bradyrhizobium species. Indeed, Bradyrhizobium species have been reported that as active nitrogen-fixing bacteria associated with rice (Chaintreuil et al. 2000), switchgrass (Bahulikar et al. 2014), and sorghum (Rodrigues Coelho et al. 2008). Furthermore, free-living Bradyrhizobium species have been shown to fix N2 in soil based on a study using the 15N2-DNA-SIP technology (Buckley et al. 2008). Our findings are consistent with Su et al. (2015), that Bradyrhizobium was the most abundant genera among the bacterial phylogenetic composition in a paddy field without leguminous crop rotation.
Besides the high abundance of Alphaproteobacteria, nifH genes were found in all the treatments. The nifH gene sequences were also found in various taxonomic groups, including Deltaprobacteria, Betaproteobacteria, and Cyanobacteria. Cyanobacteria may contribute significantly to soil nitrogen fixation even though they may not be dominating diazotrophs in the soil (Wartiainen et al. 2008). Rice paddy ecosystems are considered as favorable habitats for Cyanobacteria. Nevertheless, many studies failed to detect their present in the paddy soil (Wartiainen et al. 2008; Shu et al. 2012). Interestingly, OTU15 and OTU16, which were affiliated with the phylum Cyanobacteria, were mostly found in the CK treatment. In the present study, PolF/PolR primers covered most of the known nitrogen-fixing microorganisms (including Cyanobacteria) in various environmental samples or laboratory cultures (Demba Diallo et al. 2004; Diez et al. 2012; Estrella Alcaman et al. 2015). Therefore, these primers can give reliable information on Cyanobacteria present in paddy soils. In the CK treatment, the relatively high abundance of Cyanobacteria might be an indication that autotrophic nitrogen-fixing bacteria were favored more than heterotrophic nitrogen-fixing bacteria due to no nitrogen fertilizer inputs. This supported that the opinion that rice paddy soil can sustain moderate but constant yields without N fertilizers for thousands of years as compared with upland soils (Ladha and Reddy 2003).
Long-term fertilization altered soil nifH community composition in the paddy soils indicated by RDA results and OTU heat map (Figs. 3 and 5).The CK treatment was distinctive from other fertilization treatments. In addition, although no significant differences were found among the fertilization treatments, some genotypes were slightly higher in the NPK treatment than those in the other treatments. Furthermore, compared with the NPK+RS treatment, NPK+OM contained more OTU19 and OTU25 sequences. The changes in nifH gene composition may be related to dynamics of soil biochemical properties with long-term different fertilizer treatments. The MRT results confirmed that the difference in nifH community between CK and other treatment was related with the relatively higher pH values in CK. Soil pH is thought to be the primary factor shaping soil microbial community (Shen et al. 2013; Sun et al. 2015b; Zhalnina et al. 2015). After 33 years of different fertilization treatments, the pH values decreased from 6.47 to 6.07, which was in consistency with the previous studies that long-term fertilization decreased pH due to the nitrification or input of acidifying nitrogen fertilizers. Our results showed that decreased pH triggered by the long-term fertilization treatments could be a good predictor of nifH community composition. Similar results were also observed in other studies (Mirza et al. 2013; Liang et al. 2016; Wang et al. 2017). However, until recently, studies on drivers for diazotrophic community composition are still lack of consensus. Some studies found that soil C, N, and C/N rather than soil pH were the main factors affecting nifH community composition (Shu et al. 2012; Gonzalez Perez et al. 2014). Indeed, the effects of soil physicochemical properties on diazotrophic community composition perhaps cannot be replicated over season or year (Reardon et al. 2014), which suggested that complexity of nifH community composition to the changing environment. Therefore, multi-year investigations are necessary to identify the more detailed changes of diazotrophic communities.
Greater diversities of diazotrophic communities were found in the NPK+OM and NPK+RS treatments among the fertilized treatments, whereas the lowest diversity was observed in the NPK treatment. Interestingly, the CK treatment had greater α-diversity (as estimated by Shannon and Chao 1) and higher OTUs at 98 and 97% cut off levels, respectively, despite of the lowest sequence numbers. Similar to our results, Sun et al. (2015b) demonstrated that long-term application of chemical fertilizers decreased bacterial diversity as compared to the unfertilized and NPK manure, Hui (2012) found that NPK fertilizer also decreased the diversity of nifH community compared to NPK plus manure or manure alone in a black soil region of northeast China. Moreover, in rice paddy filed, Tan et al. (2003) found that chemical N fertilizer decreased the diversity of root-associated nifH communities. Together, our data provide evidence that long-term NPK fertilizer application leads to suppressed nifH gene diversity in rice paddy soil, and that NPK fertilizer plus straw or manure can maintain diversity of the diazotrophic community.
5 Conclusions
In conclusion, our results highlighted the effects of long-term different fertilization practices on the abundance and structure of dizaotrophic communities in rice paddy ecosystems. Chemical NPK fertilization decreased diversity of dizaotrophic community. However, NPK fertilizer combined with organic manure improved not only the diversity of dizaotrophic community, but also their abundance and nitrogen fixation rate. Importantly, the high performance of Cyanobacteria in the unfertilized soil is an evidence that diazotrophic community may change their structure to increase nitrogen fixation in a paddy rice ecosystem.
References
Bahulikar RA, Torres-Jerez I, Worley E, Craven K, Udvardi MK (2014) Diversity of nitrogen-fixing bacteria associated with switchgrass in the native tallgrass prairie of Northern Oklahoma. Appl Environ Microbiol 80:5636–5643
Bei Q, Liu G, Tang H, Cadisch G, Rasche F, Xie Z (2013) Heterotrophic and phototrophic 15N2 fixation and distribution of fixed 15N in a flooded rice–soil system. Soil Biol Biochem 59:25–31
Berthrong ST, Yeager CM, Gallegos-Graves L, Steven B, Eichorst SA, Jackson RB, Kuske CR (2014) Nitrogen fertilization has a stronger effect on soil nitrogen-fixing bacterial communities than elevated atmospheric CO2. Appl Environ Microbiol 80:3103–3112
Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH (2008) Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci U S A 105:10583–10588
Boddey RM, de Oliveira OC, Urquiaga S, Reis VM, de Olivares FL, VLD B, Döbereiner J (1995) Biological nitrogen fixation associated with sugar cane and rice: contributions and prospects for improvement. In: Ladha JK, Peoples MB (eds) Management of biological nitrogen fixation for the development of more productive and sustainable agricultural systems: extended versions of papers presented at the Symposium on Biological Nitrogen Fixation for Sustainable Agriculture at the 15th Congress of Soil Science, Acapulco, Mexico, 1994. Springer Netherlands, Dordrecht, pp 195–209
Buckley DH, Huangyutitham V, Hsu S-F, Nelson TA (2008) 15N2–DNA–stable isotope probing of diazotrophic methanotrophs in soil. Soil Biol Biochem 40:1272–1283
Chaintreuil C, Giraud E, Prin Y, Lorquin J, Bâ A, Gillis M, de Lajudie P, Dreyfus B (2000) Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl Environ Microbiol 66:5437–5447
Chinnadurai C, Gopalaswamy G, Balachandar D (2014) Long term effects of nutrient management regimes on abundance of bacterial genes and soil biochemical processes for fertility sustainability in a semi-arid tropical Alfisol. Geoderma 232-234:563–572
Demba Diallo M, Willems A, Vloemans N, Cousin S, Vandekerckhove TT, De Lajudie P, Neyra M, Vyverman W, Gillis M, Van der Gucht K (2004) Polymerase chain reaction denaturing gradient gel electrophoresis analysis of the N2-fixing bacterial diversity in soil under Acacia tortilis ssp. raddiana and Balanites aegyptiaca in the dryland part of Senegal. Environ Microbiol 6:400–415
Diez B, Bergman B, Pedros-Alio C, Anto M, Snoeijs P (2012) High cyanobacterial nifH gene diversity in Arctic seawater and sea ice brine. Env Microbiol Rep 4:360–366
Dixon R, Kahn D (2004) Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol 2:621–631
Estrella Alcaman M, Fernandez C, Delgado A, Bergman B, Diez B (2015) The cyanobacterium Mastigocladus fulfills the nitrogen demand of a terrestrial hot spring microbial mat. ISME J 9:2290–2303
Ferrando L, Fernandez Scavino A (2015) Strong shift in the diazotrophic endophytic bacterial community inhabiting rice (Oryza sativa) plants after flooding. FEMS Microbiol Ecol 91:fiv104
Fuentes-Ramírez LE, Caballero-Mellado J, Sepúlveda J, Martínez-Romero E (1999) Colonization of sugarcane by Acetobacter diazotrophicus is inhibited by high N-fertilization. FEMS Microbiol Ecol 29:117–128
Ge Y, Priester JH, Mortimer M, Chang CH, Ji Z, Schimel JP, Holden PA (2016) Long-term effects of multiwalled carbon nanotubes and graphene on microbial communities in dry soil. Environ Sci Technol 50:3965–3974
Gonzalez Perez P, Ye J, Wang S, Wang X, Huang D (2014) Analysis of the occurrence and activity of diazotrophic communities in organic and conventional horticultural soils. Appl Soil Ecol 79:37–48
Hai B, Diallo NH, Sall S, Haesler F, Schauss K, Bonzi M, Assigbetse K, Chotte JL, Munch JC, Schloter M (2009) Quantification of key genes steering the microbial nitrogen cycle in the rhizosphere of sorghum cultivars in tropical agroecosystems. Appl Environ Microbiol 75:4993–5000
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT, Nucleic acids symposium series, vol c1979-c2000. Information Retrieval Ltd, London, pp 95–98
Hsu SF, Buckley DH (2009) Evidence for the functional significance of diazotroph community structure in soil. ISME J 3:124–136
Hui T (2012) Effects of long-term fertilization on nifH gene diversity in agricultural black soil. Afr J of Microbiol Res 6:2695–2666
Izquierdo JA, Klaus N (2015) Variation in diazotrophic community structure in forest soils reflects land use history. Soil Biol Biochem 80:1–8
Keuter A, Veldkamp E, Corre MD (2014) Asymbiotic biological nitrogen fixation in a temperate grassland as affected by management practices. Soil Biol Biochem 70:38–46
Ladha JK, Reddy PM (2003) Nitrogen fixation in rice systems: state of knowledge and future prospects. Plant Soil 252:151–167
Ladha JK, de Bruijn FJ, Malik KA (1997) Introduction: assessing opportunities for nitrogen fixation in rice—a frontier project. Plant Soil 194:1–10
Lema KA, Willis BL, Bourne DG (2012) Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl Environ Microbiol 78:3136–3144
Liang Y, Pan F, He X, Chen X, Su Y (2016) Effect of vegetation types on soil arbuscular mycorrhizal fungi and nitrogen-fixing bacterial communities in a karst region. Environ Sci Pollut R 23:18482–18491
Maclean JL (2002) Rice almanac: source book for the most important economic activity on earth. International Rice Research Institute, Laguna
Mårtensson L, Díez B, Wartiainen I, Zheng W, El-Shehawy R, Rasmussen U (2009) Diazotrophic diversity, nifH gene expression and nitrogenase activity in a rice paddy field in Fujian, China. Plant Soil 325:207–218
Mirza BS, Potisap C, Nüsslein K, Bohannan BJM, Rodrigues JLM (2013) Response of free-living nitrogen-fixing microorganisms to land use change in the Amazon rainforest. Appl Environ Microbiol 80:281–288
Okubo T et al (2012) Complete genome sequence of Bradyrhizobium sp. S23321: insights into symbiosis evolution in soil oligotrophs. Microbes Environ 27:306–315
Olsen SR (1954) Estimation of available phosphorus in soils by extraction with sodium bicarbonate. United States Department Of Agriculture, Washington
Pan FX, Chapman SJ, Li YY, Yao HY (2017) Straw amendment to paddy soil stimulates denitrification but biochar amendment promotes anaerobic ammonia oxidation. J Soils Sediments 17:2428–2437
Piromyou P, Greetatorn T, Teamtisong K, Okubo T, Shinoda R, Nuntakij A, Tittabutr P, Boonkerd N, Minamisawa K, Teaumroong N (2015) Preferential association of endophytic bradyrhizobia with different rice cultivars and its implications for rice endophyte evolution. Appl Environ Microbiol 81:3049–3061
Poly F, Ranjard L, Nazaret S, Gourbiere F, Monrozier LJ (2001) Comparison of nifH gene pools in soils and soil microenvironments with contrasting properties. Appl Environ Microbiol 67:2255–2262
Reardon CL, Gollany HT, Wuest SB (2014) Diazotroph community structure and abundance in wheat–fallow and wheat–pea crop rotations. Soil Biol Biochem 69:406–412
Reed SC, Seastedt TR, Mann CM, Suding KN, Townsend AR, Cherwin KL (2007) Phosphorus fertilization stimulates nitrogen fixation and increases inorganic nitrogen concentrations in a restored prairie. Appl Soil Ecol 36:238–242
Reed SC, Cleveland CC, Townsend AR (2011) Functional ecology of free-living nitrogen fixation: a contemporary perspective. Annu Rev Ecol Evol S 42:489–512
Rodrigues Coelho MR, De Vos M, Carneiro NP, Marriel IE, Paiva E, Seldin L (2008) Diversity of nifH gene pools in the rhizosphere of two cultivars of sorghum (Sorghum bicolor) treated with contrasting levels of nitrogen fertilizer. FEMS Microbiol Lett 279:15–22
Santi C, Bogusz D, Franche C (2013) Biological nitrogen fixation in non-legume plants. Ann Bot 111:743–767
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Shen C, Xiong J, Zhang H, Feng Y, Lin X, Li X, Liang W, Chu H (2013) Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol Biochem 57:204–211
Shu W, Pablo GP, Jun Y, Danfeng H (2012) Abundance and diversity of nitrogen-fixing bacteria in rhizosphere and bulk paddy soil under different duration of organic management. World J Microbiol Biotechnol 28:493–503
Su JQ, Ding LJ, Xue K, Yao HY, Quensen J, Bai SJ, Wei WX, Wu JS, Zhou J, Tiedje JM, Zhu YG (2015) Long-term balanced fertilization increases the soil microbial functional diversity in a phosphorus-limited paddy soil. Mol Ecol 24:136–150
Sun R, Guo X, Wang D, Chu H (2015a) Effects of long-term application of chemical and organic fertilizers on the abundance of microbial communities involved in the nitrogen cycle. Appl Soil Ecol 95:171–178
Sun R, Zhang X-X, Guo X, Wang D, Chu H (2015b) Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol Biochem 88:9–18
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tan Z, Hurek T, Reinhold-Hurek B (2003) Effect of N-fertilization, plant genotype and environmental conditions on nifH gene pools in roots of rice. Environ Microbiol 5:1009–1015
Team RC (2016) A language and environment for statistical computing. R Foundation for statistical computing 2015, Vienna, Austria
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Van Nguyen N, Ferrero A (2006) Meeting the challenges of global rice production. Paddy Water Environ 4:1–9
Wang J, Zhang D, Zhang L, Li J, Raza W, Huang Q, Shen Q (2016) Temporal variation of diazotrophic community abundance and structure in surface and subsoil under four fertilization regimes during a wheat growing season. Agric Ecosyst Environ 216:116–124
Wang C, Zhou J, Liu J, Du D (2017) Responses of soil N-fixing bacteria communities to invasive species over a gradient of simulated nitrogen deposition. Ecol Eng 98:32–39
Wartiainen I, Eriksson T, Zheng W, Rasmussen U (2008) Variation in the active diazotrophic community in rice paddy—nifH PCR-DGGE analysis of rhizosphere and bulk soil. Appl Soil Ecol 39:65–75
Wegner CE, Liesack W (2015) Microbial community dynamics during the early stages of plant polymer breakdown in paddy soil. Environ Microbiol 18:2825–2842
Xie GH, Cai MY, Tao GC, Steinberger Y (2003) Cultivable heterotrophic N2-fixing bacterial diversity in rice fields in the Yangtze River Plain. Biol Fertil Soils 37:29–38
Yeoh YK, Paungfoo-Lonhienne C, Dennis PG, Robinson N, Ragan MA, Schmidt S, Hugenholtz P (2016) The core root microbiome of sugarcanes cultivated under varying nitrogen fertilizer application. Environ Microbiol 18:1338–1351
Yousuf B, Kumar R, Mishra A, Jha B (2014) Differential distribution and abundance of diazotrophic bacterial communities across different soil niches using a gene-targeted clone library approach. FEMS Microbiol Lett 360:117–125
Zhalnina K, de Quadros PD, Gano KA, Davis-Richardson A, Fagen JR, Brown CT, Giongo A, Drew JC, Sayavedra-Soto LA, Arp DJ, Camargo FA, Daroub SH, Clark IM, McGrath SP, Hirsch PR, Triplett EW (2013) Ca. Nitrososphaera and Bradyrhizobium are inversely correlated and related to agricultural practices in long-term field experiments. Front Microbiol 4:104
Zhalnina K, Dias R, de Quadros PD, Davis-Richardson A, Camargo FA, Clark IM, McGrath SP, Hirsch PR, Triplett EW (2015) Soil pH determines microbial diversity and composition in the park grass experiment. Microb Ecol 69:395–406
Zhao J, Ni T, Li J, Lu Q, Fang Z, Huang Q, Zhang R, Li R, Shen B, Shen Q (2016) Effects of organic–inorganic compound fertilizer with reduced chemical fertilizer application on crop yields, soil biological activity and bacterial community structure in a rice–wheat cropping system. Appl Soil Ecol 99:1–12
Funding information
This work was supported by the National Natural Science Foundation of China (41525002, 41471206), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB15020301), and the Ningbo Municipal Science and Technology Bureau (2015C10031).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible editor: Jizheng He
Electronic supplementary material
ESM 1
(DOCX 356 kb)
Rights and permissions
About this article
Cite this article
Liao, H., Li, Y. & Yao, H. Fertilization with inorganic and organic nutrients changes diazotroph community composition and N-fixation rates. J Soils Sediments 18, 1076–1086 (2018). https://doi.org/10.1007/s11368-017-1836-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11368-017-1836-8