Abstract
The TrAP/REn monodirectional promoter of Mungbean yellow mosaic virus (MYMV) comprises many root-specific motifs. The TrAP/REn promoter fused to the β-glucuronidase (gus) reporter gene was used to transform tobacco. Histochemical staining of various parts of seven transgenic plants showed a preferential root-specific expression pattern. Tobacco transformation with the TrAP gene under the transcriptional control of the TrAP/REn promoter yielded nine transgenic plants, of which six harboured the complete TrAP gene in the integrated T-DNAs. Transcript analysis indicated root tissue-specific expression of the TrAP gene. TrAP transgenic plants were phenotypically normal. Expression of the TrAP gene under its own promoter obviated the toxicity which was observed when the TrAP gene was expressed under the CaMV 35S promoter.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant viruses serve as a good repository for regulatory elements which can be used to drive transgene expression in plants. Promoters of Cauliflower mosaic virus (CaMV) (Odell et al. 1985), Cestrum yellow leaf curling virus (CmYLCV) (Stavolone et al. 2003), Figwort mosaic virus (FMV) (Sanger et al. 1990) and Cassava vein mosaic virus (CVMV) (Verdaguer et al. 1996) display strong, constitutive expression in transgenic plants. Generation of transgenic plants with promoter-reporter gene fusions have enabled the characterization of vascular tissue-specific expression of cinnamoyl CoA reductase (LICCR) and cinnamoyl alcohol dehydrogenase (LICAD) (Prashant et al. 2011), tuber-specific expression of granule-bound starch synthase I (GBSSI) (Bansal et al. 2011) and fruit-specific expression of 2AI2 (Wang et al. 2011). The uidA and gfp genes are the two widely used reporter genes (Mußmann et al. 2011; Ramadan et al. 2011).
Geminiviruses are plant DNA viruses characterized by one or two small, circular, single-stranded DNA genomes. Based on the genome organization, host-range and vector transmission, geminiviruses are classified into the genera, Mastrevirus, Curtovirus, Topocuvirus and Begomovirus. Geminiviruses replicate in infected plant cell nuclei by producing double-stranded replicative forms, which serve as the template for transcription (Preiss and Jeske 2003). Geminiviruses rely heavily on the host transcription machinery for viral gene expression (Hanley-Bowdoin et al. 1999). Thus, geminiviral promoter elements are well suited for transgene expression in plants. Mungbean yellow mosaic virus (MYMV) is a whitefly-transmitted, bipartite begomovirus which infects mungbean (Balaji et al. 2004; Morinaga et al. 1993) and blackgram (Karthikeyan et al. 2004). DNA A encodes the replication-associated protein (Rep) and replication enhancer protein (REn) which are required for replication. DNA A also encodes the coat protein (CP) and transcriptional activator protein (TrAP) which are needed for encapsidation and transactivation, respectively. DNA B encodes the nuclear shuttle protein (NSP) and movement protein (MP) which are required for viral movement.
The intergenic region (IGR) of DNA A and DNA B of begomoviruses share a common region (CR) of about 160–200 bp. Chromatin modification of the geminivirus genome into minichromosomes modulates gene expression. The nucleosome-free gaps are likely to be accessible for host replication and transcription factors. Fine mapping of the replicative form of Abutilon mosaic virus (AbMV) revealed that the minichromosomes comprised 11–13 nucleosomes (Pilartz and Jeske 2003). One gap in the IGR and a second gap upstream of the translation initiation codon of TrAP were identified on AbMV DNA A, which suggested the presence of promoter elements in these regions. The cis-acting elements in the CR regulate the leftward transcription of the Rep gene and rightward transcription of the CP gene. TATA box and G-box consensus elements were identified as essential elements for the transcriptional activation of the Rep gene (Eagle and Hanley-Bowdoin 1997). Rep also functions as a negative feedback regulator of its own transcription by binding to a direct repeat iteron sequence between the TATA box and the transcription start site (Behjatnia et al. 1998; Eagle et al. 1994).
The potential promoter activity of the cis-elements of the Rep gene was demonstrated in tobacco protoplasts by transient expression of the African cassava mosaic virus (ACMV) IGR regulatory element fused to the gus reporter gene (Haley et al. 1992; Hong and Stanley 1995; Zhan et al. 1991). The firefly luciferase gene fused to the ACMV IGR regulatory element expressed well in tobacco and cassava protoplasts (Frey et al. 2001). Similar transient expression studies of the Tomato golden mosaic virus (TGMV) element in Nicotiana tabacum protoplasts (Sunter et al. 1993) and the MYMV element in N. plumbaginifolia protoplasts (Shivaprasad et al. 2005) confirmed the host-dependent transcriptional activity of the bidirectional promoter.
Agroinfiltration of N. benthamiana and many legume leaves and agroinoculation of roots with Mungbean yellow mosaic India virus (MYMIV) Rep promoter-gus fusions showed a preferential GUS expression in the vascular and mesophyll cells of leaf and no expression in roots (Usharani et al. 2006). Transgenic expression from the ACMV Rep regulatory elements in N. tabacum (Haley et al. 1992) and Tomato mottle Taino virus (ToMoTV) elements in tobacco and potato (Ramos et al. 2004) showed a vascular tissue-specific expression pattern. However, the Rep regulatory elements of the monopartite begomovirus Tomato leaf curl virus (TLCV) exhibited constitutive expression in transgenic N. tabacum (Dry et al. 2000).
The expression of the virion-sense (CP) promoter is weak but is strongly upregulated by transcriptional activators. Several groups demonstrated the TrAP-mediated transactivation of the CP promoter in protoplasts (Dry et al. 2000; Frey et al. 2001; Hong et al. 1996; Shivaprasad et al. 2005; Sunter et al. 1990; Sunter and Bisaro 1991; 1992) and in transgenic plants (Rajeswaran et al. 2007). A conserved late element (CLE) in the IGR was found to be essential for the TrAP-mediated transactivation in Pepper huasteco virus (PHV) (Ruiz-Medrano et al. 1999). However, CLE was found to be dispensable in TGMV (Sunter and Bisaro 2003) and Bean golden mosaic virus (BGMV) which lacked the element (Hung and Petty 2001). The CLE motif acted as an enhancer element in PHV (Cazzonelli et al. 2005) and Beet curly top virus (BCTV) (Hur et al. 2008).
The rightward promoter of TGMV did not drive GUS expression in the phloem and mesophyll cells of transgenic N. benthamiana (Sunter and Bisaro 1997). However, the CP promoter of TLCV was active in phloem and mesophyll in transgenic plants even in the absence of TrAP (Dry et al. 2000). The transgenically expressed GUS reporter gene under the truncated CP promoters of TGMV (Sunter and Bisaro 1997) and Cabbage leaf curl virus (CaLCuV) (Lacatus and Sunter 2008) exhibited TrAP-independent activity in the phloem cells of N. benthamiana. The full-length CP promoter possessed a repressor element that repressed the TrAP-independent activity in the phloem cells. TrAP-mediated transactivation was achieved by activation of the CP promoter in the mesophyll cells and by derepression in the phloem cells (Lacatus and Sunter 2008; Sunter and Bisaro 1997, 2003). Phloem-specific expression of the virion-sense promoter of Wheat dwarf virus (WDV) was shown in various transgenic dicotyledonous plants (Dinant et al. 2004).
In addition to the strong bidirectional promoter of ACMV, a region upstream of the TrAP ORF was shown to possess moderate promoter activity in N. clevelandii protoplasts (Zhan et al. 1991). Detailed transcriptome mapping of MYMV (Shivaprasad et al. 2005) and TGMV (Shung et al. 2006) confirmed the presence of a separate monodirectional promoter which transcribed the TrAP/REn sequences. The current study focuses on the expression pattern of the monodirectional TrAP/REn promoter of MYMV in transgenic N. tabacum plants. Although this promoter transcribes both TrAP and REn open reading frames, it is referred as the TrAP promoter in the rest of the paper. We report a preferential root-specific expression pattern of the TrAP promoter for the first time. We also report that the toxicity manifested by constitutive expression of TrAP in tobacco under the 35S promoter (Rajeswaran et al. 2007) can be alleviated by the regulated, tissue-specific expression of TrAP under its native promoter.
Materials and methods
Plasmid constructs
The binary plasmid pPZP-PTrAP-gus was constructed as follows: A 359-bp sequence of the TrAP promoter (coordinates 1974 to 1615, accession number AJ132575.1) was amplified and cloned into the EcoRV site of pBSIIKS+ (Stratagene, West Cedar, USA) (pBS-PTrAP). The PstI and NcoI restriction sites were introduced in the forward and reverse primers, respectively. The TrAP promoter fragment from pBS-PTrAP was subcloned as a HincII/NcoI fragment into the corresponding sites of pRT100 (Topfer et al. 1987) (pRT-PTrAP-35S 3′). A 2.1-kb NcoI/SacI intron-gus gene fragment was subcloned into the corresponding sites of pRT-PTrAP-35S 3′ (pRT-PTrAP-gus-35S 3′). Independently, a 2.1-kb Pnos-nptII-ocs 3′ fragment was cloned as a StuI/NruI fragment in the EcoRI site of pPZP200 (Hajdukiewicz et al. 1994), in which process the EcoRI site was recreated (pPZP200-M) (Courtesy: Pradeep Burma, UDSC, New Delhi). The 2.8-kb PTrAP-gus cassette from pRT-PTrAP-gus-35S 3′was cloned as a HindIII fragment into the corresponding site of pPZP200-M (pPZP-PTrAP-gus) and the binary plasmid was mobilized by triparental mating into the Agrobacterium tumefaciens vir helper strain C58C1 (pGV2260).
The binary plasmid pGA-PTrAP-TrAP was constructed as follows: A 278-bp fragment with the CaMV 35S polyA signal was cloned as an EcoRI/XhoI fragment in pBSIIKS+ (pBS-35S 3′). An 847-bp fragment comprising the MYMV-TrAP promoter and the coding sequence (coordinates 1982 to 1135, accession number AJ132575.1) was amplified and cloned in pGEM-T (Promega, Madison, USA) (pGEM-PTrAP-TrAP). The SacI and BamHI recognition sites were introduced in the forward and reverse primers, respectively. An 850-bp SacI/BamHI fragment was excised from pGEM-PTrAP-TrAP and subcloned in the corresponding sites of pBS-35S 3′ (pBS-PTrAP-TrAP-35S 3′). The 1.1-kb PTrAP-TrAP-35S 3′ cassette was cloned as a SacI/XhoI fragment in the corresponding sites of pGA472-M (Sunitha et al. 2012) (pGA-PTrAP-TrAP) and the binary vector was mobilized by triparental mating into the A. tumefaciens strain LBA4404.
Tobacco transformation
Axenic tobacco (N. tabacum L. cv. Wisconsin 38) plants were grown in a tissue culture room under 16 h light (100 μE m−2 s−1)/8 h dark cycles at 25 ± 1°C. Tobacco leaf discs were transformed using A. tumefaciens as described by Sunilkumar et al. (1999). Transgenic shoots were selected on Murashige and Skoog (MS) medium which contained 100 mg l−1 kanamycin + 250 mg l−1 cefotaxime and were kept for root induction on the BGS medium (MS salts, 0.001 mg l−1 folic acid, 100 mg l−1 myoinositol, 0.4 mg l−1 thiamine, 0.057 μM indole-3-acetic acid, 0.14 μM kinetin, 3% [w/v] sucrose, 0.9% [w/v] agar, pH 5.7) supplemented with 250 mg l−1 cefotaxime and 100 mg l−1 kanamycin. GUS histochemical staining and GUS fluorometric assay were done as described by Sunilkumar et al. (1999).
Southern blot and RT-PCR analyses
Total plant DNA was extracted as described by Rogers and Bendich (1994). Plant DNA concentration was estimated using the Hoechst dye 33258 in the DyNA Quant 200 fluorometer (Hoefer Scientific Instruments, San Fransisco, USA). T-DNA integration was analysed by Southern blotting. Total DNA (10 μg) was digested with appropriate restriction enzymes and electrophoresed in a 0.8% agarose gel in 1× Tris–borate-EDTA buffer. Viral titre in agroinfected blackgram leaves and roots was determined by DNA blot analysis of undigested plant DNA following agarose gel electrophoresis in 1× TNE (40 mM Tris–acetate, pH 7.5, 20 mM sodium acetate, 2 mM EDTA) buffer (Karthikeyan et al. 2004). DNA was transferred to the Zeta-probe nylon membrane (Biorad, Hercules, USA) and hybridized to [α-32P]dCTP-labelled probes. RT-PCR analysis using the TrAP primers and tobacco actin primers was done as described earlier (Sunitha et al. 2011).
Results
Sequence analysis of the TrAP promoter
The 359-bp TrAP promoter sequence was submitted to the PLACE database (Higo et al. 1999) to identify the putative cis-elements. The transcription start site (TSS) nucleotide which was identified by Shivaprasad et al. (2005) was numbered as +1 (Fig. 1). A TATA box (TATAA) at the −27 position and multiple CAAT boxes at −51, −249 and −277 positions were identified (Fig. 1) upstream of the TSS. The stress-regulated motifs ABRELATERD1 (ACGTG) and MYCCONSENSUSAT (CANNTG) were identified at −266 and −231 positions, respectively. At positions −176 and −296, light-regulated motifs ASF1MOTIFCAMV (TGACG) and GATA box were identified. Cytokinin-responsive motifs CPBCSPOR (TATTAG) (at −317 position) and ARR1AT (NGATT) (at positions −122 and −306) were also found. Interestingly, several root-specific motifs ROOTMOTIFPOX1 (ATATT), SORLIP1AT (GCCAC), OSE2ROOTNODULE (CTCTT) and RHERPATEXPA7 (KCACGW) were identified at −84, −234, −32 and −268 positions, respectively (Fig. 1). The presence of four root-specific motifs in the TrAP promoter prompted us to analyse the pattern of the promoter activity in transgenic tobacco plants.
Nucleotide sequence of the 357-bp TrAP promoter and 407-bp TrAP coding sequence of MYMV. The transcription start site (TSS +1) and the translation start site (ATG) are marked with arrows and the translation termination codon (TAG) is marked in a bold line. Important cis-elements identified by the software tool PLACE for root-specific expression have been marked in boxes. Other regulatory motifs identified are marked in thin lines
Southern blot analysis of tobacco plants transformed with PTrAP-gus, histochemical and fluorometric GUS assay
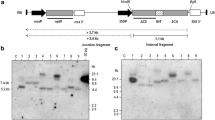
The TrAP promoter sequence fused with the gus reporter gene was placed in pPZP200-M to yield pPZP-PTrAP-gus, which harboured the neomycin phosphotransferaseII (nptII) gene as the plant selectable marker (Fig. 2a). Care was taken to avoid the presence of the CaMV 35S promoter in the T-DNA. Tobacco transformation with pPZP-PTrAP-gus yielded seven kanamycin resistant shoots which formed roots on a medium containing 100 mg l−1 kanamycin. T-DNA integration of kanamycin-resistant plants was studied by Southern blotting with the nptII probe. Digestion of DNA with HindIII and hybridization with the nptII probe will yield junction fragments longer than 2.26 kb (Fig. 2a). TG-1 displayed hybridization to four integrated T-DNA copies (9.4, 7.2, 5.8 and 5.0 kb), TG-2 to six copies (10.2, 10.0, 7.4, 6.0, 4.2 and 3.3 kb), TG-4 to four copies (10.2, 9.4, 7.2 and 4.4 kb), TG-5 to three copies (10.2, 9.4 and 5.0 kb), TG-6 to five copies (9.4, 7.2, 5.0, 4.6 and 2.9 kb), and TG-7 to two copies (7.2 and 3.3 kb) (Fig. 2b). One junction fragment of 6.6 kb hybridized weakly in TG-3.
Southern blot analysis of tobacco plants transformed with pPZP-PTrAP-gus. a The T-DNA of pPZP-PTrAP-gus. The MYMV TrAP promoter was fused to an intron-containing β-glucuronidase (gus) gene in the binary vector pPZP200-M. PTrAP MYMV TrAP/REn promoter; RB T-DNA border-right; Pnos nopaline synthase promoter; nptII neomycin phosphotransferaseII gene; ocs3′ octopine synthase polyadenylation signal; 35S 3′ CaMV 35S polyadenylation signal; LB T-DNA border-left. The probe used for Southern blotting was the nptII gene. The junction fragment size (>2.26 kb) has been marked in a dashed arrow. b The nptII probe-based Southern blot analysis of tobacco plants transformed with pPZP-PTrAP-gus. Total DNA (10 μg) from seven transgenic plants TG-1 to TG-7 (1–7) was digested with HindIII and the blot was probed with the nptII coding sequence. Total DNA from an untransformed, control tobacco plant digested with HindIII (C) was used as the negative control
Seven PTrAP-gus (TG-1 to TG-7) and two P35S-gus (CG-1 and CG-2) transgenic tobacco plants were used to study the GUS expression pattern in leaf and root tissues by histochemical staining (Fig. 3). Five of the seven PTrAP-gus plants (TG-1 to TG-5) did not show GUS staining in the leaf segments (Fig. 3a). Plants TG-6 and TG-7 exhibited very mild GUS expression in the leaf tissue (Fig. 3a). Leaf segments of CG-1 and CG-2 plants transformed with P35S-gus showed intense GUS staining (Fig. 3c). Interestingly, all seven transgenic PTrAP-gus plants (TG-1 to TG-7) exhibited GUS staining in roots (Fig. 3b) similar to the intensity found in P35S-gus transformed plant roots (Fig. 3d).
GUS histochemical analysis of PTrAP-gus transgenic plants. a, b GUS staining pattern of PTrAP-gus transformed plants TG-1 to TG-7 in leaf and root, respectively. c, d GUS staining pattern of P35S-gus transformed plants CG-1 and CG-2 in leaf and root, respectively. A GUS-positive transgenic tobacco callus was used as a positive control (P)
GUS histochemical staining pattern was analysed in other parts of the transgenic plants. The P35S-gus transgenic plant CG-1 exhibited intense staining in petals (Fig. 4a), stigma (Fig. 4b), sepals (Fig. 4c), fruit wall, immature seeds and placenta (Fig. 4d), anther (Fig. 4e) and stem (Fig. 4f). In five of the seven PTrAP-gus transgenic plants (TG-1 to TG-5) no GUS staining was observed in petals, sepals, immature seeds and stamen (Fig. 4a, c–e, respectively). Very weak GUS staining in the stem (Fig. 4f) was observed in five of the seven PTrAP-gus transgenic plants (TG-2, TG-3, TG-5, TG-6 and TG-7). Weak GUS staining was observed in the stigma of only TG-5 (Fig. 4b). Plants TG-6 and TG-7 exhibited mild GUS staining in all analysed parts (data not shown).
GUS fluorometric assay was done in leaf and root protein extracts of PTrAP-gus and P35S-gus plants to quantitate GUS activity. A high level of GUS specific activity was found in the leaves and roots of P35S-gus transformed plants (Table 1). In the PTrAP-gus transgenic plants, GUS specific activity was high in roots but was very low in leaves. The ratio of GUS specific activities in roots versus leaves was in the range of 15–110 in PTrAP-gus plants in comparison to the range of 6.3–7.0 in P35S-gus transgenic plants. Fluorometric GUS analysis confirmed a strong and preferential expression of the TrAP promoter in roots. The fluorometric GUS assays were done in duplicates which yielded similar values.
Southern blot analysis of MYMV infected blackgram plants
Since the PTrAP-gus fusion gene preferentially expressed in roots, the ability of MYMV to infect and replicate in blackgram roots was evaluated. MYMV DNA accumulation in top crop (TC), young fully expanded leaf (FL), mature leaf (ML) and roots (R) was evaluated in blackgram plants agroinoculated with MYMV DNA A and DNA B partial dimers. DNA blot analysis with the MYMV DNA A probe revealed that the top crop (comprising the shoot bud and young leaves), young fully expanded leaves and mature leaves accumulated high levels of MYMV DNA A (Fig. 5). Interestingly, blackgram roots accumulated detectable levels of DNA A, although the levels were much lower in comparison to the levels found in the leaves. Thus, MYMV infected and replicated in blackgram plants.
Southern blot analysis of leaves and roots of MYMV-infected blackgram plants. A blot with total DNA from top crop (TC), young fully expanded leaf (FL), mature leaf (ML) and roots (R) from MYMV-infected blackgram plants and leaves of uninfected blackgram plant (C) was hybridized to the full-length DNA A probe. The positions of open circular dsDNA (oc), linear dsDNA (lin), single-stranded DNA (ss) and super-coiled dsDNA (sc) of MYMV are marked
Southern blot analysis of tobacco plants transformed with the MYMV TrAP gene under its native promoter
The expression of the MYMV TrAP gene under the transcriptional control of the CaMV 35S promoter proved to be toxic in transgenic plants (Rajeswaran et al. 2007). We studied whether the toxicity of TrAP can be alleviated by expressing it under its native promoter. MYMV TrAP promoter plus its coding sequence along with the 35S polyA signal was cloned in the binary vector pGA472-M (Sunitha et al. 2012) to yield pGA-PTrAP-TrAP. There is no 35S promoter in the T-DNA. The binary vector harboured the nptII gene as the plant selectable marker (Fig. 6a). Nine transgenic plants which formed roots on a selection medium with kanamycin were obtained upon transformation with pGA-PTrAP-TrAP. Digestion of DNA with HindIII and hybridization with the nptII probe will yield junction fragments longer than 2.8 kb (Fig. 6a). Plants TT-3 (6.2 kb), TT-5 (3.4 kb), TT-7 (7.0 kb) and TT-8 (18.0 kb) displayed hybridization to single junction fragments. Plants TT-1 (8.2 and 2.9 kb), TT-2 (8.2 and 5.6 kb), TT-4 (7.4 and 2.8 kb), TT-6 (4.8 and 4.6 kb) and TT-9 (7.6 and 6.8 kb) had two integrated T-DNA copies (Fig. 6b). The presence of the complete TrAP gene in the integrated T-DNA was analysed using the TrAP probe. Upon digestion of the plant DNA with EcoRI and hybridization with the TrAP probe, six of the nine plants (TT-2, TT-3, TT-4, TT-6, TT-7 and TT-9) exhibited hybridization to the expected internal T-DNA fragment of 0.5 kb (Fig. 6c). PCR with PTrAP-TrAP-specific primers (Fig. 6a) amplified an 850-bp fragment in all these six transgenic plants (TT-2, TT-3, TT-4, TT-6, TT-7 and TT-9) (Fig. 6d). Thus, six of the nine transgenic plants harboured the complete TrAP gene.
Southern blot analysis of tobacco plants transformed with pGA-PTrAP-TrAP. a The T-DNA of pGA-PTrAP-TrAP. The MYMV TrAP promoter and TrAP coding region were cloned in the binary vector pGA472-M. RB T-DNA border-right; Pnos nopaline synthase promoter; nptII neomycin phosphotransferaseII gene; nos 3′ nopaline synthase polyadenylation signal; 35S 3′ CaMV 35S polyadenylation signal; PTrAP MYMV TrAP/REn promoter; LB T-DNA border-left. Probes used for hybridization have been marked in bold lines. The junction fragment size (>2.8 kb) and internal T-DNA fragment size (0.5 kb) have been marked in dashed arrow and line, respectively. The primers used for PCR analysis have been marked in filled arrows. b The nptII probe-based Southern blot analysis of tobacco plants transformed with pGA-PTrAP-TrAP. Total DNA (10 μg) from nine transformants TT-1 to TT-9 (1–9) was digested with HindIII and the blot was probed with the nptII coding sequence. Total DNA from the untransformed, control tobacco plant digested with HindIII (C) was used as the negative control. c The TrAP probe-based Southern blot analysis of tobacco plants transformed with pGA-PTrAP-TrAP. DNA from the transgenic plants TT-1 to TT-9 (1–9) was digested with EcoRI and the blot was probed with the TrAP coding sequence. The binary plasmid pGA-PTrAP-TrAP (50 pg) digested with EcoRI was used as the positive control (P). An internal T-DNA fragment of 0.5 kb was expected to hybridize in transgenic plants and the binary plasmid control (P). E empty lane. d PCR analysis of tobacco plants transformed with pGA-PTrAP-TrAP. DNA (100 ng) from the untransformed, control tobacco plant (C) and from nine transgenic plants TT-1 to TT-9 (1–9) was used as the PCR template. Binary plasmid DNA (50 pg) was used as the positive control (P). W water control, E empty lane, M 1 kb+ marker
Transcript and phenotypic analyses of PTrAP-TrAP transgenic tobacco plants
RT-PCR was done using the TrAP-specific primers in the six plants which harboured the complete TrAP gene. RNA was extracted from leaf and root tissues and analysed for TrAP transcript accumulation. No amplification was observed in control and transgenic leaf tissues (Fig. 7a). Interestingly, the expected 400-bp fragment was amplified in the roots of four of the six transgenic plants (TT-4, TT-6, TT-7 and TT-9) (Fig. 7a). No amplification was observed in the control reaction in which the reverse transcription step was omitted (data not shown), thus ruling out DNA contamination. RT-PCR with tobacco actin primers amplified a 467-bp fragment in leaf and root tissues of control and all six transgenic plants (Fig. 7a).
Expression and phenotypic analyses of PTrAP-TrAP transgenic plants. a RT-PCR of the PTrAP-TrAP transgenic plants with the TrAP-specific primers. Total RNA extracted from the leaf and root tissues of untransformed, control tobacco plant (C) and from six transgenic plants TT-2, TT-3, TT-4, TT-6, TT-7 and TT-9 harbouring the TrAP gene (2, 3, 4, 6, 7 and 9) was used as the RT-PCR template. A Southern blot-positive transgenic plant expressing a TrAP mutant gene (Rajeswaran et al. 2007) was used as the positive control (P). The bottom panel presents an equal loading control with samples amplified with the tobacco actin gene-specific primers. W water control. b Phenotype of PTrAP-TrAP transgenic plants. Six transgenic tobacco plants TT-2, TT-3, TT-4, TT-6, TT-7 and TT-9 which harboured the TrAP gene and untransformed, control tobacco plant were established in the greenhouse. Six-week-old plants were photographed
Silencing suppressors are known to cause phenotypic abnormalities upon expression in transgenic plants. TrAP of MYMV was reported to suppress RNA silencing (Trinks et al. 2005). The transgenic PTrAP-TrAP plants were established in the greenhouse. All plants were phenotypically normal (Fig. 7b). Thus, the expression of PTrAP-TrAP did not cause any phenotypic abnormality.
Discussion
TrAP/REn transcripts are made from monodirectional promoters in ACMV (Zhan et al. 1991), MYMV (Shivaprasad et al. 2005) and TGMV (Shung et al. 2006). The constitutively expressed TrAP promoter was further activated by TrAP in N. plumbaginifolia protoplasts (Shivaprasad et al. 2005). In contrast, the ACMV TrAP promoter displayed only constitutive expression (Haley et al. 1992). The autoregulation of Rep enhanced the TrAP and REn expression in TGMV (Shung and Sunter 2007). A 9-bp conserved site in the TGMV TrAP promoter, which binds to host nuclear proteins, is essential for TrAP/REn expression and viral replication (Tu and Sunter 2007). Interestingly, the leftward transcription of the bidirectional promoter inhibited the TrAP promoter activity, since the regulatory sequence was within the Rep coding region.
Cis-elements in the MYMV TrAP promoter suggest root-specific expression
The TrAP promoter sequence harboured four motifs associated with root-specific expression (Fig. 1). ROOTMOTIFPOX1 was reported in the root-specific rol D promoter from Agrobacterium rhizogenes (Elmayan and Tepfer 1995). SORLIP1AT upregulated many root-specific genes (Jiao et al. 2005). The motifs OSE2ROOTNODULE2 and RHERPATEXPA7 controlled expression in root nodule (Fehlberg et al. 2005) and root hair (Kim et al. 2006), respectively. Interestingly, the cytokinin-dependent elements CPBCSPOR and ARR1AT also were attributed to root-specific expression (Fusada et al. 2005; Ross et al. 2004).
Root-specific expression of the MYMV TrAP promoter in transgenic tobacco
PTrAP-gus displayed a preferential root-specific expression pattern of GUS in five out of seven transgenic plants (Fig. 3b). No GUS staining was observed in leaf (Fig. 3a), petals, sepals, immature seeds and stamen (Fig. 4) in those transgenic plants. The mild GUS staining observed in stem sections of five PTrAP-gus plants (TG-2, TG-3, TG-5, TG-6 and TG-7) could be attributed to the presence of the bean GRP 1.8 gene stem elements 1 and 2 (SE1, SE2) (Keller and Baumgartner 1991) with 75 and 71% nucleotide sequence similarity, respectively, in the MYMV TrAP promoter (data not shown). The weak GUS staining exhibited by the plants TG-6 and TG-7 in all analysed parts of the plants may be due to the impact of flanking plant DNA sequences at the sites of T-DNA integration. Integration of multiple T-DNA copies did not have an adverse effect on GUS expression in roots. TG-3, a plant in which a single T-DNA copy was integrated and the plant TG-6, in which five copies of the T-DNA were integrated had comparable levels of GUS activity (Fig. 2b; Table 1). As a majority of the transgenic plants (TG-1 to TG-5) expressed PTrAP-gus only in roots, it is inferred that the TrAP promoter displays preferential root-specific expression. The ability of MYMV to invade and replicate in the roots (Fig. 5) lends significance to the expression of the TrAP promoter in roots. It is important to note that Shivaprasad et al. (2005) reported accumulation of the TrAP/REn transcript in the MYMV-infected blackgram leaves. Two explanations may be given for the preferential root-specific expression of the PTrAP-gus gene in transgenic tobacco: (1) The promoter sequence we studied in this report might lack a leaf-specific regulatory sequence element, which may be present in the complete MYMV DNA A. (2) A protein encoded in another part of the viral genome may alter the spatial expression pattern of the TrAP promoter during MYMV infection in blackgram. The transgenic tobacco plants which expressed the C2-gus or C3-gus translational fusions of TLCV showed GUS activity in vascular tissues of leaf, stem and root (Dry et al. 2000). However, it is not very clear from this report whether the C2 and C3 proteins were translated from the Rep transcript initiated from the bidirectional promoter or from the C2/C3 transcript initiated from the monodirectional TrAP promoter. The root versus leaf GUS specific activity ratio was in the range of 15–110 in PTrAP-gus plants (Table 1) in comparison to the range of 6.3–7.0 displayed by P35S-gus transgenic plants. These observations substantiate the preferential root-specific expression pattern of the PTrAP-gus plants. Thus, the MYMV TrAP promoter is useful to achieve root-specific expression of transgenes.
Expression of MYMV TrAP under its own promoter alleviated its toxicity
Begomovirus TrAP acts as a transcriptional activator of late genes (Haley et al. 1992; Shivaprasad et al. 2005; Sunter and Bisaro 1991, 1992), as a suppressor of gene silencing (Bisaro 2006; Trinks et al. 2005) and as a pathogenicity determinant (Hong et al. 1996, 1997). Constitutive expression of MYMV-TrAP resulted in truncated T-DNA integrations which were attributed to TrAP-mediated toxicity (Rajeswaran et al. 2007). This lead to the intriguing question on how MYMV infection in blackgram circumvented the TrAP-mediated toxicity. Shung et al. (2006) demonstrated that the TGMV TrAP expression is tightly regulated. Of the three complementary-sense transcripts AL62, AL1629 and AL1935, AL1629 is the only transcript that encodes TrAP. Rajeswaran et al. (2007) proposed that TrAP may accumulate at low, subtoxic levels in MYMV infected plants. Six of the nine (67%) transgenic plants with the PTrAP-TrAP gene (TT-2, TT-3, TT-4, TT-6, TT-7 and TT-9) harboured the complete TrAP portion of the T-DNA (Fig. 6). This should be compared with the previous report (Rajeswaran et al. 2007) in which six out of seven (86%) transgenic plants which were raised with the P35S-TrAP gene were found to harbour truncated T-DNAs without the complete TrAP gene. Transgenic expression of the ACMV TrAP gene under the 35S promoter caused severe and moderate phenotypic abnormalities in N. tabacum and N. benthamiana, respectively (Siddiqui et al. 2008). No phenotypic abnormality was observed in any of the MYMV PTrAP-TrAP transgenic tobacco plants (Fig. 7b) although four transgenic plants accumulated the TrAP transcript in the roots (Fig. 7a). Thus, expression of MYMV-TrAP, under its own promoter was not toxic to tobacco plants.
Abbreviations
- Rep:
-
Replication-associated protein
- TrAP:
-
Transcriptional activator protein
- REn:
-
Replication enhancer protein
- CP:
-
Coat protein
- MP:
-
Movement protein
- NSP:
-
Nuclear shuttle protein
- IGR:
-
Intergenic region
References
Balaji V, Vanitharani R, Karthikeyan AS, Anbalagan S, Veluthambi K (2004) Infectivity analysis of two variable DNA B components of Mungbean yellow mosaic virus-Vigna in Vigna mungo and Vigna radiata. J Biosci 29:297–308
Bansal A, Kumari V, Taneja D, Sayal R, Das N (2011) Molecular cloning and characterization of granule-bound starch synthase I (GBSSI) alleles from potato and sequence analysis for detection of cis-regulatory motifs. Plant Cell Tiss Organ Cult (in press). doi:10.1007/s11240-011-0090-9
Behjatnia SAA, Dry IB, Rezaian MA (1998) Identification of the replication-associated protein binding domain within the intergenic region of tomato leaf curl geminivirus. Nucleic Acid Res 26:925–931
Bisaro DM (2006) Silencing suppression by geminivirus proteins. Virology 344:158–168
Cazzonelli CI, Burke J, Velten J (2005) Functional characterization of the geminiviral conserved late element (CLE) in uninfected tobacco. Plant Mol Biol 58:465–481
Dinant S, Ripoll C, Pieper M, David C (2004) Phloem specific expression driven by wheat dwarf geminivirus V-sense promoter in transgenic dicotyledonous species. Physiol Plantarum 121:108–116
Dry I, Krake L, Mullineaux P, Rezaian A (2000) Regulation of tomato leaf curl viral gene expression in host tissues. Mol Plant-Microbe Interact 13:529–537
Eagle PA, Hanley-Bowdoin L (1997) Cis elements that contribute to geminivirus transcriptional regulation and the efficiency of DNA replication. J Virol 71:6947–6955
Eagle PA, Orozco BM, Hanley-Bowdoin L (1994) A DNA sequence required for geminivirus replication also mediates transcriptional regulation. Plant Cell 6:1157–1170
Elmayan T, Tepfer M (1995) Evaluation in tobacco of the organ specificity and strength of the rolD promoter, domain A of the 35S promoter and the 35S2 promoter. Transgenic Res 4:388–396
Fehlberg V, Vieweg MF, Dohmann EM, Hohnjec N, Puhler A, Perlick AM, Kuster H (2005) The promoter of the leghaemoglobin gene VfLb29: functional analysis and identification of modules necessary for its activation in the infected cells of root nodules and in the arbuscule-containing cells of mycorrhizal roots. J Exp Bot 56:799–806
Frey PM, Scharer-Hernandez NG, Futterer J, Potrykus I, Puonti-Kaerlas J (2001) Simultaneous analysis of the bidirectional African cassava mosaic virus promoter activity using two different luciferase genes. Virus Genes 22:231–242
Fusada N, Masuda T, Kuroda H, Shimada H, Ohta H, Takamiya K (2005) Identification of a novel cis-element exhibiting cytokinin-dependent protein binding in vitro in the 5′-region of NADPH-protochlorophyllide oxidoreductase gene in cucumber. Plant Mol Biol 59:631–645
Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25:989–994
Haley A, Zhan X, Richardson K, Head K, Morris B (1992) Regulation of the activities of African cassava mosaic virus promoters by the AC1, AC2, and AC3 gene products. Virology 188:905–909
Hanley-Bowdoin L, Settlage S, Orozco BM, Nagar S, Robertson D (1999) Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit Rev Plant Sci 18:71–106
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acid Res 27:297–300
Hong Y, Stanley J (1995) Regulation of African cassava mosaic virus complementary-sense gene expression by N-terminal sequences of the replication-associated protein. J Gen Virol 76:2415–2422
Hong Y, Saunders K, Hartley MR, Stanley J (1996) Resistance to geminivirus infection by virus-induced expression of dianthin in transgenic plants. Virology 220:119–127
Hong Y, Saunders K, Stanley J (1997) Transactivation of dianthin transgene expression by African cassava mosaic virus AC2. Virology 228:383–387
Hung H-C, Petty ITD (2001) Functional equivalence of late gene promoters in bean golden mosaic virus with those in tomato golden mosaic virus. J Gen Virol 82:667–672
Hur J, Choi E, Buckley KJ, Lee S, Davis KR (2008) Identification of a promoter motif involved in Curtovirus sense-gene expression in transgenic Arabidopsis. Mol Cells 26:131–139
Jiao Y, Ma L, Strickland E, Deng XW (2005) Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17:3239–3256
Karthikeyan AS, Vanitharani R, Balaji V, Anuradha S, Thillaichidambaram P, Shivaprasad PV, Parameswari C, Balamani V, Saminathan M, Veluthambi K (2004) Analysis of an isolate of Mungbean yellow mosaic virus (MYMV) with a highly variable DNA B component. Arch Virol 149:1643–1652
Keller B, Baumgartner C (1991) Vascular specific expression of the bean GRP l.8 gene is negatively regulated. Plant Cell 3:1051–1061
Kim DW, Lee SH, Choi SB, Won SK, Heo YK, Cho M, Park YI, Cho HT (2006) Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18:2958–2970
Lacatus G, Sunter G (2008) Functional analysis of bipartite begomovirus coat protein promoter sequences. Virology 376:79–89
Morinaga T, Ikegami M, Miura K (1993) The nucleotide sequence and genome structure of mung bean yellow mosaic geminivirus. Microbiol Immunol 37:471–476
Mußmann V, Serek M, Winkelmann T (2011) Selection of transgenic Petunia plants using the green fluorescent protein (GFP). Plant Cell Tiss Organ Cult 107:483–492
Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313:810–812
Pilartz M, Jeske H (2003) Mapping of Abutilon mosaic geminivirus minichromosomes. J Virol 77:10808–10818
Prashant S, Sunita MSL, Sirisha VL, Bhaskar VV, Rao AM, Narasu ML, Kishor PBK (2011) Isolation of cinnamoyl CoA reductase and cinnamyl alcohol dehydrogenase gene promoters from Leucaena leucocephala, a leguminous tree species, and characterization of tissue-specific activity in transgenic tobacco. Plant Cell Tiss Organ Cult (in press). doi:10.1007/s11240-011-0053-1
Preiss W, Jeske H (2003) Multitaskng in replication is common among geminiviruses. J Virol 77:2972–2980
Rajeswaran R, Sunitha S, Shivaprasad PV, Pooggin MM, Hohn T, Veluthambi K (2007) The mungbean yellow mosaic begomovirus transcriptional activator protein transactivates the viral promoter-driven transgene and causes toxicity in transgenic tobacco. Mol Plant-Microbe Interact 20:1545–1554
Ramadan AM, Eissa HF, El-Domyati FM, Saleh OM, Ibrahim NE, Salama M, Mahfouz MM, Bahieldin A (2011) Characterization of inhibitor(s) of β-glucuronidase enzyme activity in GUS-transgenic wheat. Plant Cell Tiss Organ Cult 107:373–381
Ramos PL, Fuentes AD, Quintana Q, Castrillo G, Guevara-González RG, Peral R, Rivera-Bustamante RF, Pujol M (2004) Identification of the minimal sequence required for vascular-specific activity of Tomato mottle Taino virus replication-associated protein promoter in transgenic plants. Virus Res 102:125–132
Rogers SO, Bendich AJ (1994) Extraction of total cellular DNA from plants, algae and fungi. In: Gelvin SB, Schilperoort RA (eds) Plant molecular biology manual. Kluwer Academic Publishers, Dordrecht, pp D1:1–D1:8
Ross EJ, Stone JM, Elowsky CG, Arredondo-Peter R, Klucas RV, Sarath G (2004) Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J Exp Bot 55:1721–1731
Ruiz-Medrano R, Guevara-Gonzalez RG, Arguello-Astorga GR, Monsalve-Fonnegra Z, Herrera-Estrella LR, Rivera-Bustamante RF (1999) Identification of a sequence element involved in AC2-mediated transactivation of the Pepper Huasteco virus coat protein gene. Virology 253:162–169
Sanger M, Daubert S, Goodman RM (1990) Characterization of a strong promoter from figwort mosaic virus: comparison with the analogous 35S promoter from cauliflower mosaic virus and the regulated mannopine synthase promoter. Plant Mol Biol 14:433–443
Shivaprasad PV, Akbergenov R, Trinks D, Rajeswaran R, Veluthambi K, Hohn T, Pooggin MM (2005) Promoters, transcripts, and regulatory proteins of mungbean yellow mosaic geminivirus. J Virol 79:8149–8163
Shung C-Y, Sunter G (2007) AL1-dependent repression of transcription enhances expression of Tomato golden mosaic virus AL2 and AL3. Virology 364:112–122
Shung C-Y, Sunter J, Sirasanagandla SS, Sunter G (2006) Distinct viral sequence elements are necessary for expression of Tomato golden mosaic virus complementary sense transcripts that direct AL2 and AL3 gene expression. Mol Plant-Microbe Interact 19:1394–1405
Siddiqui SA, Sarmiento C, Truve E, Lehto H, Lehto K (2008) Phenotypes and functional effects caused by various viral RNA silencing suppressors in transgenic Nicotiana benthamiana and N. tabacum. Mol Plant Microbe Interact 21:178–187
Stavolone L, Kononova M, Pauli S, Ragozzino A, de Haan P, Milligan S, Lawton K, Hohn T (2003) Cestrum yellow leaf curling virus (CmYLCV) promoter: a new strong constitutive promoter for heterologous gene expression in a wide variety of crops. Plant Mol Biol 53:703–713
Sunilkumar G, Vijayachandra K, Veluthambi K (1999) Preincubation of cut tobacco leaf explants promotes Agrobacterium-mediated transformation by increasing vir gene induction. Plant Sci 141:51–58
Sunitha S, Marian D, Hohn B, Veluthambi K (2011) Antibegomoviral activity of the agrobacterial virulence protein VirE2. Virus Genes 43:445–453
Sunitha S, Shivaprasad PV, Sujata K, Veluthambi K (2012) High frequency of T-DNA deletions in transgenic plants transformed with intron-containing hairpin RNA genes. Plant Mol Biol Rep 30:158–167
Sunter G, Bisaro DM (1991) Transactivation in a geminivirus: AL2 gene product is needed for coat protein expression. Virology 180:416–419
Sunter G, Bisaro DM (1992) Transactivation of geminivirus AR1and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 4:1321–1331
Sunter G, Bisaro DM (1997) Regulation of a geminivirus coat protein promoter by AL2 protein (TrAP): evidence for activation and derepression mechanisms. Virology 232:269–280
Sunter G, Bisaro DM (2003) Identification of a minimal sequence required for activation of the tomato golden mosaic virus coat protein promoter in protoplasts. Virology 305:452–462
Sunter G, Hartitz MD, Hormuzdi SG, Brough CL, Bisaro DM (1990) Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology 179:69–77
Sunter G, Hartitz MD, Bisaro DM (1993) Tomato golden mosaic virus leftward gene expression: autoregulation of geminivirus replication protein. Virology 195:275–280
Topfer R, Matzeit V, Gronenborn B, Schell J, Steinbiss HH (1987) A set of plant expression vectors for transcriptional and translational fusions. Nucleic Acid Res 15:5890
Trinks D, Rajeswaran R, Shivaprasad PV, Akbergenov R, Oakeley EJ, Veluthambi K, Hohn T, Pooggin MM (2005) Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J Virol 79:2517–2527
Tu J, Sunter G (2007) A conserved binding site within the Tomato golden mosaic virus AL-1629 promoter is necessary for expression of viral genes important for pathogenesis. Virology 367:117–125
Usharani KS, Periasamy M, Malathi VG (2006) Studies on the activity of a bidirectional promoter of Mungbean yellow mosaic India virus by agroinfiltration. Virus Res 119:154–162
Verdaguer B, de Kochko A, Beachy RN, Fauquet C (1996) Isolation and expression in transgenic tobacco and rice plants of the cassava vein mosaic virus (CVMV) promoter. Plant Mol Biol 31:1129–1139
Wang L, Xin M, Qin Z, Liu HY (2011) Functional analysis of an iaaM gene in parthenocarpic fruit development in transgenic Physalis pubescens L. plants. Plant Cell Tiss Organ Cult 107:333–340
Zhan X, Haley A, Richardson K, Morris B (1991) Analysis of the potential promoter sequences of African cassava mosaic virus by transient expression of the β-glucuronidase gene. J Gen Virol 72:2849–2852
Acknowledgments
We are thankful to Dr. K. Dharmalingam, Madurai Kamaraj University for permitting us to use the Liquid Scintillation Counter. This study was financially supported by the Department of Biotechnology (Project Number-BT/PR7866/AGR/02/379/2006), Government of India. S.S acknowledges the Council of Scientific and Industrial Research, Government of India for the research fellowship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sunitha, S., Mahajan, N. & Veluthambi, K. The TrAP/REn monodirectional promoter of Mungbean yellow mosaic geminivirus (MYMV) displays root-specific expression in transgenic tobacco. Plant Cell Tiss Organ Cult 109, 535–545 (2012). https://doi.org/10.1007/s11240-012-0120-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-012-0120-2