Abstract
Hydrolysis is one of the most important processes of transformation of organic chemicals in water. The rates of reactions, final chemical entities of these processes, and half-lives of organic chemicals are of considerable interest to environmental chemists as well as authorities involved in the controlling the processing and disposal of such organic chemicals. In this study, we have proposed QSPR models for the prediction of hydrolysis half-life of organic chemicals as a function of different pH and temperature conditions using only two-dimensional molecular descriptors with definite physicochemical significance. For each model, suitable subsets of variables were elected using a genetic algorithm method; next, the elected subsets of variables were subjected to the best subset selection with a key objective to determine the best combination of descriptors for model generation. Finally, QSPR models were constructed using the best combination of variables employing the partial least squares (PLS) regression technique. Next, every final model was subjected for strict validation employing the internationally accepted internal and external validation parameters. The proposed models could be applicable for data gap filling to determine hydrolysis half-lives of organic chemicals at different environmental conditions. Generally, presence of aliphatic ether and ether functional groups, high percentage of oxygen content in the molecule and presence of O–Si pairs of atoms at topological distance one, results in a shorter hydrolysis half-life of organic chemicals. On the other hand, higher unsaturation content and high percentage of nitrogen content in molecules lead to higher hydrolysis half-life. It is also found that branched and compact molecules will have a lower half-life while straight chain analogues will have a higher half-life. To the best of our knowledge, the presented models are the first reported QSPR models for hydrolysis half-lives of organic chemicals at different pH values.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
The monetary value of worldwide-produced organic chemicals is in the range of trillions of US dollars. These organic chemicals are widely used in several products as well as processes such as food and beverages, pharmaceuticals, agrochemicals, water-treatment chemicals, biocides, personal care products and cosmetics, house hold products, petroleum industry, polymers industry, and ceramics. In a simple word, organic chemicals play a pivotal role in human daily life. The widespread use and release of organic chemicals may produce direct or indirect toxic effects on the living organisms and the environmental health. Over the years, an exponential increase has been observed in the release, discharge, or introduction of organic chemicals into several water streams, river, lake, ponds, and seas (Mill and Mabey 1988). Large number of organic chemicals react with water (OH- and O+ components) to get metabolized into new chemical entities with half-lives accounting from few seconds to years (Mill and Mabey 1988). The half-life of organic chemicals is defined as the time required for them to reduce their initial concentrations to about half. The process of transformation of organic chemicals into new chemical entities different from its precursors in the aquatic media is known as hydrolysis. Hydrolysis is one the most important chemical reactions to determine the stability of chemicals and is considered as one of the major pathways for transformation of organic chemicals into the environment (EPA-OPPTS 1998; OECD 2004). Therefore, hydrolysis is a major transformation process for different varieties of organic chemicals in water, and the rates of reactions and final chemical entities of these processes are of great concern to the environmental chemists as well as regulatory authorities involved in controlling the processing and disposal of such organic chemicals in the environment. Conversion of initial chemical compounds into new chemical entities may alleviate potential hazardous effects on the living being as well as environment health (Mill and Mabey 1988). Hence, it is essential to understand and predict the chemical transformation process and the final products as quickly as possible. The significance of abiotic hydrolysis in the aquatic media can be measured quantitatively from the data of hydrolysis rate constant and half-lives of organic chemicals (EPA-OPPTS 1998). Usually, organic chemicals may exist in different environmental conditions. Therefore, it is an essential task to understand or examine the hydrolysis behavior of organic chemicals at different pH and temperature conditions (OECD 2004).
There are several chemometric software tools available to predict probable transformation products of organic chemicals in environmental as well as biological systems such as METEOR (Marchant et al. 2008) (a knowledge-based expert system developed to predict the probable metabolic fate of a chemical based on its chemical structure), TIMES (Dimitrov et al. 2011) (to predict abiotic and microbial transformation of chemicals in water and soil), enviPath (Wicker et al. 2016) (useful to determine biotransformation pathway of environment pollutants), META expert system (Sedykh et al. 2001) (used to predict transformation pathways of organic compounds under UV light, i.e., phototransformation), Zeneth software tool (Kleinman et al. 2014) (to predict transformation products of active pharmaceutical ingredients under several conditions used in stability studies), and the Chemical Transformation Simulator (Tebes-Stevens et al. 2017) (a web-based software tool based on the abiotic hydrolysis reaction library). All the aforementioned software tools are mostly used to predict the transformation pathway or transformation product of organic chemicals; however, to the best of our knowledge, there are no previously reported quantitative structure-property relationship (QSPR) models or chemometric software tools to predict the hydrolysis half-life of organic chemicals in aquatic media.
Over the years, chemometric approaches such as QSPRs have been proved to be successful for the prediction of diverse properties of organic chemicals and helpful in data gap filling for safety assessment of organic chemicals (Roy et al. 2015). QSPR is a mathematical model which provides a quantitative correlation between independent (numerical molecular descriptors of obtained from chemical structures) and dependent (response endpoint, i.e., property) variables. In the current study, we have generated a number of QSPR models for hydrolysis half-life as a function of different pH and temperature conditions of organic chemicals employing only two-dimensional variables. The suitable subsets of variables were chosen employing a genetic algorithm method; then, the relevant subsets of variables were subjected to the best subset selection with an objective to identify best combination of descriptors for the model building; next the best combination of descriptors was used for the final model development using partial least squares (PLS) regression technique. The final models were strictly validated using the internationally accepted internal and external validation parameters. The final validated models of hydrolysis half-life as a function of different pH and temperature conditions of organic chemicals may be helpful to predict their environmental persistence (hydrolysis half-life as a function of different pH and temperature) quickly, based on the knowledge of chemical structures only, thus providing a better alternative to the experimental testing methods, which are costly and time-consuming.
Materials and methods
Dataset preparation
The response data of the hydrolysis half-life of organic chemicals as a function of different pH and temperature values were extracted from the Ambit database (Ambit 2019) (accessed in March 2019), and it refers to the ECHA registration dossiers. The extracted data were cleaned covering the key and supporting studies with experimental data obtained according with the following guidelines: OECD 111; Method C.7—degradation—abiotic degradation hydrolysis as a function of pH. annex v: consolidated version of dir 67/548/EEC (and similar); EEC directive 67/548, annex v, part c, test c10-preliminary test (OECD 2004) (and similar); EPA OPPTS 835.2110 (hydrolysis as a function of pH); EPA OPPTS 835.2120 (hydrolysis of parent and degradates as a function of pH at 25 °C) (EPA-OPPTS 1998); EPA OTS 796.3500 (hydrolysis as a function of pH at 25 °C) (CFR 2012). In addition, we eliminated the compounds with a qualifier and/or without a measure unit, the inorganics, and the mixtures. The response values of the collected raw data were present in different units such as minutes, hours, days, and years. For unification of the units of the response endpoint, we have transformed the hydrolysis half-life values in the day unit, and subsequently, the hydrolysis half-life values were transformed into logarithmic scale (log10). The chemical structures were cautiously drawn manually using Marvin sketch ChemAxon tool (http://www.chemaxon.com/) and cross-verified from the PubChem small molecule database (https://pubchem.ncbi.nlm.nih.gov/) (Kim et al. 2016). The curated chemical structures were prepared by cleaning and adding explicit hydrogen atoms and then saved in the MDL .mol format, a suggested format for descriptor estimation software tools such as PaDEL-Descriptor (Yap 2011) and Dragon (Mauri et al. 2006; Todeschini et al. 2004).
The final QSPR analysis of hydrolysis half-life of organic chemicals was performed using the data points of 45, 63, and 68 organic compounds for pH 4, 7, and 9 at 25 °C respectively and 27, 34, and 36 organic compounds for pH 4, 7, and 9 at 50 °C, respectively. The temperatures and pH were selected according to the OECD 111 guideline (OECD 2004).
Descriptor calculation and dataset division
The prepared chemical structures of organic chemicals were subjected to descriptor calculation employing the PaDEL-Descriptor (Yap 2011) and Dragon (Mauri et al. 2006; Todeschini et al. 2004) software tools. We used Dragon (Version 7) software to compute a few selected classes of 2D descriptors such as connectivity indices, ring descriptors, atom type E-state indices, 2D atom pairs, functional group counts, atom-centered fragment, constitutional indices, and molecular property descriptors (Mauri et al. 2006; Todeschini et al. 2004). Only the extended topochemical atom (ETA) descriptors were computed from the PaDEL-Descriptor (Version 2.20) program (Yap 2011). The computed descriptors were easily interpreted and had definite physicochemical significance. The initial pools of estimated variables for the datasets for pH 4, 7, and 9 at 25 °C comprise 510, 587, and 596 respectively, while 403, 474, and 473 molecular variables for pH 4, 7, and 9 at 50 °C, respectively, excluding the redundant variables and constant, intercorrelated descriptors (|r| > 0.9).
The main objective of this analysis was to generate robust QSPR models for the predicting the hydrolysis half-life of organic chemicals as a function of various pH and temperature conditions. In order to meet the primary objective, we have divided each dataset into a training set (employed for model training) and a test set (for model rigorous validation) in approximately 70:30 ratio (Roy et al. 2015) employing three techniques in-built in the dataset division (ver. 1.2) software tool (available at http://dtclab.webs.com/software-tools); these techniques are Kennard-Stone (Kennard and Stone 1969), Euclidean distance (Golmohammadi et al. 2012) and sorted response. However, in the current study, statistically significant models for pH 4, 7, and 9 at 25 °C and pH 4 and 9 at 50 °C were obtained using the sorted response based division, while those for pH 7 at 50 °C were obtained employing the Euclidean distance based division (Golmohammadi et al. 2012). The final models for pH 4, 7, and 9 at 25° for the hydrolysis half-life prediction of organic chemicals were obtained using 34, 47, and 51 compounds in the training set (Ntrain) and 11, 15, and 17 molecules in the test set (Ntest) respectively. Again, for pH 4, 7, and 9 at 50 °C, the “training sets” comprise 21, 26, and 29 compounds and the “test sets” comprise 6, 8, and 7 organic chemicals, respectively.
Model development and validation
In the process of model generation, selection of suitable features from the large pool of descriptors was considered as an essential step. There are a number of feature selection techniques available for this purpose such as step-by-step selection (with stepping criteria of F-for-inclusion and F-for-exclusion based on the partial F statistics), genetic algorithm (GA), and all possible subset selection (Khan and Roy 2018; Roy et al. 2015), which can be used to select relevant subsets of features. In the present work, we have employed the GA method (tool available at http://teqip.jdvu.ac.in/QSAR_Tools/) for the feature selection process. The numbers of variables in the selected subset for hydrolysis half-life modeling for pH 4, 7, and 9 at 25 °C are 19, 24, and 26 respectively, while those for pH 4, 7, and 9 at 50 °C are 22, 25, and 21, respectively. The selected subsets of variables were separately subjected to best subset selection (BSS) (tool available at http://teqip.jdvu.ac.in/QSAR_Tools/) with a goal of identifying the best combination of descriptors for model development. Finally, the best combination of descriptors was used for model generation using the partial least squares (PLS) regression (Wold et al. 2001) (tool available at http://teqip.jdvu.ac.in/QSAR_Tools/).
In the present study, we have reported PLS models (Wold et al. 2001) because it offers numerous advantages over the MLR technique such as it can deal with strongly collinear, correlated, noisy data, and it is helpful in modelling with a large number of X-variables and more than one response endpoints. The PLS algorithm extracts meaningful information from the original descriptors into the lower number variables knows as latent variables (LVs). Please note that MLR is only a special case of PLS. Unlike MLR models, determination of standard errors of PLS regression coefficients is not straightforward. However, the relative importance of the descriptors in a PLS model can be shown by a variable importance plot (VIP). The optimum number of latent variables was selected based on a sequential basis. For example, the initial PLS model was generated with a single latent variable, and the corresponding Q2 value of the generated model was noted; the next LV variable was then added based on the 5% rule (Roy et al. 2015). This suggests that the addition of a latent variable is permitted only when it results in an increase in the value of Q2 by 5% or more.
The final best models were strictly validated on the basis of internal and external validation metric criteria which are accepted internationally. The quality and validation metrics included numerous training set parameters such as determination coefficient (R2), leave-one-out (LOO) cross-validation (Q2), \( {r}_{m\left(\mathrm{LOO}\right)}^2 \) (Roy and Mitra 2011; Roy et al. 2012), and mean absolute error of training set (MAEtrain100%), and the test set metrics such as external predictive variance R2pred, \( {r}_{m\left(\mathrm{test}\right)}^2 \); mean absolute error of test set (MAEtest100%). Finally, the consistency of each model was tested on the basis of MAE criteria as proposed by Roy et al. (2016) in terms of “Bad”, “Moderate”, and “Good” (Roy et al. 2016). The overview of the methodology followed in the present work is depicted in Fig. 1.
Applicability domain assessment
The generated models were subjected to the applicability domain (AD) analysis, with an objective to define hypothetical domain of each model in the chemical space. The AD of any model presents essential information whether the prediction of each compound obtained by the respective model is reliable or not. In the current study, we have used the DModX (distance to model in X space) (Wold et al. 2001) approach embedded in the SIMCA-P software to define the AD of the generated models in the chemical space. A compound with a DModX value greater than the 2.5 times the total SD of the X residuals shall be known as an outlier in case of the training set and outside AD in case of the test set.
Results and discussion
QSPR modelling of hydrolysis half-life of organic chemicals as a function of pH at 25 °C
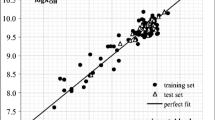
To model hydrolysis half-life as a function of pH, we have employed experimental hydrolysis half-life data of organic chemicals estimated at pH 4, 7, and 9 and 25 °C. The datasets comprise 45, 63, and 68 organic compounds for pH 4, 7, and 9, respectively. The curated data of each dataset were divided into a “training set” and a “test set” prior to model generation using the different data division approaches. At pH 4, the “training set” included 34 compounds and the “test set” included 11 compounds (Ntest = 11), at pH 7 the “training set” comprised 47 compounds (Ntraining = 47) and the “test set” 15 compounds (Ntest = 15), and at pH 9, the “training set” was composed of 51 compounds (Ntraining = 51) and the “test set” 17 compounds (Ntest = 17). The training dataset was used for model training, while the test dataset was involved in model validation in each case. All the final models were obtained by using the partial least squares regression (PLS) (Wold et al. 2001) algorithm with different latent variables (LV). The scatter plot (Fig. 2) shows that observed hydrolysis half-life (log(d)) values of organic chemicals are well correlated with the predicted hydrolysis half-life (log(d)) values at pH 4, 7, 9, and 25 °C. The chosen model for each endpoint shows significant and promising internal and external prediction quality, as shown by below-mentioned equations:
Model for pH 4
Model for pH 7
Model for pH 9
To identify the relative importance of each descriptor appearing in the final QSPR model derived from data at pH 4 to predict hydrolysis half-life of organic chemicals, we have performed the VIP analysis (UMETRICS, S-P 2005), which revealed that O-059 (presence or absence of Al–O–Al functional group) (Todeschini and Consonni 2008) and ETA_Shape_X (Roy and Das 2017) variables with higher VIP score were the most essential descriptors for model development, while other descriptors such as Uc (unsaturation count) (Todeschini and Consonni 2008), nO (C=O)2 (number of anhydrides (-thio) functional group) (Todeschini and Consonni 2008) and ETA_Shape_P (Roy and Das 2017) were of lower importance in the final model development (Figure S1 in Supplementary Information). Subsequently we have also performed loading plot analysis to identify the most influential variables for the response (UMETRICS, S-P 2005). The loading plot analysis also suggests that O-059 and ETA_Shape_X variables with negative correlation towards response endpoint are situated far from the origin and considered as most influential variables in the final model, while the rest of least influential variables (situated close to the origin) are Uc, nO(C=O)2, and ETA_Shape_P (Figure S2 in Supplementary Information).
The QSPR equation (Eq. 1) comprises five unique two-dimensional descriptors. Out of the five descriptors, two (Uc and ETA_Shape_P) show positive contributions towards the prediction of hydrolysis half-life of organic chemicals, indicating that higher unsaturation content in the molecules lead to an increase in the hydrolysis half-life of a particular compound and vice versa. For example, compounds 83 and 114 show longer hydrolysis half-lives due to the presence of unsaturated rings such as benzophenone and tri-phenyl moiety, respectively, in their chemical structures. On the other hand, compound 67 shows a shorter hydrolysis half-life due to absence of any kind of unsaturation in the chemical structure (Fig. 3).
On the other hand, the remaining three descriptors (O-059, nO(C=O)2 and ETA_Shape_X) show negative contributions towards the response endpoint prediction, suggesting that the presence of the fragment Al-O-Al (where Al represents aliphatic carbon) and the anhydride functional group in the molecule result in the smaller hydrolysis half-life. For example, compound 88 (tris-(2-methoxyethoxy) vinylsilane, which hydrolyzes to vinyl-silanetriol and 2-methoxyethanol) and 67 (tetra-ethyl orthosilicate, which transforms to silicon tetra-hydroxide and ethanol) show lower hydrolysis half-lives due to the presence of a repeated number of Al–O–Al fragments in the compounds. The hydrolysis of such compounds is based on selectivity rule, which specifies that the hydrolysis is initiated with the carbon atom with the highest electrophilicity attached to leaving group (O, S, N) and so on (Tebes-Stevens et al. 2017) (Figs. 4 and 5).
The next descriptor with a negative contribution in the QSPR equation is nO(C=O)2, which provides information about the presence or absence of anhydrides functional group in the molecule and indicate that presence of such fragment in the molecule leads to a decrease of the half-life of the organic compound. For example, compound 50 (hexanoic anhydride easily hydrolyzes into the hexanoic acid) shows lower hydrolysis half-life due to the presence of the anhydride functional group, which can easily be converted into the acid (Fig. 6).
The last descriptor with a negative effect on half-life estimation is ETA_Shape_X, which offers an aspect on the molecular shape of the molecule based on the sum of core count of vertices that are joined with four other nonhydrogen vertices in the molecules (Roy and Ghosh 2003). For example, compound 94 displays a high value of the descriptor due to presence of vertices that are bound to four other nonhydrogen vertices that results in a shorter half-life. On the other hand, the descriptor ETA_Shape_P with a positive contribution towards the response endpoint also offers an information about the molecular shape of the molecule depending on the sum of core count of vertices that are connected to only another nonhydrogen vertex in the molecules (Roy and Ghosh 2003). For example, compound 69 shows a longer half-life due to presence of a vertex which is connected to only another nonhydrogen vertex (Fig. 7). This indicates that branched and compact molecules will have a lower half-life while straight chain analogues will have a higher half-life.
Finally, we have performed applicability domain (AD) study of the final model using DModX (distance to model in X space) technique. It was found that none of organic chemicals was an outlier (training set) or outside AD (test set). Figures S3 and S4 (Supplementary Information) give the graphical overview of the applicability domain of the final model for the training and test set substances, respectively for prediction of hydrolysis half-life of organic chemicals at pH 4 and 25°.
For the model derived for data at pH 7, we have also performed the VIP analysis with an objective to determine the most essential features among the descriptor appearing in the final QSPR model to predict hydrolysis half-life of organic chemicals. The study suggests that nO(C=O)2 and ETA_BetaP_s (which provide information about the electronegative atom count of the molecule relative to the molecular size) descriptors (Roy and Das 2017) with VIP scores more than one were the most relevant variables for explaining the response endpoint, while other descriptors with VIP score lower than one include N% (percentage of N atoms in the molecule) (Todeschini and Consonni 2008), Uc (unsaturation count) (Todeschini and Consonni 2008), nR = Cs (presence or absence of number of aliphatic secondary C (sp2) in the molecule) (Todeschini and Consonni 2008), and B10[C–O] (presence/absence of C–O at the topological distance 10) (Todeschini and Consonni 2008) are considered as less relevant variables in the final model (Figure S5 in Supplementary Information). Next, we have also performed loading plot analysis to identify the most significant features towards the response. The loading plot analysis also showed that nO(C=O)2 and ETA_BetaP_s variables with negative contributions to the response are situated far from the origin, and they are the most significant descriptors in the final model, while least significant descriptors are N%, Uc, nR = Cs, and B10[C–O], which are positioned close to the origin (Figure S6 in Supplementary Information).
Two descriptors in the QSPR equation (Eq. 2) show negative contributions towards the response, while the remaining four descriptors have positive regression coefficients. The variables with a negative impact on the hydrolysis half-life include nO(C=O)2 and ETA_BetaP_s which provide information, respectively, about the presence or absence of an anhydride functional group and a measure of electronegative atom count in the molecules relative to the molecular size. For example, molecule 37 (1, 2, 3, 6-tetrahydromethyl-3, 6-methanophthalic anhydride) shows a shorter half-life due to the presence of an anhydride functional group, which easily hydrolyzes to an acid and 94 (hexamethylcyclotrisiloxane, which initially hydrolyses to 1,1,3,3,5,5-hexamethyltrisiloxane-1,5-diol) shows a shorter half-life due to the presence of a repeated number of electronegative atoms (O and Si) in the molecule (Figs. 8 and 9).
Again, the variables with positive contributions towards the response include Uc (unsaturation count), N% (percentage of nitrogen atom in the compound), B10[C–O] (presence or absence of C-O at topological distance 10), and nR = Cs (number of aliphatic secondary carbon), indicating that higher values of these variables result in higher hydrolysis half-life of organic chemicals and vice versa. For example, compound 73 shows a longer hydrolysis half-life due to the presence of four nitrogen atoms (higher nitrogen content) and unsaturation in the molecule.
Similarly, compounds 84 and 56 have higher hydrolysis half-lives due to the presence of C–O at the topological distance ten and the presence of sp2 carbon atom in the molecule, respectively (Fig. 10). According to the AD analysis, only one molecule (comp 50) with a higher threshold value than the D critical value was deemed as an outlier (training sample); none of the molecules in the test group were found to be outside AD. Figures S7 and S8 (Supplementary Information) provide the schematic overview of the AD of the final model for the training and test set compounds respectively for prediction of hydrolysis half-life of organic chemicals at pH 7 and 25 °C.
At pH 9, we have again performed the VIP analysis, the study proposed that nO(C=O)2 and B01[O–Si] (presence/absence of O–Si at the topological distance 1) descriptors were considered as the most prominent descriptors with VIP scores more than one. On the other hand, descriptors with a VIP score lower than one are O% (percentage of oxygen atoms in the molecule), ETA_dBeta (providing information about relative unsaturation content of molecule) (Roy and Das 2017), nROH (presence or absence of hydroxyl function group in the molecule), and B09[O–O] (presence or absence of O–O atom pairs at the distance 9 edges) were less important towards the prediction of the response endpoint (Figure S9 in Supplementary Information). Further, we have also generated a loading plot for the final model in order to identify the most influential descriptors among the final descriptors. The loading plot observation revealed that nO(C=O)2 and B01[O–Si] descriptors with negative impact on the response endpoint are located far away from the origin and considered as the most influential descriptors in the final model, while the least influential descriptors O%, ETA_dBeta, nROH, and B09[O–O], which are located at a smaller distance from the origin (Figure S10 in Supplementary Information).
The QSPR equation (Eq. 3) comprises six unique 2D descriptors, two of them show positive contributions (nROH and ETA_dBetaP), indicating that higher values of these descriptors result in longer hydrolysis half-lives of organic chemicals. For example, compound 15 (presence of two hydroxyl functional groups) and 96 (high value of the ETA_dBetaP variable) show longer half-lives (Fig. 11).
On the other hand, the variable with negative contributions include O%, B09[O–O], B01[Si–O], and nO(C=O)2, suggesting that higher values of these variables result in lower hydrolysis half-lives. For example, compound 85 (due to a high percentage of the oxygen atom content in the molecule) and 37 (due to presence of anhydride functional group labile to hydrolysis) show shorter hydrolysis half-lives. Similarly, compound 94 and 5 show lower hydrolysis half-lives due to Si–O bonds and presence of two oxygen atoms at the topological distance nine in the molecules, respectively (Fig. 12). Lastly, we have performed the AD study of the final QSPR model. The analysis suggests that compounds neither in the training set nor in the test set are outliers or outside AD. Figures S11 and S12 (Supplementary Information) provide the schematic representation of the applicability domain of the final model for the training and test set compounds respectively for the prediction of hydrolysis half-lives of organic chemicals at pH 9 and 25 °C.
QSPR modelling of hydrolysis half-life of organic pollutants as a function of pH at 50 °C
At temperature 50 °C, we have generated QSPR models for the prediction of hydrolysis half-lives of organic chemicals at pH 4, 7, and 9 using the datasets of 27, 34, and 36 organic compounds, respectively. With the aid of data division software tool, each dataset was divided into a “training set” and a “test set” prior to the model development. The training set contains 21, 26, and 29 compounds and the test set consists of 6, 8, and 7 organic chemicals, for pH 4, 7, and 9, respectively. The models were generated by employing only the training set compounds using the PLS regression technique with different latent variables. The scatter plot (Fig. 13) shows that observed hydrolysis half-life (log(d)) values of organic chemicals are well correlated with the predicted hydrolysis half-life (log(d)) values at pH 4, 7, 9, and 50 °C. All final models have been rigorously validated by using different internationally accepted internal and external metrics, as shown below.
Model for pH 4
Model for pH 7
Model for pH 9
To identify the most relevant descriptors from descriptor appearing in the final QSPR model derived from data at pH 4 to predict the hydrolysis half-life of organic chemicals, we have generated the VIP plot using SIMCA-P software tool; the plot revealed that the nR06 (number of 6-membered rings) descriptor was the most essential descriptor for model development with a VIP score more than one. On the other hand, other descriptors such as TPSA (tot) (topological polar surface area using N, O, S, and P polar contributions), nCIC (number of rings (cyclomatic number)), and nRCO (number of ketones (aliphatic)) were the least relevant descriptors with VIP scores less than one (Figure S13 in Supplementary Information). Subsequently, we have also generated loading plot with an objective to determine the most influential variable in the final QSPR model. The loading plot analysis also recommends that nR06 with a positive correlation towards the response is situated far from the origin and considered the most influential variable, while the rest of the descriptors such as nRCO, TPSA (tot), and nCIC were located at a lower distance from the center of the plot and considered the least influential variables (Figure S14 in Supplementary Information).
The QSPR model (Eq. 4) comprises four unique independent variables with either positive or negative contribution towards the response. The variables with a positive impact on hydrolysis half-life include nR06and nRCO while descriptors with negative impact include nCIC and TPSA (tot). To understand the mechanistic importance of each variable, we have analyzed the data carefully and observed that compound 4 shows higher hydrolysis half-life due to the presence of a four six-member ring (nR06 variable) in its chemical structure. Similarly, compounds 11 and 77 show longer half-lives due to the presence two aliphatic carbonyl functional groups (nRCO variable) in their chemical structures. However, the half-life difference of approximately three days of these two compounds (11 and 77) is due to a difference in their total polar surface area (Fig. 14). On the other hand, two negatively correlating descriptors are nCIC and TPSA (tot), suggesting that presence of high number of cyclomatic ring (providing information about presence of all type of alicyclic as well as aromatic rings in the molecules) as well as total polar surface area of any compound result in lower half-life and vice versa. For example, compounds 2 and 105 result into smaller half-lives due to the presence of four cyclomatic rings (three epoxide rings and one phenyl ring) in the compounds and due to the larger total polar surface area of the molecule, respectively (Fig. 14). Finally, we performed the AD study of the final QSPR model using the DModX approach. The results show that compounds neither in the training set nor in the test set were outliers and outside the AD, suggesting that the generated model was robust with significant predictive quality. Figures S15 and S16 (Supplementary Information) provide the schematic representation of the applicability domain of the final model for the training and test set compounds respectively for the prediction of hydrolysis half-life of organic chemicals at pH 4 and 50 °C.
For the model derived from data at pH 7, again we have performed the VIP analysis using SIMCA-P software tool. The analysis suggests that NssCH2 (number of atoms of type ssCH2) variable was the most crucial descriptor for model development with a higher VIP score. On the other hand, other variables such as H-048 (H attached to C2(sp3)/C1(sp2)/C0(sp)), Uc, and nO (number of oxygen atoms) were the least essential descriptor with a lower VIP score (Figure S17 in Supplementary Information). Further, we have also performed a loading plot analysis, which also revealed that the NssCH2 descriptor with a positive contribution towards the response is located away from the center and considered as most influential variable, while the rest of the variables such as H-048, Uc, and nO were located at a lower distance from the center of the plot and labeled as the least influential descriptors for prediction of hydrolysis half-life of organic chemicals (Figure S18 in Supplementary Information).
The final QSPR model (Eq. 5 for pH 7) was obtained using the dataset of 34 organic compounds at three latent variables (extracting vital information from four unique variables). All the variables appearing in the final equation show negative contributions towards the hydrolysis half-life prediction at 50 °C except one variable (Uc). The descriptor with a positive contribution (Uc) provides information about the unsaturation content in the molecule. For example, compound 114 shows a longer half-life value due to high unsaturation (due to presence of three aromatic rings in the chemical structure). On the other hand, the variables with negative contribution include H-048, NssCH2, and nO suggesting that higher values of the descriptors result in shorter half-lives and vice versa. For example, compounds 107 and 62 show shorter half-lives at pH 7 due to the presence of hydrogen atoms attached to sp2 carbon atom and higher number of –CH2-fragments in the chemical structures, respectively. Similarly, compound 105 shows a lower half-life due to the presence of higher number of oxygen atoms in the chemical structure (Fig. 15). Lastly, we have subjected the final QSPR model to the AD study using the DModX approach. The study suggests that a single compound (compound 108) was an outlier, while all the test set compounds were found to be within the applicability domain. Figures S19 and S20 (Supplementary Information) provide the schematic representation of the applicability domain of the final model for the training and test set compounds, respectively for prediction of hydrolysis half-life of organic chemicals at pH 7 and 50 °C.
We have also performed the VIP analysis for the model obtained from data at pH 9. The analysis suggests that out of five variables appearing in the final model, three variables with VIP scores more than one were most essential for QSPR model development and these are B02[O–O], X4sol, and O-060. On the other hand, other two variables (T(N … .S) and X5Av, see Figure S21 in Supplementary Information) with lower VIP scores were considered less important ones for hydrolysis half-life prediction of organic chemicals. Additionally, we have also performed the loading plot analysis, which also shows that B02[O–O] and O-060 are situated away from the origin and thus considered as most prominent variables, while the rest of the variables such as X4sol, T(N … .S) and X5Av were positioned at a lower distance from the center and considered as less important descriptors for the prediction of hydrolysis half-lives of organic chemicals (Figure S22 in Supplementary Information).
The best QSPR model (Eq. 6) was derived using the dataset of 37 organic pollutants at pH 9. The final QSPR equation comprises five 2D descriptors calculated using Dragon and PaDEL-descriptor software tools. The descriptors appearing in the model and showing a negative impact towards the prediction of the half-life of organic compounds at pH 9 and 50 °C are O-060 (presence of Al–O–Ar/Ar-O-Ar/R..O..R/R–O–C = X functional group, here Al and Ar represent aliphatic and aromatic substitutions), T(N..S) (sum of topological distances between N..S), X5Av (average valence connectivity index of order 5) and B02[O–O] (presence/absence of O–O at topological distance 2), which suggest that the presence of these specific fragments/functional groups or combination of atoms at a specific distance in the chemical structures leads to a decrease in half-life of the compounds. For example, 62, 108, and 66 show low half-lives due to the presence of carbonate functional group, a large sum of the topological distance between nitrogen and sulfur atoms and presence two oxygen atoms at a distance of two bonds, respectively (Fig. 16). Again, the descriptor with a positive coefficient in the final equation is X4sol (solvation connectivity index of order 4), which indicates that a higher value of solvation connectivity index of order 4 variable leads to an increase in half-life of organic chemicals. For example, compound 53 shows higher value of X4sol variable resulting into a higher hydrolysis half-life of the molecule (Fig. 16). At the end, we have carried out an AD study. The analysis showed that not a single compound in the training and the test set behaved as an outlier or outside AD. Figures S23 and S24 (Supplementary Information) provide the schematic representation of the applicability domain of the final model for the training and test set compounds respectively for prediction of hydrolysis half-lives of organic chemicals at pH 9 and 50 °C.
Conclusion
In the current work, we have proposed QSPR models for the prediction of hydrolysis half-life of organic chemicals as a function of different pH at different temperature conditions employing only two-dimensional molecular descriptors. For every model, the appropriate subsets of descriptors were elected using a genetic algorithm method; next, the appropriate subsets of descriptors were subjected to the best subset selection with a key objective to determine the best combination of descriptors for the model generation. Finally, the QSPR models were built using the best combination of variables employing the partial least squares (PLS) regression technique. Next, every final model was subjected to strict validation employing the internationally accepted internal and external validation parameters. As per the QSPR models developed at pH 4, 7, and 9 and 25 °C, in general, presence of aliphatic ether and ethereal functional groups, high percentage of oxygen content in the molecules, and presence of O–Si pair of atoms at topological distance one result in shorter hydrolysis half-life of organic chemicals. On the other hand, higher unsaturation content and presence of high percentage of nitrogen content in a molecule result in higher hydrolysis half-life. It is also found that branched and compact molecules will have a lower half-life while the straight chain analogues will have a higher half-life. As per the QSPR models at pH 4, 7, and 9 and 50 °C, we also found that the few key features for prediction of hydrolysis half-lives of organic chemicals are higher number of –CH2 groups, oxygen atoms, and presence of carbonate functional groups and presence of O–O atom pair at topological distance 2 resulting in shorter half-life. On the other hand, high unsaturation content, presence of six-member rings (imparting unsaturation), and presence of aliphatic ketone result in higher hydrolysis half-life of organic chemicals. The final models can be useful for quickly determining or predicting the environmental persistence of organic chemical compounds, based solely on knowledge of chemical structures, thus providing a better alternative to the costly and time consuming experimental testing methods.
References
Ambit (2019) https://ambitlri.ideaconsult.net/tool2/substance/. Accessed Mar 2019
CFR (2012) Title 40- protection of environment, chapter I - environmental protection agency. 33:94–100. https://www.govinfo.gov/content/pkg/CFR-2012-title40-vol33/pdf/CFR-2012-title40-vol33-chapI.pdf. Accessed Mar 2019
Dimitrov S, Pavlov T, Dimitrova N, Georgieva D, Nedelcheva D, Kesova A, Vasilev R, Mekenyan O (2011) Simulation of chemical metabolism for fate and hazard assessment. II CATALOGIC simulation of abiotic and microbial degradation. SAR QSAR Environ Res 22:719–755
EPA-OPPTS (1998) Fate, transport and transformation test guidelines OPPTS 835.2130 hydrolysis as a function of pH and temperature U.S Enviromental Protection Agency, Washington DC:1-13
Golmohammadi H, Dashtbozorgi Z, Acree WE Jr (2012) Quantitative structure-activity relationship prediction of blood-to-brain partitioning behavior using support vector machine. Eur J Pharm Sci 47:421–429
Kennard RW, Stone LA (1969) Computer aided design of experiments. Technometrics. 11:137–148
Khan PM, Roy K (2018) Current approaches for choosing feature selection and learning algorithms in quantitative structure-activity relationships (QSAR). Expert Opin Drug Discov 13:1075–1089
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA (2016) PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213
Kleinman MH, Baertschi SW, Alsante KM, Reid DL, Mowery MD, Shimanovich R, Foti C, Smith WK, Reynolds DW, Nefliu M (2014) In silico prediction of pharmaceutical degradation pathways: a benchmarking study. Mol Pharm 11:4179–4188
Marchant CA, Briggs KA, Long A (2008) In silico tools for sharing data and knowledge on toxicity and metabolism: Derek for windows, meteor, and vitic. Toxicol Mech Method 18:177–187
Mauri A, Consonni V, Pavan M, Todeschini R (2006) Dragon software: an easy approach to molecular descriptor calculations. Match. 56(2):237–248
Mill T, Mabey W (1988) Hydrolysis of organic chemicals, reactions and processes. Springer, pp 71-111
OECD (2004) OECD guidelines for the testing of chemicals: hydrolysis as a function of pH. OECD Paris pp 1-15
Roy K, Das RN (2017) The “ETA” Indices in QSAR/QSPR/QSTR Research, Pharmaceutical Sciences: Breakthroughs in Research and Practice. IGI Global, pp. 978-1011
Roy K, Ghosh G (2003) Introduction of extended topochemical atom (ETA) indices in the valence electron mobile (VEM) environment as tools for QSAR/QSPR studies. Internet Electron J Mol Des 2:599–620
Roy K, Mitra I (2011) On various metrics used for validation of predictive QSAR models with applications in virtual screening and focused library design. Comb Chem High Throughput Screen 14:450–474
Roy K, Mitra I, Ojha PK, Kar S, Das RN, Kabir H (2012) Introduction of rm2 (rank) metric incorporating rank-order predictions as an additional tool for validation of QSAR/QSPR models. Chemom Intell Lab Syst 118:200–210
Roy K, Kar S, Das RN (2015) A primer on QSAR/QSPR modeling: fundamental concepts. Springer
Roy K, Das RN, Ambure P, Aher RB (2016) Be aware of error measures. Further studies on validation of predictive QSAR models. Chemometr Intell Lab sys 152:18–33
Sedykh A, Saiakhov R, Klopman G (2001) META V. A model of photodegradation for the prediction of photoproducts of chemicals under natural-like conditions. Chemosphere 45:971–981
Tebes-Stevens C, Patel JM, Jones WJ, Weber EJ, technology (2017) Prediction of hydrolysis products of organic chemicals under environmental pH conditions. Environ Sci 51:5008–5016
Todeschini R, Consonni V (2008) Handbook of molecular descriptors. Wiley
Todeschini R, Consonni V, Mauri A, Pavan M (2004) DRAGON-Software for the calculation of molecular descriptors. Web version 3
UMETRICS, S-P (2005) User guide and Tutorial. Société Umetrics
Wicker J, Lorsbach T, Gütlein M, Schmid E, Latino D, Kramer S, Fenner K (2016) enviPath–The environmental contaminant biotransformation pathway resource. Nucleic Acids Res 44:D502–D508
Wold S, Sjöström M, Eriksson L (2001) PLS-regression: a basic tool of chemometrics. Chemom Intell Lab Syst 58:109–130
Yap CW (2011) PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32:1466–1474
Funding
PMK thanks the Department of Pharmaceuticals, Ministry of Chemicals and Fertilizers, Govt. of India for a fellowship. KR thanks Science and Engineering Research Board (SERB), New Delhi for financial assistance under the MATRICS scheme (File number MTR/2019/000008). AL and EB thank the financial contribution of the project LIFE-VERMEER contract (LIFE16 ENV/ES/000167).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Responsible editor: Marcus Schulz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 449 kb)
Rights and permissions
About this article
Cite this article
Khan, P.M., Lombardo, A., Benfenati, E. et al. First report on chemometric modeling of hydrolysis half-lives of organic chemicals. Environ Sci Pollut Res 28, 1627–1642 (2021). https://doi.org/10.1007/s11356-020-10500-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11356-020-10500-0