Abstract
Cardiac arrhythmia has been identified as a type of cardiovascular diseases (CVDs) that causes approximately 12% of all deaths globally. The development of Internet-of-Things has spawned novel ways for heart monitoring but also presented new challenges for manual arrhythmia detection. An automated method is highly demanded to provide support for physicians. Current attempts for automatic arrhythmia detection can roughly be divided as feature-engineering based and deep-learning based methods. Most of the feature-engineering based methods are suffering from adopting single classifier and use fixed features for classifying all five types of heartbeats. This introduces difficulties in identification of the problematic heartbeats and limits the overall classification performance. The deep-learning based methods are usually not evaluated in a realistic manner and report overoptimistic results which may hide potential limitations of the models. Moreover, the lack of consideration of frequency patterns and the heart rhythms can also limit the model performance. To fill in the gaps, we propose a framework for arrhythmia detection from IoT-based ECGs. The framework consists of two modules: a data cleaning module and a heartbeat classification module. Specifically, we propose two solutions for the heartbeat classification task, namely Dynamic Heartbeat Classification with Adjusted Features (DHCAF) and Multi-channel Heartbeat Convolution Neural Network (MCHCNN). DHCAF is a feature-engineering based approach, in which we introduce dynamic ensemble selection (DES) technique and develop a result regulator to improve classification performance. MCHCNN is deep-learning based solution that performs multi-channel convolutions to capture both temporal and frequency patterns from heartbeat to assist the classification. We evaluate the proposed framework with DHCAF and with MCHCNN on the well-known MIT-BIH-AR database, respectively. The results reported in this paper have proven the effectiveness of our framework.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cardiac arrhythmia is a type of cardiovascular diseases (CVDs) that threatens millions of people’s lives around the world. The easiest way to identify arrhythmia is to perform a manual inspection on 24 to 72 hours electrocardiograms (ECG). Traditionally, to have such long-term ECG recordings, patients need to wear a Holter Monitor for a continuous time period, which is a very uncomfortable experience. The rapid growth of Internet-of-Things (IoT) techniques has spawned novel ways, like Fitbit, Apple Watch, or Android Wear, for heart status tracking [47]. In comparison to the Holter Moniter, the IoT-based devices are more human-friendly because they have fewer cords and smaller-sizes, and cause fewer disruptions to patient’s daily routines. However, on the other hand, the prevalence of IoT-based devices has also resulted in a dramatic increase of ECG data, posing a great challenge to the ECG interpretation. Manual inspections become time-consuming and error-prone, which is no longer possible. An automated method is highly demanded to provide a cost-effective screening for arrhythmia and allow at-risk patients to receive timely treatments.
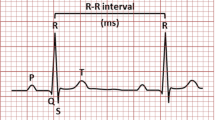
Heartbeat classification plays a crucial role in identification of arrhythmia. Basically, heartbeats can be classified into five classes: Normal(N), Supra-ventricular (S) ectopic, Ventricular (V ) ectopic, Fusion (F) and Unknown (Q) beats [6]. Particularly, most arrhythmias are found in S and V beats. Figure 1 presents a sample ECG segment, where the problematic heartbeats are highlighted by circles. It can be seen that the S beat exhibits a great morphological similarity in temporal dimension to the normal heartbeats. Since ECG recordings are mostly dominated by normal heartbeats for the majority of patients [22], the similarity brings a great difficulty in distinguishing the S beats from the normal ones.
Many research attempts have been made to provide solutions for automated heartbeat classification. The existing methods are roughly divided as feature-engineering based and deep-learning based methods. However, none of these methods has achieved a clinical significance. Most feature-engineering methods are facing a bottleneck of applying a standalone classifier and using a static feature set to classify all heartbeat samples [11, 15, 16, 31, 50]. This has been shown to cause huge impacts on identification of the problematic heartbeats. The deep-learning based methods are commonly limited by learning temporal patterns from the raw ECG heartbeats only. The frequency patterns and the RR-intervals have not been well considered to assist the classification. Moreover, to supply sufficient training data for driving the deep neural networks, many works [2, 3, 26, 49, 51, 54] followed a biased evaluation procedure, in which they synthesized heartbeat samples from the whole dataset and then randomly split all heartbeats for model training, validation and test. Consequently, heartbeats from the same patient are likely to appear in both the training and test datasets, leading to an over estimation of the model performance. The overoptimistic results may hide potential limitations of the neural networks.
Besides, data quality also presents challenges for an IoT-based arrhythmia detection method. First, the IoT-based heart rate sensors may vary the rate of measurement for battery preservation [7]. Second, the collected ECG recordings are likely interrupted by background noises and baseline wonders (the effect that the base axis (X-axis) of individual heartbeats appear to move up or down rather than being straight all the time).
To solve these problems, we propose a framework for arrhythmia detection from IoT-based ECGs. The framework consists of a data cleaning module and a heartbeat classification module. Specifically, we provide two novel solutions for the heartbeat classification task. The first one is a feature-engineering based method, in which we introduce the Dynamic Ensemble Selection (DES) technique and specially design a result regulator to improve the problematic heartbeats detection. The other one is a deep neural network that performs multi-channel convolutions in parallel to manage both temporal and frequency patterns to assist the classification. To remedy the impact brought by the lack of consideration of heart rhythms, the proposed network accepts heart rhythms (RR-intervals) as part of the input. In order to reveal the performance of the proposed methods in real-world practices, we evaluate the models on the benchmark MIT-BIH arrhythmia database following the inter-patient evaluation paradigm proposed in [16]. The paradigm divides the benchmark database into a training and a test dataset at patient level, making the heartbeat classification a significantly more difficult task.
The rest of this paper is structured as follows. Section 2 reviews current methods in heartbeat classification. Section 3 presents the proposed framework and the two embedded solutions for heartbeat classification. The experiment results and discussion are presented in Section 4. Section 5 concludes this paper and discusses the future work.
2 Related work
This section provides a comprehensive review of current methods for heartbeat classification. As mentioned before, the existing methods can be roughly allocated to either the feature-engineering based or the deep-learning based category. The differences between them are summarized in Table 1.
The feature-engineering based methods focus on signal feature extraction and classifier selection. Commonly used features includes RR-intervals [4, 11, 52], samples or segments of ECG curves [35], higher-order statistics [4, 17], wavelet coefficients [15, 20, 37], and signal energy [50]. They are mostly extracted from cardiac rhythm, or time/frequency domains. Feature correlation and effectiveness are important concerns for this type of methods. To avoid negative impacts of noisy data, techniques, like the floating sequential search [29] and the weighted LD model [18], must be employed to reduce the feature space. Regarding the selection of classifiers, the support vector machine (SVM) is the most widely used for its robustness, good generalization and computationally efficiency [1, 14]. Besides, the nearest neighbors (NN) and artificial neural networks (ANN) are also frequently found in the literature. The performances of current feature-engineering based methods are mainly limited by the application of single classifiers and the use of fixed features to classify all heartbeat types. On one hand, in consideration of the intra- and inter-subjects variations of the feature values, it is difficult for a single classifier to well handle a wide region of the feature space [53]. Although some ensemble methods, such as random forest [4] and ensemble of support vector machine [24], have been employed to remedy the disadvantages, the problem is still open because the diversity of the traditional ensembles is relatively low. On the other hand, using fixed features tends to make sporadically occurred S beats be wrongly classified as V beats because both heartbeats types exhibits anomalies in heart rhythms.
By contrast, the deep-learning based methods are more straightforward and integrated, in which features and classifiers are not concerns. They provide end-to-end solutions to the heartbeat classification task. The existing deep learning models are mainly extensions of convolution neural network (CNN) [2, 3, 26, 38] or combinations of CNN and recurrent neural network (RNN) [49, 51]. However, most of the CNN models are limited by the lack of consideration of frequency patterns and the heart rhythm to assist the classification. Moreover, in order to provide enough training data, many of them are evaluated in an ideal experimental setting where heartbeats from the same patient are allowed to appear in both training and test sets. The results can not reveal the true performances of the models in real-world practices and also may hide potential limitations of the methods. As compared to the feature-engineering based methods, both the results and the intermediate process of deep neural networks are less explainable. This is a potential impediment that prevents deep learning models from being widely applied in practices because explainability is important for clinicians to justify and rationalize the model outcome.
3 The proposed framework for arrhythmia detection
The proposed framework for arrhythmia detection from IoT-based ECGs is presented in this section. Figure 2 shows the framework architecture and the whole life-cycle of arrhythmia detection from IoT-based ECGs. The framework consists of a data cleaning module and a heartbeat classification module. It accepts raw ECG signals that collected from different IoT devices as input and outputs predictions for individual heartbeats.
Architecture of the proposed framework. The whole life-cycle of arrhythmia detection from IoT-based ECGs includes 4 phases: data collection, storage, analysis and results notification. Specifically, ECG sensing network generates ECG recordings for patients and transmits the produced data to the IoT cloud, where fast access storage are conducted. The proposed framework is deployed in the IoT cloud to provide data analysis. Results from the framework will be pushed to patients’ ends via Internet
To reduce the impact of noisy data on the prediction accuracy, the input signals are performed a series of preprocessing, such as frequency calibration, baseline correction, and noise reduction, before heartbeat classification. We propose two solutions, namely Dynamic Heartbeat Classification with Adjusted Features (DHCAF) and Multi-channel Heartbeat Convolution Neural Network (MCHCNN), for the heartbeat classification task. DHCAF is a feature-engineering based method, whereas MCHCNN is a deep-learning based method.
Details of the data cleaning module and two heartbeat classification solutions are presented below.
3.1 Data cleaning module
Frequency calibration
To avoid the possible bias in sampling frequency caused by different ECG collectors, we develop a frequency calibration component to re-sample all incoming ECG recordings to 360 Hz at the input of the system.
Baseline correction
To correct the baseline wanders, we process each ECG recording with a 200-ms width median filter followed by a 600-ms median filter to obtain the recording baseline, and then subtract the baseline from the raw ECG recording to get the baseline corrected data.
Noise reduction
For noise reduction, we apply discrete wavelet transform [39] with Daubechies-4 mother wavelet function to remove recordings’ Gaussian white noise. The Daubechies-4 function has short vanishing moment, which is ideal for analyzing signals like ECG with sudden changes. Concretely, in the noise reduction component, the baseline corrected recordings are decomposed to different frequency bands with various resolutions. The coefficients of detail information (cDx) in each frequency band is then processed by a high-pass filter with a threshold value
where n indicates the length of the input recording. Coefficients that blocked by the filter are set to zero. Finally, the clean recordings are obtained by employing inverse discrete wavelet transform on all the coefficients.
Heartbeat segmentation
The clean signals are segmented to individual heartbeats by taking advantage of the R peak locations that detected by the Pan-Tompkins algorithm [36]. For each R peak, 90 samples (250 ms) before R peak and 144 samples (400 ms) after R peak are taken to represent a heartbeat, which is long enough to catch samples to represent the re-polarization of ventricles and short enough to exclude the neighbor heartbeats [4].
3.2 Dynamic heartbeat classification with adjusted features
Architecture of the proposed DHCAF is shown in Figure 3. The model contains 4 processing stages: Feature Extraction, Classifier Pool Training, Classifier Selection and Prediction, and Result Refinement.
Feature extraction
In this stage, three types of features are extracted to represent individual heartbeats: RR-intervals, higher order statistics and wavelet coefficients.
As experimentally proven in [52], the RR-interval is one of the most indispensable features for heartbeat classification and it has great capacity to tell both the S and V beats from the normal beats. In this work, four types of RR-intervals are extracted from ECG signals: pre_RR, post_RR, local_RR and global_RR [30]. The RR-intervals can significantly vary with patients. To reduce the negative impact of the variation, we normalize the RR-intervals in the way below:
where ds.pre_RR denotes the average of all pre_RRs in the ds that the heartbeat belongs to, and so on.
Regarding the higher order statistics (HOS), it is reported being useful in catching subtle changes in ECG data [32]. In this work, the skewness (3rd order statistics) and kurtosis (4th order statistics) are calculated for each heartbeat. They can be mathematically defined as follows, where X1..., N denotes all the data samples in a signal, \(\bar {X}\) is the mean and s is the standard deviation.
The wavelet coefficients provide both time and frequency domain information of a signal, which is claimed to be the best features of ECG signal [30]. The choice of the mother wavelet function used for coefficients extraction is crucial to the final classification performance. In this work, the Haar wavelet function is chosen because of its simplicity and that it has been demonstrated as the ideal wavelet for short time signal analysis [50].
Classifier pool training
In this stage, a collection of classifiers, including multi-layers perceptron, support vector machine (SVM), linear SVM, Bayesian model with Gaussian kernel, decision tree, and K-nearest neighbors model, are trained using the extracted features, to create an accurate and diverse classifier pool.
Classifier selection and prediction
This stage plays a core role in the model. The Dynamic Ensemble Selection (DES) [13] technique is introduced in this stage to select the most competent classifiers for making predictions of the test samples. It helps to solve both the intra- and inter-subjects variations of the feature values.
In DES, the competence of a classifier in the pool is measured by its performance over a local region of the feature space where the testing sample is located. Methods for defining a local region includes clustering [28], k-nearest neighbors [40], potential function model [44, 45] and decision space [9]. The criterion for measuring the performance of a base classifier can be divided as individual-based and group-based criterion. In the individual-based criterion, each base classifier is independently measured by evaluation metrics such as ranking, accuracy, probabilistic, behavior [9], meta-learning [12]. In the group-based criterion, the performance of a base classifier relates to its iterations with other classifiers in the pool. For example, diversity, data handling [46] and ambiguity [19] are widely used group-based performance metrics.
Once the candidates classifiers are selected, aggregation of results from these classifiers is then performed to give a united decision. There are three main strategies for results combination: static combiner, trained combiner and dynamic weighting. The majority voting scheme is a representative static combiner, which is also commonly used in the traditional ensemble methods. In trainable combiners, the outputs of the selected based classifiers are used as the input features for another learning algorithm, such as [8, 33]. In dynamic weighting, higher weight value will be allocated to the most competent classifier and then the outputs of all the weighted classifiers are aggregated to give the united decision.
Currently, prevalent DES techniques that can be used in this stage include DES-KL [45], DES-KNN [41], KNORA-E [27], KNORA-U [27], KNOP [9], DES-P [45], DES-RRC [44], and META-DES [12]. Extensive experiments have been done to evaluate these DES techniques in our previous work [21]. The results showed that there is no significant difference of their performances. We adopt the META-DES in this work since it has reported superior performances in a wider range of datasets.
Result refinement
The aggregated result from the previous stage will be refine in this stage by our adjusted features strategy. Specifically, we train an SVM classifier with only HOS and wavelet coefficients (the RR-intervals are removed) to improve the results of S and V beats. The rationale of such a classification strategy is that the sensitivities to certain feature varies with heartbeat types [52] . For instance, the RR-intervals are indispensable to for identifying disease heartbeats from the normal ones. However, the RR-intervals can also cause troubles to make a distinction between different kinds of disease heartbeats, such as S and V beats.
3.3 Multi-channels heartbeat convolution neural network
Architecture of the proposed Multi-channels Convolution Neural Network (MCHCNN) is presented in Figure 4. The network accepts two inputs: raw ECG heartbeat and heart rhythm (RR-intervals). As motivated by an electroencephalogram (EEG) processing network [42] which uses different sizes of convolution filters to capture temporal and frequency patterns from EEG signals, the proposed MCHCNN perform 3 channels of convolutions in parallel on the input ECG heartbeats to extract the temporal and frequency information. The convolution filter size varies with channels, where the smaller filter is used to capture temporal patterns and the larger filter is used to capture frequency patterns. We denote the convolution process as Conv(x, y) in Figure 4, where x is the convolution filter size and y is the amount of the output feature maps. Each convolution operation is followed by a batch normalization and a ReLu activation. The batch normalization normalizes the output of the convolution by subtracting the batch mean and dividing by the batch standard deviation, which reduces the problem of internal covariate shift [25] and overfitting. The introduction of a ReLu activation is to allow the network to extract nonlinear features.
Every three stacked convolutions are wrapped into a building block and bypassed by a shortcut connection. The learned features are added to the shortcut at the end of each building block. Such a design helps to reduce the network degradation problem [23]. Each channel contains 3 building blocks. Learned features from the three channels are integrated by addition before a pooling layer. The pooling layer is used to reduce feature dimensions, after which the learned features are reduced to half-size. It helps to reduce the number of parameters in the following fully connected layer and lower the risk of overfitting.
A Rhythm Integration layer is specially designed to concatenate the learned features and the input heart rhythms. It reduces the impact brought by the lack of consideration of heart rhythms on identification of disease heartbeats in many existing network models.
Next, the dense layer is used to learn non-linear combinations of the learned features. The softmax layer gives the probabilities of the each heartbeat type.
4 Evaluation
In this section, we evaluate the proposed framework equipped with DHCAF and with MCHCNN, respectively. The MIT-BIH-AR database [34] is used as the benchmark database. It is the most representative database for arrhythmia detection and it has been used for most of the published research [16]. Details of the database are given below.
4.1 The MIT-BIH-AR database
The MIT-BIH-AR database contains 48 two-leads ambulatory ECG records from 47 patients (22 females and 25 males). Each record has approximately 30 minutes in length. These recordings were digitized at 360Hz. For most of them, the first lead is modified limb lead II (except for the recording 114). The second lead is a pericardial lead (usually V 1, sometimes are V 2, V 4 or V 5, depending on subjects).
In order to reveal the performance of the proposed framework, we follow the evaluation paradigm proposed in [16] to divide the database into a training and a test dataset. The paradigm avoids heartbeats of the same patient appearing in both training and test stages, ensuring a fair evaluation. Table 2 shows the division details, where DS1 is the training set and DS2 denotes the test set.
Noticing that DS1 is extremely imbalanced and dominated by N beats, we apply the SMOTEENN technique [5, 10, 43] on DS1 to over-sample the minority heartbeats (S and V ) to the same amount of N beats.
4.2 Evaluation metrics
Evaluation metrics used in this work are sensitivity (Se), positive predictive value (+ P) and accuracy value (Acc),as formulated below,
where TP, TN, FP and FN denotes true positive, true negative, false positive and false negative, respectively, and \(\sum \) represents the amount of instances in the data set. According to the AAMI standard [16], penalties would not be applied for the misclassification of F and Q beats, as they are naturally unclassifiable.
4.3 Results of the proposed framework
Confusion matrixs of the proposed framework with DHCAF and with MCHCNN on DS2 are presented in Table 3. We summarize the results and compare our framework with multiple state-of-the-art methods in Table 4. All results reported in Table 4 are obtained under the same evaluation paradigm on DS2 of MIT-BIH-AR database.
It is clear that the proposed framework with DHCAF achieves the best sensitivity of both class S and V, and maintain a good performance in overall accuracy and classification of class N. Shan’s model [11] obtains the highest accuracy and class N sensitivity. However, it fails in the detection of class S, with the sensitivity of class S being merely 29.5%, which limits the model’s practical significance. The proposed framework with MCHCNN outperforms DHCAF in terms of the overall accuracy, sensitivity of N beats, and the positive predictive value of S beat, but its performance on sensitivity of S beat is less satisfied. In fact, it can be found that the positive predictive values of S beats for most listed works in Table 4 are relatively low, as compared to other metrics. This is mainly caused by some N beats being misclassified as S beats. As mentioned in the Introduction section, the similar QRS complex and the data imbalance problem have introduced a great difficulty in distinguishing the S from the N beats. We compare the proposed framework with MCHCNN to another deep-learning based method by Sellami et al. [38], which reports model performance under the same unbiased evaluation. The results show that Sellami’s work has achieved a promising performance on identification of both the problematic S and V beats, being close to that of the proposed framework with DHCAF. However, this is at the cost of the overall accuracy and sensitivity of normal beats. In real-world practices, the large amount of misclassification of normal heartbeats as the disease heartbeats will result in an unnecessary waste in medical resources.
From the above analysis, the proposed framework with DHCAF is believed to be a more appropriate choice than other listed works for cardiac arrhythmia detection, because it achieves the best identification performance on disease heartbeats while maintaining a good overall accuracy and classification performance on the normal heartbeats.
4.4 Ablative analysis
We perform ablative analysis for the proposed DHCAF and MCHCNN to demonstrate the effectiveness of the model architectures. The results are summarized in Table 5 and Table 6, respectively.
Two baselines are used in the ablative analysis for DHCAF. One is DHCAF with the result refinement stage removed. The other one is DHCAF with the dynamic ensemble selection classification of DHCAF replaced by ensemble of SVM classification. It is apparent that the result regulator has made unique contributions to DHCAF, with which the overall accuracy, sensitivity of class S, and positive predictive of class S and V are visibly increased. On the other hand, the poor classification performance of the SVM ensemble has demonstrated the importance of the introduction of dynamic ensemble selection to the proposed method.
As discussed in Sections 1 and 2, many existing deep neural network models have not taken the heart rhythms into account for heartbeat classification, but this limitation is hidden by the over-optimistic results obtained in a biased evaluation paradigm. In the ablative test of MCHCNN, we want to know the actual impact of heart rhythms on model performances. Therefore, we construct a baseline MCHCNN which only take raw ECG heartbeats as input. The results, as seen in Table 6, indicate that heart rhythm (RR-intervals) are necessary for identification of the disease heartbeats. Without consideration of heart rhythm, the baseline can hardly detect S beats. The detection on V beats is also affected. The outcome is in line with the medical fact. As we can see in Figure 1, most V beats present a huge morphological difference with other heartbeats. That is why the baseline can still maintain 73.8% sensitivity on V beats. However, for S beats, the heart rhythm is essential for distinguishing them from the normal heartbeats.
Although heartbeat rhythms has been part of the input to the proposed MCHCNN, the S beats detection performance is still less satisfied. This indicates that the raw heartbeat rhythms provide limited assistance to our MCHCNN in identification of S beats. A possible explanation is that the heartbeat rhythms are not integrated well to the network and also easily affected by other learned features. A future study is needed to investigate this issue.
5 Conclusion
Millions of people around the world are suffering from cardiac arrhythmia. In this work, we propose a framework for automated arrhythmia detection from IoT-based ECGs. The framework consists of two modules: a data cleaning module to tackle the challenges presented by IoT-based ECGs, and a heartbeat classification module for identification the disease heartbeats. Specifically, we proposed two solutions, DHCAF and MCHCNN, for the heartbeat classification task. DHCAF is a feature-engineering based method which introduces the dynamic ensemble selection techniques and uses an adjust-feature strategy to assist disease heartbeats identification. By contrast, MCHCNN is an end-to-end solution that performs multi-channel convolutions to capture both the temporal and frequency information from the raw heartbeats to improve the classification performance. We evaluate the proposed framework on the MIT-BIH-AR database under the inter-patient evaluation paradigm. The results show that the proposed framework with DHCAF is a qualified candidate for automated arrhythmia detection from IoT-based ECGs. Besides, although the S beats detection performance of MCHCNN is less satisfied, the network still provide some insights to our future study.
This work is a first step to provide a solution for the automated arrhythmia detection in the era of Internet-of-Things. In our next study, we aim to investigate a more effective way for integration of the heart rhythms into a neural network.
References
Abawajy, J.H., Kelarev, A.V., Chowdhury, M.: Multistage approach for clustering and classification of ecg data. Comput. Meth. Program. Biomed. 112(3), 720–730 (2013)
Acharya, U.R., Fujita, H., Oh, S.L., Hagiwara, Y., Tan, J.H., Adam, M.: Application of deep convolutional neural network for automated detection of myocardial infarction using ecg signals. Inform. Sci. 415, 190–198 (2017)
Acharya, U.R., Oh, S.L., Hagiwara, Y., Tan, J.H., Adam, M., Gertych, A., San Tan, R.: A deep convolutional neural network model to classify heartbeats. Comput. Biol. Med. 89, 389–396 (2017)
Afkhami, R.G., Azarnia, G., Tinati, M.A.: Cardiac arrhythmia classification using statistical and mixture modeling features of ecg signals. Pattern Recogn. Lett. 70, 45–51 (2016)
Alejo, R., Sotoca, J.M., Valdovinos, R.M., Toribio, P.: Edited nearest neighbor rule for improving neural networks classifications. In: International Symposium on Neural Networks, pp 303–310. Springer (2010)
ANSI/AAMI: Testing and reporting performance results of cardiac rhythm and st segment measurement algorithms. Association for the Advancement of Medical Instrumentation -AAMI ISO EC57 (Unknown Month 1998)
Ballinger, B., Hsieh, J., Singh, A., Sohoni, N., Wang, J., Tison, G.H., Marcus, G.M., Sanchez, J.M., Maguire, C., Olgin, J.E., et al: Deepheart: semi-supervised sequence learning for cardiovascular risk prediction. In: Thirty-Second AAAI Conference on Artificial Intelligence (2018)
Britto, A.S. Jr, Sabourin, R., Oliveira, L.E.: Dynamic selection of classifiers—a comprehensive review. Pattern Recogn. 47(11), 3665–3680 (2014)
Cavalin, P.R., Sabourin, R., Suen, C.Y.: Dynamic selection approaches for multiple classifier systems. Neural Comput. Appl. 22(3-4), 673–688 (2013)
Chawla, N.V., Bowyer, K.W., Hall, L.O., Kegelmeyer, W.P.: Smote: synthetic minority over-sampling technique. J. Artificial Intell. Res. 16, 321–357 (2002)
Chen, S., Hua, W., Li, Z., Li, J., Gao, X.: Heartbeat classification using projected and dynamic features of ecg signal. Biomed. Signal Process. Control 31, 165–173 (2017)
Cruz, R.M., Sabourin, R., Cavalcanti, G.D.: Meta-des. oracle: meta-learning and feature selection for dynamic ensemble selection. Information Fusion 38, 84–103 (2017)
Cruz, R.M., Sabourin, R., Cavalcanti, G.D.: Dynamic classifier selection: recent advances and perspectives. Information Fusion 41, 195–216 (2018)
Daamouche, A., Hamami, L., Alajlan, N., Melgani, F.: A wavelet optimization approach for ecg signal classification. Biomed. Signal Process. Control 7(4), 342–349 (2012)
De Albuquerque, V.H.C., Nunes, T.M., Pereira, D.R., Luz E.J.d.S., Menotti, D., Papa, J.P., Tavares, J.M.R.: Robust automated cardiac arrhythmia detection in ecg beat signals. Neural Comput. Appl. 29(3), 679–693 (2018)
De Chazal, P., O’Dwyer, M., Reilly, R.B.: Automatic classification of heartbeats using ecg morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 51 (7), 1196–1206 (2004)
De Lannoy, G., François, D., Delbeke, J., Verleysen, M.: Weighted conditional random fields for supervised interpatient heartbeat classification. IEEE Trans. Biomed. Eng. 59(1), 241–247 (2012)
Doquire, G., De Lannoy, G., François, D., Verleysen, M.: Feature selection for interpatient supervised heart beat classification. Comput. Intell. Neurosci. 2011, 1 (2011)
Dos Santos, E.M., Sabourin, R., Maupin, P.: A dynamic overproduce-and-choose strategy for the selection of classifier ensembles. Pattern Recogn. 41(10), 2993–3009 (2008)
Güler, İ., ÜBeylı, E.D.: Ecg beat classifier designed by combined neural network model. Pattern Recogn. 38(2), 199–208 (2005)
He, J., Rong, J., Sun, L., Wang, H., Zhang, Y., Ma, J.: D-Ecg: a dynamic framework for cardiac arrhythmia detection from Iot-Based Ecgs. In: International Conference on Web Information Systems Engineering, pp 85–99. Springer (2018)
He, J., Sun, L., Rong, J., Wang, H., Zhang, Y.: A pyramid-like model for heartbeat classification from ecg recordings. Plos one 13(11), e0206593 (2018)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp 770–778 (2016)
Huang, H., Liu, J., Zhu, Q., Wang, R., Hu, G.: A new hierarchical method for inter-patient heartbeat classification using random projections and rr intervals. Biomed. Eng. Online 13(1), 90 (2014)
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. arXiv:1502.03167 (2015)
Kiranyaz, S., Ince, T., Gabbouj, M.: Real-time patient-specific ecg classification by 1-d convolutional neural networks. IEEE Trans. Biomed. Eng. 63(3), 664–675 (2016)
Ko, A.H., Sabourin, R., Britto, A.S. Jr: From dynamic classifier selection to dynamic ensemble selection. Pattern Recogn. 41(5), 1718–1731 (2008)
Lin, C., Chen, W., Qiu, C., Wu, Y., Krishnan, S., Zou, Q.: Libd3c: ensemble classifiers with a clustering and dynamic selection strategy. Neurocomputing 123, 424–435 (2014)
Llamedo, M., Martínez, J.P.: Heartbeat classification using feature selection driven by database generalization criteria. IEEE Trans. Biomed. Eng. 58(3), 616–625 (2011)
Luz, E.J.D.S., Schwartz, W.R., Cámara-Chávez, G., Menotti, D.: Ecg-based heartbeat classification for arrhythmia detection: a survey. Comput. Meth. Programs Biomed. 127, 144–164 (2016)
Ma, J., Sun, L., Wang, H., Zhang, Y., Aickelin, U.: Supervised anomaly detection in uncertain pseudoperiodic data streams. ACM Trans. Internet Technol. (TOIT) 16(1), 4 (2016)
Martis, R.J., Acharya, U.R., Ray, A.K., Chakraborty, C.: Application of higher order cumulants to ecg signals for the cardiac health diagnosis. In: 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Embc, pp 1697–1700. IEEE (2011)
Masoudnia, S., Ebrahimpour, R.: Mixture of experts: a literature survey. Artif. Intell. Rev. 42(2), 275–293 (2014)
Moody, G.B., Mark, R.G.: The impact of the mit-bih arrhythmia database. IEEE Eng. Med. Biol. Mag. 20(3), 45–50 (2001)
Özbay, Y., Tezel, G.: A new method for classification of ecg arrhythmias using neural network with adaptive activation function. Digital Signal Processing 20 (4), 1040–1049 (2010)
Pan, J., Tompkins, W.J.: A real-time qrs detection algorithm. IEEE Trans. Biomed. Eng 32(3), 230–236 (1985)
Sahoo, S., Kanungo, B., Behera, S., Sabut, S.: Multiresolution wavelet transform based feature extraction and ecg classification to detect cardiac abnormalities. Measurement 108, 55–66 (2017)
Sellami, A., Hwang, H.: A robust deep convolutional neural network with batch-weighted loss for heartbeat classification. Expert Syst. Appl. 122, 75–84 (2019)
Shensa, M.J.: The discrete wavelet transform: wedding the a trous and mallat algorithms. IEEE Trans. Signal Process. 40(10), 2464–2482 (1992)
Sierra, B., Lazkano, E., Irigoien, I., Jauregi, E., Mendialdua, I.: K nearest neighbor equality: giving equal chance to all existing classes. Inform. Sci. 181 (23), 5158–5168 (2011)
Soares, R.G., Santana, A., Canuto, A.M., de Souto, M.C.P.: Using accuracy and diversity to select classifiers to build ensembles. In: International Joint Conference on Neural Networks, 2006. IJCNN’06, pp 1310–1316. IEEE (2006)
Supratak, A., Dong, H., Wu, C., Guo, Y.: Deepsleepnet: a model for automatic sleep stage scoring based on raw single-channel eeg. IEEE Trans. Neural Syst. Rehabil. Eng. 25(11), 1998–2008 (2017)
Wilson, D.L.: Asymptotic properties of nearest neighbor rules using edited data. IEEE Trans. Sys. Man Cyber. SMC-2(3), 408–421 (1972)
Woloszynski, T., Kurzynski, M.: A probabilistic model of classifier competence for dynamic ensemble selection. Pattern Recogn. 44(10-11), 2656–2668 (2011)
Woloszynski, T., Kurzynski, M., Podsiadlo, P., Stachowiak, G.W.: A measure of competence based on random classification for dynamic ensemble selection. Information Fusion 13(3), 207–213 (2012)
Xiao, J., Xie, L., He, C., Jiang, X.: Dynamic classifier ensemble model for customer classification with imbalanced class distribution. Expert Syst. Appl. 39 (3), 3668–3675 (2012)
Yang, Z., Zhou, Q., Lei, L., Zheng, K., Xiang, W.: An iot-cloud based wearable ecg monitoring system for smart healthcare. J. Med. Sys. 40(12), 286 (2016)
Ye, C., Kumar, B.V., Coimbra, M.T.: Heartbeat classification using morphological and dynamic features of ecg signals. IEEE Trans. Biomed. Eng. 59(10), 2930–2941 (2012)
Yildirim, Ö.: A novel wavelet sequence based on deep bidirectional lstm network model for ecg signal classification. Comput. Biol. Med. 96, 189–202 (2018)
Yu, S.N., Chen, Y.H.: Electrocardiogram beat classification based on wavelet transformation and probabilistic neural network. Pattern Recogn. Lett. 28(10), 1142–1150 (2007)
Zhang, C., Wang, G., Zhao, J., Gao, P., Lin, J., Yang, H.: Patient-specific ecg classification based on recurrent neural networks and clustering technique. In: 2017 13th IASTED International Conference on Biomedical Engineering (Biomed), pp 63–67. IEEE (2017)
Zhang, Z., Dong, J., Luo, X., Choi, K.S., Wu, X.: Heartbeat classification using disease-specific feature selection. Comput. Biol. Med. 46, 79–89 (2014)
Zhu, X., Wu, X., Yang, Y.: Dynamic classifier selection for effective mining from noisy data streams. In: Fourth IEEE International Conference On Data Mining, 2004. ICDM’04, pp 305–312. IEEE (2004)
Zubair, M., Kim, J., Yoon, C.: An automated ecg beat classification system using convolutional neural networks. In: 2016 6th International Conference on IT Convergence and Security (ICITCS), pp 1–5. IEEE (2016)
Acknowledgments
This work is supported by the NSFC (No. 61672161)and the National Natural Science Foundation of China (Grants No. 61702274).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article belongs to the Topical Collection: Special Issue on Web Information Systems Engineering 2018
Guest Editors: Hakim Hacid, Wojciech Cellary, Hua Wang and Yanchun Zhang
Rights and permissions
About this article
Cite this article
He, J., Rong, J., Sun, L. et al. A framework for cardiac arrhythmia detection from IoT-based ECGs. World Wide Web 23, 2835–2850 (2020). https://doi.org/10.1007/s11280-019-00776-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11280-019-00776-9