Abstract
Viral promoters can be used to drive heterologous gene expression in transgenic plants. As part of our quest to look for new promoters, we have explored, for the first time, the promoters of okra enation leaf curl virus (OELCuV), a begomovirus infecting okra (Abelmoschus esculentus). The Rep and CP promoters of OELCuV fused with the gfp reporter gene, were expressed transiently in the natural host okra and the laboratory host cotton and Nicotiana benthamiana. The expression levels of the promoters were quantified through confocal laser scanning microscopy and GFP assay in N. benthamiana and okra. The results indicated that the Rep promoter was more active than the CP promoter, whose activity was similar to that of CaMV 35S promoter. Additionally, the Rep and CP promoters showed increase of expression, probably due to transactivation, when assayed following inoculation of OELCuV and betasatellite DNAs in cotton plants. A moderate increase in promoter activity in N. benthamiana was also seen, when assayed following the inoculation of the heterologous begomovirus Sri Lankan cassava mosaic virus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ability to produce transgenic plants with improved agronomic traits represents an important breakthrough in crop improvement [1]. Transgene expression is controlled by promoters, defined by DNA present 5' to the gene's coding region that contains specific sequences recognized by the proteins involved in initiating transcription. Hence, the optimal expression of the transgene is largely determined by the promoter present in the DNA construct used during genetic transformation [1,2,3,4].
The cauliflower mosaic virus (CaMV) 35S promoter is most frequently used for the expression of transgenes in plants. Apart from CaMV 35S, several other plant viral promoters, taken from single-stranded DNA viruses (geminiviruses) have been identified and characterized [3]. Although, most of the geminiviral promoters are weaker than CaMV 35S, some have been reported to express transgenes to levels comparable or better than the CaMV 35S promoter. The geminiviral promoters investigated include those derived from African cassava mosaic virus (ACMV) [5], blechum interveinal chlorosis virus [6], cotton leaf curl Burewala virus (CLCuBuV) [7, 8], cotton leaf curl Kokhran virus [9], cotton leaf curl Multan virus (CLCuMuV) [10], maize streak virus [11], mungbean yellow mosaic virus (MYMV) [12, 13], mungbean yellow mosaic India virus [14], tomato golden mosaic virus (TGMV) [15], tomato leaf curl virus [16], and wheat dwarf virus [17].
The family Geminiviridae, which includes plant-infecting viruses having single-stranded circular DNA genome, consists of fourteen genera, the largest of which is Begomovirus that has over 400 species [18,19,20]. Begomoviruses are either bipartite having two genomic components (DNA-A and DNA-B) or monopartite having single genomic component. Six or seven open reading frames (ORFs) are encoded by DNA-A, of which four or five (Rep/AC1, TrAP/AC2, REn/AC3, AC4 and AC5) are in complementary-sense orientations and two (CP/AV1 and AV2) are in virion-sense orientation [20, 21]. Virion-sense and complementary sense genes are separated by an intergenic region (IR) present between 5' ends of the first complimentary and virion-sense ORFs [8, 20]. DNA-B encodes two ORFs NSP/BV1 and MP/BC1 in virion-sense and complimentary sense directions respectively. Monopartite begomoviruses have a single genomic component that is similar to the DNA-A of bipartite begomoviruses [20].
Functional analysis of the IR of monopartite and common region of bipartite begomoviruses showed that these regions possess promoter activity [7, 8, 10, 13]. These promoters were mapped between 5ʹ ends of the first complementary-sense gene (encoding the replication-associated protein: Rep) and virion-sense gene (encoding the coat protein: CP/AV1), or the V2/AV2 ORF present upstream and overlapping AV1 [8, 12, 22,23,24]. The rep promoter of ACMV, TGMV, CLCuMuV and CLCuBuV showed stronger activities than that of CP promoter in the absence of transcriptional activator protein (TrAP) [5, 8, 10, 25].
Transactivator protein (encoded by AC2/TrAP) regulates the activity of the virion-sense promoter for which, a conserved late element (CLE) has been found to be important [6, 26, 27]. Both complementary-sense and virion-sense genes are transactivated by the TrAP, but the degree of transactivation varies. TrAP transactivates the late viral genes AV1 (CP) of DNA-A and BV1 of DNA-B of bipartite MYMV [12]. The late promoters driving AV1, AV2, BV1, and BC1 exhibit significant (15- to 300-fold) TrAP-mediated transactivation, whereas the early promoters driving AC1, AC4, AC2, and AC3 exhibit much less prominent (two to fourfold) activation [12]. Several previous reports have suggested the involvement of specific sequence elements, including CLE (GTGGTCCC), TATA box-associated Composite Elements (TACE, ACTT-N7-AAGT) and Abscisic acid Responsive Elements (ABRE, ACGTGG) in the above transactivation [6, 27,28,29] in various geminiviruses.
Okra enation leaf curl virus (OELCuV) is a monopartite begomovirus infecting hosts such as okra (Abelmoschus esculentus) and cotton (Gossypium hirsutum) in the Indian subcontinent [30,31,32,33]. OELCuV is associated with smaller molecules known as betasatellites [31, 34]. The infectivity of partial dimer clones of OELCuV and associated bhendi yellow vein mosaic betasatellite (BYVMB) has been demonstrated recently in the natural host okra as well as in the experimental host Nicotiana benthamiana [33]. Sri Lankan cassava mosaic virus (SLCMV) is a bipartite begomovirus infecting cassava (Manihot esculenta) in India and Sri Lanka, which can also infect N. benthamiana under laboratory conditions [35].
In this study, the approximately 430 bp of DNA fragment upstream of the translation initiation codons of the Rep/C1 and V1/CP genes from the IR of OELCuV were isolated and their ability to express reporter gene (gfp) were characterized. The accumulation of GFP driven by the above promoters was quantified and compared to CaMV 35S promoter transiently in N. benthamiana, cotton and okra plants. Subsequently, the accumulation of GFP was checked in cotton in the presence and absence of OELCuV (cognate virus) to determine a possible transactivation of the 35S and OELCuV V1/CP and C1/Rep promoters following viral infection. Finally, the above experiment was conducted in N. benthamiana in the presence and absence of SLCMV, to study possible transactivation following infection with an unrelated (non-cognate) virus.
Materials and methods
Designing and development of C1/Rep and V1/CP promoter constructs
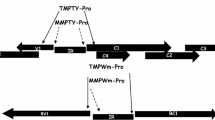
The IR of OELCuV (GenBank Accession No. MK069433) containing Rep and CP promoters was PCR amplified using ProRep For and ProRep Rev, and ProCP For and Pro CP Rev primers, respectively (Table 1). The amplified DNA fragments of approximately 430 bp was digested with NcoI and XbaI and cloned into 5' upstream of gfp in pCAMBIA1302 vector. Cloning was confirmed by PCR, restriction digestion and sequencing. The constructs representing gfp under the control of Rep and CP promoters were named as pRep-GFP and pCP-GFP, respectively. The gfp under the control of CaMV 35S promoter as in pCAMBIA1032 (p35S-GFP) and gfp without promoter pGFP0029 [8] were used as positive and negative controls, respectively (Fig. 1).
Schematic representation of OELCuV Rep and CP promoter constructs, CaMV35S-GFP and promoter-less GFP (pGFP0029) constructs (OELCuV: Okra enation leaf curl virus; Rep: Replication initiation protein; CP: Coat protein; CaMV 35S: Cauliflower mosaic virus 35S promoter; HygR: Hygromycin resistance gene; KanR: Kanamycin resistance gene; MCS: Multiple cloning site; GFP: Green fluorescent protein; Ter: Terminator; NOSTer: Nopaline synthase terminator; LB: Left-border sequence of T-DNA; RB: Right-border sequence of T-DNA)
Transient expression of GFP under control of C1/Rep, V1/CP and 35S promoters in N. benthamiana
The individual promoter constructs viz. pRep-GFP, pCP-GFP, p35S-GFP and pGFP0029 (along with helper plasmid pSoup) were mobilized into Agrobacterium tumefaciens GV3101 using freeze–thaw method [36]. Four-week-old N. benthamiana leaves were agroinfiltrated using these promoter constructs, following the procedure described by Khan et al. (2015) [8] with modifications. A single colony of A. tumefaciens GV3101 with individual gene constructs grew in 10 ml Luria–Bertani (LB) broth (50 μg/ml kanamycin and 50 μg/ml rifampicin) at 28 °C for 48 h (200 rpm). The primary culture (500 µl) was inoculated into 50 ml LB medium (100 μM acetosyringone, 50 μg/ml kanamycin and 50 μg/ml rifampicin) and was grown until OD600 reached 0.6. Bacterial cells were harvested, resuspended in 10 mM MES pH 5.6, 10 mM MgCl2, 100 μM acetosyringone, and incubated for 4 h before infiltrating 100 µl into the abaxial side of each intact leaf using a 1 ml needleless syringe. A total of 60 plants were infiltrated, i.e., 15 plants per construct and the experiment was repeated thrice. At 3 days post-infiltration (DPI) the inoculated leaves were harvested for GFP assay.
Localization of GFP in agroinfiltrated N. benthamiana using confocal laser scanning microscopy
The expression of gfp under the control of individual promoters were localized in N. benthamiana leaves at 3 DPI using Confocal Laser Scanning Microscopy (CLSM, Leica Microsystems Germany) as described earlier [8]. The agroinfiltrated portion of leaves were cut in 1 cm2 and put on slide and the fluorescent images were captured through CLSM with fluorescence emissions between 501 and 598 nm.
Transient expression of GFP under control of Rep, CP and 35S promoters in okra and cotton
Seeds of A. esculentus L. N-568 (Nirmal seeds Pvt. Ltd., Maharashtra, India) and Gossypium hirsutum HS6 (CICR, Sirsa, India) were sown in pots containing sterilized soil and kept at 28–30 °C for germination and allow to grow in growth chamber (Conviron, Canada) with a 16 h light and 8 h dark cycle [33]. Agrobacterium tumefaciens cultures of individual promoter constructs, grown to O.D. 1.0 at 600 nm were prepared as described earlier [33]. The cotyledonary leaves of two-week-old cotton and cotyledonary and true leaves of two-week-old okra plants were infiltrated with the agrobacterium cultures (agroinfiltrated) using 1 ml syringe without needle. The expression of gfp was monitored using UV lamp (UVP, Black-Ray®, USA) in intact leaves at 3 DPI.
GFP assay
GFP quantification was done at 3 DPI following the method described earlier [37] with some modifications. About 100 mg agroinfiltrated leaves were ground in liquid nitrogen and transferred into protein extraction buffer (50 mM Tris pH 8.0, 150 mM NaCl, 0.1% Triton X-100 and 0.1% β-mercaptoethanol), centrifuged at 13,000×g for 15 min at 4 °C (Eppendorf, Germany) and the supernatants were transferred into fresh microcentrifuge tube. The GFP fluorescence was measured using spectrofluorometer (infinite M200 PRO, TECAN) at excitation and emission wavelengths of 490 nm and 520 nm, respectively. The fluorescence units were converted into ng GFP per mg fresh weight using a GFP standard curve (rAcGFP1, Clonetech, Takara Bio, USA).
Transactivation of CP and Rep promoters upon begomovirus inoculation
To study the transactivation of OELCuV V1/CP and C1/Rep promoters upon infection with OELCuV and BYVMB, the following constructs in agrobacterium were used for infiltration in cotton plants: OELCuV1.5, BYVMB1.1 [33], pRep-GFP, pCP-GFP, p35S-GFP and pGFP0029. The agroinfiltration of infectious clones was performed by inoculating a 1:1 combination of OELCuV1.5 and BYVMB1.1 containing agrobacterial cultures in cotton following the method used for okra inoculation [33]. The infection was confirmed by PCR using CP/V1-specific primers as described earlier [33]. At 15 DPI, 5 plants each were agroinfiltrated by pRep-GFP, pCP-GFP, p35S-GFP and pGFP0029 constructs; and 5 plants were kept uninoculated. At 3 days following inoculation of the above constructs, GFP accumulation was visually monitored in intact leaves using UV lamp (UVP, Black-Ray®, USA) and processed for GFP assay.
To check the possible transactivation of the CP- and Rep promoters by SLCMV, infectious clones of the same (SLCMV DNA-A and SLCMV DNA-B), as described earlier [38] were used to initiate infection in N. benthamiana. At 10 DPI the promoter constructs (pRep-GFP, pCP-GFP and p35S-GFP) were individually infiltrated into the systemic leaves (newly emerged leaves) of SLCMV-inoculated N. benthamiana. The tissue sampling for quantification of GFP accumulation was done at 3 DPI. GFP quantification was done as described in the previous section.
NightShade GFP imaging in cotton with and without begomovirus inoculation
The NightShade LB 985 in vivo plant imaging system (Berthold technologies, Germany) was used for GFP localization and quantification in inoculated cotton plants at 3 DPI. The cotton plants that were inoculated for GFP quantification as described in Sect. "Transactivation of CP and Rep promoters upon begomovirus inoculation" were used for imaging before tissue sampling. The built-in software Indigo was used for creating templates for imaging. For fluorescence imaging, GFP filter with excitation filter (475 nm) and emission filter (520 nm) was used with exposure time of 10 s. The imaging report was then exported for analysis.
Results
Sequence analysis of OELCuV promoter
The sequence analysis of IR of OELCuV using Plant CARE database [39] showed that it contained several putative cis-acting regulatory elements viz. CAAT-box, TATA-box, G-box, GATA-motif and MYB binding site, implicated in controlling transcription in plant genes (Fig. 2 and Supplementary Table 1). On careful examination, an element closely resembling CLE (GTGGTGGG instead of the consensus GTGGTCCC, with mis-matches in the last three nucleotides) was found at position − 247 from the ATG of the gene encoding V1/CP. In addition, an element resembling TACE (ACTA-N7-AATT, instead of the consensus ACTT-N7-AAGT, with a two nucleotide mis-matches) was found at position − 166 from the ATG of the gene encoding V1/CP. Finally, an element resembling ABRE (AGGTGG, instead of the consensus ACGTGG) with a single nucleotide mis-match was found at position -155 from the ATG of V1.
The nucleotide sequence of the intergenic region of okra enation leaf curl virus. Putative cis-acting elements including TATA box, G-box, AAAG motif, GC-rich sequence, stem-loop motif/replication initiation site, are indicated. The putative translation starts sites ATG and CAT are highlighted in bold italics
Transient expression of promoters of OELCuV in N. benthamiana, cotton and okra
At 3 DPI, GFP was localized in the leaf tissues of N. benthamiana plants infiltrated with pRep-GFP, pCP-GFP and CaMV 35S promoter-GFP (p35S-GFP) as positive control under CLSM, showing that both V1/CP- and C1/Rep promoters brought about the expression of GFP as did the CaMV 35S promoter, the promoter-less construct showing no expression (Fig. 3). This indicated that the IR-derived DNA fragment indeed contained functional promoter elements, which could be driving the viral- and complementary-sense genes in OELCuV. Similarly, in the cotyledonary leaves (in intact plants) of cotton and cotyledonary and true leaves of okra infiltrated with agrobacterium containing pRep-GFP, pCP-GFP and p35S-GFP, fluorescent patches were visible when visually examined under UV illumination in the dark (Fig. 4 and Supplementary Fig. S1). However, none of the uninoculated leaves and the leaves inoculated with promoter-less GFP construct (pGFP0029), showed any fluorescence. This indicated that, as in N. benthamiana, the CP- and the Rep- promoters of OELCuV were active in both cotton as well as the natural host okra under the conditions employed.
Visualization of GFP accumulation by confocal laser scanning microscope in Nicotiana benthamiana leaves agroinfiltrated with OELCuV pRep-GFP, OELCuV pCP-GFP and p35S-GFP promoter constructs, and GFP without promoter (pGFP0029) was used as negative control. GFP: Green fluorescent protein; BF: Bright field
Visualization of GFP in infiltrated cotton plants under handheld UV lamp at 3 days post-infiltration: Leaves infiltrated with OELCuV pCP-GFP, OELCuV pRep-GFP and p35S-GFP showing GFP expression as green fluorescence around the infiltration zone. Mock (pGFP0029) infiltrated leaf was used as negative control. The infiltrated zones are indicated by arrows
Quantitative assays for promoter activities in N. benthamiana, cotton and okra
At 3 DPI, the extent of accumulation of GFP in the N. benthamiana leaves agroinfiltrated with pRep-GFP, pCP-GFP and p35S-GFP promoter constructs, were 38, 25 and 20 ng per mg of fresh weight, respectively. The experiment was repeated thrice with three replicates. The promoter-less construct (pGFP0029) did not exhibit any GFP accumulation, as anticipated. In cotton, the average GFP accumulation with pRep-GFP, pCP-GFP and p35S-GFP promoter constructs were 38, 32 and 45 ng per mg of fresh weight, respectively. Similarly, in okra plants, the values were 80, 56, and 47 ng per mg of fresh weight, respectively (Fig. 5). The results show that all the three promoters were active in the three plants tested. The levels of expression of the reporter gene driven by three promoters were generally comparable.
Monitoring of GFP accumulation at 3 days post infiltrated Nicotiana benthamiana, cotyledonary leaves of cotton and okra plants. The data represent average ± SD (n = 3) of each construct: CaMV35S promoter (p35S-GFP), OELCuV Rep promoter (pRep-GFP), OELCuV CP promoter (pCP-GFP) and mock (pGFP0029). Experiment was repeated twice. Error bars indicate SE. (n: number of samples; p: p values)
Transactivation of CP- and Rep promoters upon begomovirus infection
The transactivation by the CP/V1- and the Rep/C1-promoters by the homologous virus OELCuV was tested in cotton and by the heterologous virus SLCMV, in N. benthamiana. The accumulation of GFP was determined in cotton following inoculation with OELCuV and BYVMB. PCR amplification of 700 bp DNA fragment confirmed the presence of virus in the inoculated cotton plants (Supplementary Fig. S2). The average accumulation of GFP in cotton leaves inoculated with OELCuV + BYVMB followed by agroinfiltration with pRep-GFP, pCP-GFP and p35S-GFP promoter constructs, were 158, 189 and 96 ng per mg of fresh weight, whereas the corresponding values without the OELCuV + BYVMB were 38, 32 and 45 ng per mg of fresh weight, respectively (Fig. 6). This indicated that V1/CP and C1/Rep promoters of OELCuV were transactivated up to 5.9- and 4.2-fold, respectively in the presence of OELCuV + BYVMB. The fluorescence was seen as green patches in the inoculated portions of leaves infiltrated with positive control p35S-GFP, and the test constructs, pCP-GFP and pRep-GFP (Supplementary Fig. S3), the negative control (promoter-less GFP construct pGFP0029) showing no fluorescence.
GFP quantification in transient expression assay in cotton cotyledonary leaves at 3 days post-infiltration to test the inducibility of OELCuV CP and Rep promoter following agroinoculation with cloned DNAs of OELCuV and BYVMB. The data represent average ± SD (n = 3; p value ** = 0.05–0.01, when compared with CaMV35S promoter by student’s t test) of each construct: p35S-GFP, pRep-GFP, pCP-GFP, and mock (pGFP0029). Error bars indicate SE. (n: number of samples; p: p values)
Upon inoculation of N. benthamiana plants with SLCMV DNA-A + SLCMV DNA-B, the plants showed intense leaf curling and vein thickening (Supplementary Fig. S4). In the presence of SLCMV DNA-A and SLCMV DNA-B, the GFP accumulation was 79, 68 and 42 ng per mg of fresh weight for pRep-GFP, pCP-GFP and p35S-GFP promoter constructs, respectively in N. benthamiana. Thus, in the presence of SLCMV DNA-A + SLCMV DNA-B, the accumulation of GFP increased 3.0-fold, 3.3-fold and 2.3-fold under the control of C1/Rep, V1/CP and 35S promoters, respectively, as compared to the plants without SLCMV infection (Supplementary Fig. S5).
NightShade GFP imaging
The cotton cotyledonary leaves infiltrated with promoter-less GFP construct (pGFP0029), did not show any fluorescence, as the leaves appeared totally grey (Fig. 7a). On the other hand, false colour, generated by the software could be seen in leaves infiltrated with p35S-GFP, pCP-GFP and pRep-GFP, indicating the accumulation of GFP in the region of infiltration. Inoculation of OELCuV + BYVMB to the plants substantially enhanced gfp expression in p35S-GFP, pCP-GFP and pRep-GFP infiltrated cotyledonary leaves, visible as bright green patches, with small spots showing red in the false colour-generated image of the pCP-GFP leaf (Fig. 7b), indicating relatively high levels of gfp expression in certain infiltrated areas.
Discussion
Geminiviruses, because of their DNA genome, represent a potential source of new promoters for expression of transgenes in plants. The transcriptional strength of several geminiviral promoters have been investigated earlier [6, 8, 12, 27, 29, 40, 41]. This study represents an investigation on the promoters derived from the recently characterized infectious clone of OELCuV [33], and their transactivation by virus infection.
The results presented in Fig. 5 indicated that the expression of GFP under OELCuV V1/CP- and C1/Rep promoters in okra, one of its natural hosts were higher than in N benthamiana and its other natural host, cotton. This partly differed from an earlier report on the transcriptional activities of V1/CP and C1/Rep promoters of CLCuBV [7] wherein, the promoter activities were comparable between N. benthamiana and cotton, its natural host. Also, unlike the findings with the OELCuV promoters in this study, CLCuBV V1/CP promoter was threefold less active and C1/Rep promoter was twofold more active than 35S promoter, in both the plants. Other examples of reports indicating higher activities of the Rep promoters include those of begomoviruses ACMV and TGMV, about 22-fold more than the CP promoters [5] and a mastrevirus, wheat dwarf virus [17]. The reason for the comparable activities of both the OELCuV promoters and the 35S promoters, indicated in this study, needs to be explored further.
In the presence of viral infection, the V1/CP promoter showed stronger transactivation (5.9-fold) than the C1/Rep promoter (4.2-fold). Upon inoculation with a heterologous virus SLCMV, V1/CP promoter was transactivated 3.3-fold. The results strongly indicated the inducible nature of OELCuV promoters upon the cognate virus infection, as reported earlier for other begomoviruses, such as TGMV [42] and by a heterologous or non-cognate begomovirus. The result correlates with several previous studies, using transient and transgenic expression systems, showing the degree of transactivation of both virion- and complementary-sense strand genes in begomoviruses by AC2/TrAP. In a study based on transient expression, using N. plumbaginifolia leaf protoplast, the promoters of the complimentary sense genes in MYMV showed less transactivation (two to fourfold) than the promoters of the viral-sense genes (15- to 300-fold) [12] by viral infection or AC2. Similarly, using a transgenic system, Rajeswaran et al., (2007) [40] reported the fourfold transactivation of MYMV viral-sense/CP promoter in the presence of homologous AC2 in tobacco plants.
As mentioned earlier, the transactivation of the CP promoters has been correlated with the presence and the number of CLEs. This was demonstrated for the first time in the begomovirus pepper huasteco virus by addition of extra copies of CLE or their deletion [26]. More recently, a similar report has appeared using the CP promoter of TGMV [6], where the AC2-mediated transactivation was shown to be dependent upon the number of CLE units present. There is a possibility that the transactivation of OELCuV CP promoter may also be dependent on the potential CLE-like element present at position − 247 position from the ATG of V1/CP.
There also has been a report of the ABRE in bringing about AC2-mediated transactivation of viral promoter. The stimulation of MYMV viral-sense BV1 promoter by AC2 in a transient system in N. benthamiana, was dependent upon ABRE [29]. As shown in Fig. 2, an element strongly resembling ABRE was indeed detected in the OELCuV V1/CP promoter. The possibility that AC2 recruiting ABA-responsive transcription factors being responsible for AC2-mediated transactivation, taking place in OELCuV CP promoter cannot be ruled out.
An unexpected observation in this study was the mild increase in the activity of 35S promoter upon viral infection, about twofold, as compared to about fivefold for the V1/CP promoter (Fig. 6). There could be at least three explanations for this enhancement. First, several viral proteins, which includes AC2, are known to suppress RNA interference (RNAi)-mediated anti-viral defence responses of the plant [43]. The increase in the activity of the 35S promoter may reflect the potential suppression of the RNAi-mediated degradation of the above transcripts by AC2 [44]. There is a possibility that all ectopic transcripts, including those generated by the V1/CP promoter and the C1/Rep promoter are stabilized due to the above suppression of RNAi brought about by the OELCuV AC2, and the actual levels of transactivation may be two-fold lower than suggested by the data in Fig. 6. The second explanation could be related to the reports of the global transcriptome changes in Arabidopsis upon infiltration with MYMV AC2 [44, 45]. A similar phenomenon maybe taking place upon viral infection in the three plant systems tested in this study, some of which could contribute towards promoter activation. Finally, a third explanation could be based on the report of Babu et al. (2018) [45], which indicate potential interaction of a large number of plant proteins with geminiviral AC2. Some of these proteins may also contribute towards the activation of promoters such as CaMV 35S.
The results of this work suggest that the OELCuV C1/Rep and V1/CP promoters may be used for ectopic gene expression, particularly for malvaceous plants. The CP promoter can be used as a virus inducible promoter for specific applications.
Conclusion
This is the first study of transient expression of the OELCuV V1/CP- and C1/Rep promoters in three host plants; cotton, N. benthamiana and okra. The DNA fragments used drove the expression of the reporter gene gfp in all three hosts, indicating strongly the presence of functional promoters. Quantitative estimation of the product of the reporter gene (gfp) indicated comparable levels of expression of both the promoters, which were comparable to the levels of expression of the standard CaMV 35S promoter. In cotton plants, inoculation of OELCuV + BYVMB DNA enhanced the expression levels of V1/CP promoter 5.9-fold and the C1/Rep promoter 4.2-fold, respectively. Similarly, in N. benthamiana plants, in the presence of the heterologous virus SLCMV, the expression of V1/CP and C1/Rep promoter increased 3.3-fold and 3.0-fold, respectively.
Data Availability
No data were generated in the article.
References
Porto MS, Pinheiro MPN, Batista VGL, dos Santos RC, de Albuquerque Melo Filho P, de Lima LM, (2014) Plant promoters: an approach of structure and function. Mol Biotechnol 56:38–49
Potenza C, Aleman L, Sengupta-Gopalan C (2004) Targeting transgene expression in research, agricultural, and environmental applications: promoters used in plant transformation. In Vitro Cell Dev Biol Plant 40:1–22
Kummari D, Palakolanu SR, Kishor PBK, Bhatnagar-Mathur P, Singam P, Vadez V et al (2020) An update and perspectives on the use of promoters in plant genetic engineering. J Biosci 45:119. https://doi.org/10.1007/s12038-020-00087-6
Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217:109–119
Zhan X, Haley A, Richardson K, Morris B (1991) Analysis of the potential promoter sequences of African cassava mosaic virus by transient expression of the β-glucuronidase gene. J Gen Virol 72:2849–2852
Cantú-Iris M, Pastor-Palacios G, Mauricio-Castillo JA, Bañuelos-Hernández B, Avalos-Calleros JA, Juárez-Reyes A et al (2019) Analysis of a new begomovirus unveils a composite element conserved in the CP gene promoters of several Geminiviridae genera: clues to comprehend the complex regulation of late genes. PLoS ONE 14:e0210485
Ashraf MA, Shahid AA, Rao AQ, Bajwa KS, Husnain T (2014) Functional characterization of a bidirectional plant promoter from cotton leaf curl Burewala virus using an Agrobacterium-mediated transient assay. Viruses 6:223–242
Khan ZA, Abdin MZ, Khan JA (2015) Functional characterization of a strong bi-directional constitutive plant promoter isolated from cotton leaf curl Burewala virus. PLoS ONE 10:e0121656
Rasul F, Asad S, Zafar Y, Mansoor S (2014) Characterization of a strong constitutive promoter from cotton leaf curl Kokhran virus for high level gene expression in monocotyledonous and dicotyledonous plants. Int J Agric Biol 16:342–346
Xie Y, Liu Y, Meng M, Chen L, Zhu Z (2003) Isolation and identification of a super strong plant promoter from cotton leaf curl Multan virus. Plant Mol Biol 53:1–14
Mazithulela G, Sudhakar D, Heckel T, Mehlo L, Christou P, Davies JW et al (2000) The maize streak virus coat protein transcription unit exhibits tissue-specific expression in transgenic rice. Plant Sci 155:21–29
Shivaprasad PV, Akbergenov R, Trinks D, Rajeswaran R, Veluthambi K, Hohn T et al (2005) Promoters, transcripts, and regulatory proteins of mungbean yellow mosaic geminivirus. J Virol 79:8149–8163
Sunitha S, Mahajan N, Veluthambi K (2012) The TrAP/REn monodirectional promoter of mungbean yellow mosaic geminivirus (MYMV) displays root-specific expression in transgenic tobacco. Plant Cell Tissue Organ Cult 109:535–545
Usharani KS, Periasamy M, Malathi VG (2006) Studies on the activity of a bidirectional promoter of mungbean yellow mosaic India virus by agroinfiltration. Virus Res 119:154–162
Brough CL, Sunter G, Gardiner WE, Bisaro DM (1992) Kinetics of tomato golden mosaic virus DNA replication and coat protein promoter activity in Nicotiana tabacum protoplasts. Virology 187:1–9
Dry I, Krake L, Mullineaux P, Rezaian A (2000) Regulation of tomato leaf curl viral gene expression in host tissues. Mol Plant Microbe Interact 13:529–537
Hofer JM, Dekker EL, Reynolds HV, Woolston CJ, Cox BS, Mullineaux PM (1992) Coordinate regulation of replication and virion sense gene expression in wheat dwarf virus. Plant Cell 4:213–223
Roumagnac P, Lett J-M, Fiallo-Olivé E, Navas-Castillo J, Zerbini FM, Martin DP et al (2022) Establishment of five new genera in the family Geminiviridae: Citlodavirus, Maldovirus, Mulcrilevirus, Opunvirus, and Topilevirus. Arch Virol 167:695–710
Varsani A, Roumagnac P, Fuchs M, Navas-Castillo J, Moriones E, Idris A et al (2017) Capulavirus and Grablovirus: two new genera in the family Geminiviridae. Arch Virol 162:1819–1831
Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J et al (2017) ICTV virus taxonomy profile: Geminiviridae. J Gen Virol 98:131
Fiallo-Olivé E, Lett J-M, Martin DP, Roumagnac P, Varsani A, Zerbini FM et al (2021) ICTV virus taxonomy profile: Geminiviridae 2021. J Gen Virol 102:001696
Eagle PA, Hanley-Bowdoin L (1997) cis elements that contribute to geminivirus transcriptional regulation and the efficiency of DNA replication. J Virol 71:6947–6955
Fenoll C, Black DM, Howell SH (1988) The intergenic region of maize streak virus contains promoter elements involved in rightward transcription of the viral genome. EMBO J 7:1589–1596
Morris-Krsinich BAM, Mullineaux PM, Donson J, Boulton MI, Markham PG, Short MN et al (1985) Bidirectional transcription of maize streak virus DNA and identification of the coat protein gene. Nucleic Acids Res 13:7237–7256
Petty ITD, Coutts RHA, Buck KW (1988) Transcriptional mapping of the coat protein gene of tomato golden mosaic virus. J Gen Virol 69:1359–1365
Ruiz-Medrano R, Guevara-Gonzalez RG, Argüello-Astorga GR, Monsalve-Fonnegra Z, Herrera-Estrella LR, Rivera-Bustamante RF (1999) Identification of a sequence element involved in AC2-mediated transactivation of the pepper huasteco virus coat protein gene. Virology 253:162–169
Borah BK, Zarreen F, Baruah G, Dasgupta I (2016) Insights into the control of geminiviral promoters. Virology 495:101–111
Hur J, Choi E, Buckley KJ, Lee S, Davis KR (2008) Identification of a promoter motif involved in Curtovirus sense-gene expression in transgenic Arabidopsis. Mol Cells 26:131–139
Sun R, Han J, Zheng L, Qu F (2020) The AC2 protein of a bipartite geminivirus stimulates the transcription of the BV1 gene through abscisic acid responsive promoter elements. Viruses 12:1403
Hameed U, Zia-Ur-Rehman M, Herrmann H-W, Haider MS, Brown JK (2014) First report of Okra enation leaf curl virus and associated cotton leaf curl Multan betasatellite and cotton leaf curl Multan alphasatellite infecting cotton in Pakistan: a new member of the cotton leaf curl disease complex. Plant Dis 98:1447
Naresh M, Khan ZA, Kumar R, Kale SP, Patil VM, Rajput JC et al (2019) Occurrence and variability of begomoviruses associated with bhendi yellow vein mosaic and okra enation leaf curl diseases in south-western India. Virusdisease 30:511–525
Emmanuel CJ, Manohara S, Shaw MW (2020) Molecular characterization of begomovirus–betasatellite–alphasatellite complex associated with okra enation leaf curl disease in Northern Sri Lanka. 3 Biotech 10:506
Gupta K, Rishishwar R, Khan ZA, Dasgupta I (2022) Agrobacterium-mediated co-inoculation of okra plants with cloned okra enation leaf curl virus DNA and bhendi yellow vein mosaic beta-satellite DNA furthers Koch’s postulates for enation leaf curl disease. J Virol Methods 300:114413
Venkataravanappa V, Reddy CNL, Jalali S, Briddon RW, Reddy MK (2015) Molecular identification and biological characterisation of a begomovirus associated with okra enation leaf curl disease in India. Eur J Plant Pathol 141:217–235
Saunders K, Salim N, Mali VR, Malathi VG, Briddon R, Markham PG et al (2002) Characterisation of Sri Lankan cassava mosaic virus and Indian cassava mosaic virus: evidence for acquisition of a DNA B component by a monopartite begomovirus. Virology 293:63–74
Holsters M, de Waele D, Depicker A, Messens E, van Montagu M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163:181–187
Remans T, Schenk PM, Manners JM, Grof CPL, Elliott AR (1999) A protocol for the fluorometric quantification of mGFP5-ER and sGFP (S65T) in transgenic plants. Plant Mol Biol Report 17:385–395
Mittal D, Borah BK, Dasgupta I (2008) Agroinfection of cloned Sri Lankan cassava mosaic virus DNA to Arabidopsis thaliana, Nicotiana tabacum and cassava. Arch Virol 153:2149–2155
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, van de Peer Y et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Rajeswaran R, Sunitha S, Shivaprasad PV, Pooggin MM, Hohn T, Veluthambi K (2007) The mungbean yellow mosaic begomovirus transcriptional activator protein transactivates the viral promoter-driven transgene and causes toxicity in transgenic tobacco plants. Mol Plant Microbe Interact 20:1545–1554
Khan ZA, Khan JA (2022) Geminivirus promoters: a breakthrough in transgenic research. Geminivirus: detection, diagnosis and management. Elsevier, Amsterdam, pp 357–366
Sunter G, Bisaro DM (1991) Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology 180:416–419
Csorba T, Kontra L, Burgyán J (2015) Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 479:85–103
Trinks D, Rajeswaran R, Shivaprasad PV, Akbergenov R, Oakeley EJ, Veluthambi K et al (2005) Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J Virol 79:2517–2527
Babu KSD, Manoharan P, Pandi G (2018) Computational studies on begomoviral AC2/C2 proteins. Bioinformation 14:294
Acknowledgements
Authors thank Science and Engineering Research Board (SERB), Department of Science and Technology (DST), Government of India, for funding this study under J. C. Bose Fellowship (SB/S2/JCB-057/2016). ZAK and KG acknowledge SERB (PDF/2016/003514) and DST (INSPIRE-2015/IF150761), respectively, for research fellowships under NPDF and INSPIRE. ID acknowledges Senior Scientist Fellowship (INSA/SP/SS/2022/479) of Indian National Science Academy, New Delhi.
Funding
Funds available under the J.C. Bose Fellowship (SB/S2/JCB-057/2016) awarded by Science and Engineering Research Board, Government of India to ID were utilized for this study. KG acknowledges DST-INSPIRE fellowship (2015/IF150761) by Department of Science and Technology, Government of India. ZAK acknowledges National Postdoctoral Fellowship (PDF/2016/003514) by the Science and Engineering Research Board, Department of Science and Technology, Government of India. ID acknowledges the support of Senior Scientist Fellowship (INSA/SP/SS/2022/479) awarded by the Indian National Science Academy, New Delhi.
Author information
Authors and Affiliations
Contributions
Zainul A. Khan, Kanika Gupta: Conceptualization, Methodology, Formal analysis, Writing—original draft, Visualization. Indranil Dasgupta: Conceptualization, Resources, Writing—review & editing, Supervision, Funding acquisition.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Edited by Maija Pollari.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11262_2024_2074_MOESM1_ESM.jpg
Supplementary Fig. S1. Visualization of GFP in agroinoculated okra plants under handheld UV lamp at 3 days post-infiltration: Leaves infiltrated with OELCuV pCP-GFP, OELCuV pRep-GFP and p35S-GFP showing GFP expression as green fluorescence around the infiltration zone. Mock (pGFP0029) infiltrated leaf was used as negative control. The site of the infiltration is indicated by an arrow (JPG 160 KB)
11262_2024_2074_MOESM2_ESM.jpg
Supplementary Fig. S2. Gel electrophoresis pattern showing the PCR amplification products of the coat protein (CP) gene of OELCuV (Lane 1-10) using CP-specific primers in cotton plants agroinoculated with OELCuV and BYVMB at 15 dpi (M: 1 kb ladder, -ve: PCR negative control) (JPG 48 KB)
11262_2024_2074_MOESM3_ESM.jpg
Supplementary Fig. S3. Visualization of GFP in agroinoculated cotton plants under handheld UV lamp at 3 days following agroinoculation with cloned DNAs of OELCuV and BYVMB. Leaves infiltrated with OELCuV pCP-GFP, OELCuV pRep-GFP and p35S-GFP showing GFP expression as green fluorescence around the infiltration zone. The mock (pGFP0029) infiltrated leaf was used as negative control. The site of the infiltration is indicated by an arrow (JPG 169 KB)
11262_2024_2074_MOESM4_ESM.jpg
Supplementary Fig. S4. Nicotiana benthamiana plants infiltrated with (A) SLCMV DNA-A and DNA-B, and (B) mock at 10 days post-infiltration. Arrows indicate typical leaf curling (JPG 98 KB)
11262_2024_2074_MOESM5_ESM.jpg
Supplementary Fig. S5. GFP quantification in transient expression assay in Nicotiana benthamiana leaves at 3 days post-infiltration to test the inducibility of OELCuV CP and Rep promoters upon SLCMV DNA-A and SLCMV DNA-B infection. The data represent average ± SD (n = 3) of each construct: p35S-GFP, pRep-GFP, pCP-GFP, and mock (pGFP0029). Error bars indicate SE (JPG 74 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khan, Z.A., Gupta, K. & Dasgupta, I. Transient expression analysis of promoters of okra enation leaf curl virus in Nicotiana benthamiana, cotton and okra plants. Virus Genes 60, 412–422 (2024). https://doi.org/10.1007/s11262-024-02074-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11262-024-02074-7