Abstract
Genetically engineered plants have varied applications in agriculture for enhancing the values of food and feed. Genetic engineering aims to introduce selected genetic regions with desirable traits into target plants for both spatial and temporal expressions. Promoters are the key elements responsible for regulating gene expressions by modulating the transcription factors (TFs) through recognition of RNA polymerases. Based on their recognition and expression, RNA polymerases were categorized into RNA pol II and pol III promoters. Promoter activity and specificity are the two prime parameters in regulating the transgene expression. Since the use of constitutive promoters like Cauliflower mosaic virus (CaMV) 35S may lead to adverse effects on non-target organisms or ecosystem, inducible/tissue specific promoters and/or the RNA pol III promoters provide myriad opportunities for gene expressions with controlled regulation and with minimum adverse effects. Besides their role in transgene expression, their influence in synthetic biology and genome editing are also discussed. This review provides an update on the importance, current prospects, and insight into the advantages and disadvantages of promoters reported thus far would help to utilize them in the endeavour to develop nutritionally and agronomically improved transgenic crops for commercialization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
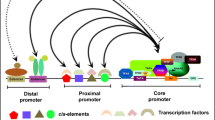
Promoters are gene switches located upstream of gene coding regions, which turn on and off the functional activity of genes and contain specific cis-acting elements which are binding targets for proteins involved in the initiation and regulation of transcription. Promoters are molecular biological clocks crucial for choice of the targeted gene expression (Potenza et al. 2004), that act as key regulatory check points for transcription of genes that are recognized by transcription factors (TFs) (Smale and Kadonaga 2003). TFs bind to specific cis-acting elements present on the respective promoter sequences through RNA polymerase and regulate expression of the downstream genes (Hernandez-Garcia and Finer 2014). Promoters of coding genes often contain core, proximal and distal regions. Further, proximal and distal regions of the promoter contain different regulatory sequences such as enhancers, silencers, insulators, and cis-elements that contribute to fine regulation of gene expression at the transcriptional level (Hernandez-Garcia and Finer 2014). Detailed structural properties of promoters and gene regulatory elements were discussed earlier by Hernandez-Garcia and Finer (2014), Porto et al. (2014) and Shah et al. (2015). Promoter sequences that regulate the controlled transgene expression in plants are useful for developing the genetically modified (GM) crops with improved agronomical and nutritional traits (Mittler and Blumwald 2010). There are few promoter prediction bioinformatics tools; one among them is TSSPlant, a novel tool that predicts both TATA and TATA-less promoters in sequences of a wide spectrum of plant genomes (Shahmuradov et al. 2017). As complete genome sequences of most of the plant species are becoming available, several promoters are being identified, isolated and evaluated, while many more are likely to be elucidated in the near future. Predicted regulatory sequences may or may not be functionally active or necessitating the confirmation of the role of specific elements for promoter activity. Promoters can be isolated through different methods some of which being: genome walking, inverse PCR (IPCR) plasmid rescue, screening of genomic DNA library constructed from mutant plant, the thermal asymmetric interlaced PCR TAIL-PCR (Reddy et al. 2008; Liu and Whittier 1995). Quick, efficient, predictable, and high-throughput analysis of gene expressions and their promoters will be crucial for validating the functional regulatory element sequences.
Promoters can be selected to develop transgenic plants based on the type of trait and target tissue to be regulated (Bilas et al. 2016a, b). A variety of plant promoters that regulate the degree of expression of a transgene can be obtained from various sources. These can be categorized into pol II and pol III that are activated upon recognition by the RNA polymerases II and III. Pol II promoters can in turn be classified into constitutive, tissue-specific, stress-inducible and synthetic (Bilas et al. 2016a, b), whereas pol III promoters are U3 and U6 (Marshallsay et al. 1992). CaMV35S (commonly referred to as 35S) constitutive promoter is the most widely used for gene expressions in transgenic plants and in basic functional genomic studies (Porto et al. 2014). Hence, specific or inducible promoters not only provide increased gene expression in a tissue of interest or at specific developmental stage(s), but also provide more predictable gene expression, with minimal or no penalties on final yield. The discovery of proteins with programmable DNA-binding specificities has triggered an array of applications in synthetic biology, regulation of transcription, including genome editing, and epigenetic modifications. Among those, transcription activator-like effectors (TALENs) and the clustered regularly interspaced short palindromic repeats (CRISPR/Cas9) system played high attention due to their in-built function as transcription regulators for the control of gene expression. Recent studies have shown that the expression of Cas9/gRNA with the help of specific promoters is important for highly efficient genome editing in plants (Hyun et al. 2015; Wang et al. 2015; Yan et al. 2015; Mao et al. 2016). Here, we attempt to provide a comprehensive overview of promoters in plants, their selection, specificity, cross activity, and how they can be useful in maximizing the transgene expression for potential applications in crop improvement programs.
2 Types of promoters
Based on the type of recognition of RNA polymerases, they are mainly classified into RNA polymerase (pol II and pol III) promoters. Detailed information and study of these promoters are provided in the subsequent sections of this review.
2.1 RNA pol II promoters
RNA pol II promoters are the shortest sequences where RNA pol II polymerase binds to the DNA to initiate transcription with help of the TATA box, a common component located at -35bp upstream of the transcription start codon. Based on the type of expression, pol II promoters are further categorized into constitutive, organ- or tissue-specific, stress-inducible and synthetic (figure 1).
A flow chart showing different types of RNA Pol II promoters categorized based on their mode of expression into four major classes: constitutive, tissue-specific, stress-inducible and synthetic promoters. Each of these promoters are further categorized into different types; constitutive promoters (viral, bacterial, plant), tissue-specific promoters (root, fruit, seed), stress-inducible promoters (abiotic {light, wound, heat, drought, salt, cold, hormone and chemical} and biotic {pathogen, pest, weed}).
2.1.1 Constitutive promoters
Constitutive promoters express constitutively throughout the plant life cycle, irrespective of the external and developmental factors which is beneficial for expression of the industrial enzymes, insect resistance and selectable marker genes (Jiang et al. 2018). Based on the source of origin, they are further classified into viral, bacterial and plant constitutive promoters.
2.1.1.1 Viral:
In 1980s, Chua and collaborators isolated a promoter, responsible for the transcription of the whole genome of a Cauliflower mosaic virus (CaMV) infecting turnips. Later, this was named as CaMV35S promoter due to the sedimentation coefficient of the viral transcript whose expression is driven by CaMV35S promoter and used for constitutive overexpression of genes (Odell et al., 1985). There is not much variation in the activities between the full-length CaMV35S promoter (−941 to +9 bp) (Odell et al. 1985) and −343bp deletion fragment (Fang et al. 1989). Although the CaMV35S promoter and its derivatives can drive high levels of transgene expression in dicotyledonous plants (Benfey et al. 1990a, 1990b), their activities are substantially lower in monocotyledonous plants (Gupta et al. 2001). The expression of uidA gene under CaMV35S promoter was observed in different parts of transgenic tobacco with maximum levels in leaf and root tissues (Malik et al. 2002). In contrast, uidA gene expression under the CaMV35S promoter in transgenic canola and Arabidopsis showed expression in all plant tissues (Malik et al. 2002). Sharma et al. (2006) demonstrated a higher activity of the cry1Ab gene under the constitutive CaMV35S promoter in flowers of pigenopea, although other parts like leaves, seeds and pod walls also showed expression of the transgenes to varying degrees.
Apart from CaMV35S, peanut chlorotic streak caulimovirus (PC1SV) and figwort mosaic virus (FMV) have also been shown to be very useful for generating GM plants. PC1SV is a strong constitutive promoter and comparative analysis with FMV promoter in GM plants showed similar expression pattern. Although functionally analogous, the nucleotide sequence of PClSV promoter has limited homology with other caulimovirus promoters (Maiti and Shepherd 1998). The FMV Sgt (Figwort mosaic virus sub-genomic transcript) promoter is another constitutive promoter whose expression was observed in all tissues (Bhattacharyya et al. 2002). A comparative analysis of the FMV Sgt promoter with that of CaMV35S promoter showed that it was 2-folds stronger than the latter, and was also less active in monocots like maize where its expression was about 27.5-folds lower than in tobacco (Bhattacharyya et al. 2002). Gemini viral promoters are in general located within the intergenic region, though promoters have also been identified within the genes. In the same way, Gemini virus-associated beta satellite harbors a promoter element for driving the expression of its only ORF, which can be useful in the regulation of plant genes (Borah et al. 2016). TaB virus-derived promoters may be useful for the high level constitutive expression of transgenes either in monocotyledonous or dicotyledonous species (Yang et al. 2003), thus proving its versatility. Rep promoter from cotton leaf curl Burewala virus (CLCuBuV) showed consistent strong transient expression in tobacco and cotton leaves as compared to the CaMV35S promoter, whereas other CP promoter showed lower expression (Khan et al. 2015). Promoters originating from the tomato leaf curl virus (TLCV) drive both constitutive as well as tissue-specific expression in transgenic tobacco (Seemanpillai et al., 2003). But it is not widely used and accepted from the research community. Comparative analysis of the CaMV35S and the enhanced CaMV35S (E35S), CsVMV, FMV, and the Strawberry vein banding virus (SVBV2) promoters indicated that the FMV promoter facilitated a strong expression of target genes in soybean hairy roots and root nodules (Govindarajulu et al. 2008). Transgenic maize with aryloxyalkanoate dioxygenase (aad-1) expression under the influence of different promoters demonstrated that viral promoters such as CaMV35S and SCBV produce lower transformation efficiencies, but higher percentages of low copy number events contrary to plant constitutive promoters like OsAct1 and ZmUbi1 (Beringer et al. 2017). These promoters are recommended for wide use since they promote low copy number transgenic events. Various constitutively expressed viral promoters used in the development of transgenic plants are listed in table 1.
2.1.1.2 Bacterial:
Bacterial-origin promoters such as nopaline synthase (nos), octopine synthase (ocs) and mannopine synthase (mas) have been isolated from Agrobacterium tumefaciens and used for the control the transgene expression. Although their level of expression and activity can be affected by hormones and wounding, they are repeatedly being used for the transformation of plants. mas promoters can be used as enhancers or silencers, since they have the ability to bind with the factors of nuclear protein from different plants (Shah et al. 2015). Thus, it appears that both mas and CaMV35S promoters can be used for the construction of improved plant transformation vectors. Tn5 neomycin phosphotransferase (nptH) gene was expressed with either of these promoters (Kevin et al. 1989). Sequence analysis of the AV3 promoter from Ageratum yellow vein virus (AYVV) showed that it might be a remnant of prokaryotic ancestors that could be related to certain promoters of bacteria from marine or freshwater environments (Wang et al. 2013). Bacterial promoters show differential expression in transgenic plants, and regulation of rol gene expression plays a role in the biological effects that are caused by the rol A, B, and C genes containing the uidA reporter gene under the control of rol A, B, and C promoters of Agrobacterium rhizogenes (Schmulling et al. 1989). However, their use for generating transgenic crop plants is limited.
2.1.1.3 Plant:
Promoters derived from plant origin are used for the constitutive transgene expressions in plants (Dhankher et al. 2002). Plant constitutive promoters such as rice actin (OsAct1) (McElroy et al. 1991), maize alcohol dehydrogenase1 (ZmAdh1) (Fromm et al. 1990), and maize ubiquitin (ZmUbi1and ZmUbi2) (Christensen et al. 1992) are the most commonly used in crop plants. Act2 promoter was obtained from the actin gene family, which is the cytoskeletal component and expressed in every plant cell (An et al. 1996). Similarly, the OsActin1 promoter also displayed expression in almost all tissues when transformed back into rice (McElroy et al. 1991). The OsAct promoter was shown to drive high levels of expression of HVA1 gene in the leaf and root tissues of rice, thereby conferring salt and water stress tolerance (Xu et al. 1996). ZmAdh is another constitutive promoter expressed in specific tissues like root, shoot meristems, endosperms and pollen (Kyozuka et al. 1991). When MtHP promoter from Medicago was fused to a uidA gene, it showed expression in various plant parts in Medicago and Arabidopsis with an expression pattern similar to that of the CaMV35S promoter (Xiao et al. 2005). Expression of uidAb gene driven by the CaMV35S and MpEF1α promoters were compared throughout plant development. While CaMV35S promoter resulted in in adequate expression in the meristematic tissues and a strong expression in the callus, the MpEF1α-promoter caused a strong meristematic uidA gene expression and was more active in female sexual tissues. It appears therefore, MpEF1α- promoter is a better option for obtaining strong and ubiquitous transgene expression compared to the CaMV35S promoter as has also been pointed out by Althoff et al. 2014.
Expression of uidA under the influence of UBQ1or UBQ2 rice ubiquitin promoters were 8 to 35-folds higher in transgenic rice plants, respectively, when compared to CaMV35S (Wang and Oard 2003). This indicates that ubiquitin promoters are superior for activating the transgenes. Expression of the gfp gene mediated by a GmUbi showed high levels of constitutive expression in soybean tissues, thereby providing an alternative to viral promoters for driving gene expression in soybean (Hernandez-Garcia et al. 2009). The native ubi1 promoter is a promising genetic element for constitutive expression of any gene in rice tissues (Bhattacharyya et al. 2011). OsCon1 promoter exhibited comparable activity with OsCc1, OsAct1 or ZmUbi promoters in most tissues, and even more active than the CaMV35S promoter in roots, seeds and calluses indicating that it is a novel constitutive promoter which could potentially be used for developing transgenics (Li et al. 2014). Incorporation of AtTCTP promoter into creeping bent grass showed that it can be used as a plant-derived constitutive promoter for the expression of selectable marker genes, and as an alternative to the CaMV35S promoter for developing GM crops (Han et al. 2015). The plant-derived JcUEP promoter could be another alternative to the CaMV35S promoter for directing constitutive transgene expression in Jatropha and other plants (Tao et al. 2015). Pineapple SUI1 and L36 promoters seem to drive uidA expression in all tissues of Arabidopsis at levels comparable to that of the CaMV35S promoter (Koia et al. 2013). Studies on rice APX promoter show edits constitutive expression in the seed, root, blade, flower and leaf of transgenic rice (Park et al. 2010). The inducible activities of PvUbi1 and PvUbi2 promoters in switchgrass, rice and tobacco are strong constitutive promoter candidates that can be used in genetic transformation of both monocots and dicots (Mann et al. 2011). Promoters such as pBdEF1α and pBdUBI10 are constitutively expressed and highly active in maize, whereas pBdGLU1 isolated from Brachypodium was clearly endosperm-specific, thereby indicating that it is an excellent resource for promoters for transgenic research in heterologous cereal species (Coussens et al. 2012). The KST1 partial promoter was shown to drive constitutive expression in the guard cells of monocots and dicots, as well as in both annual and perennial plants (Kelly et al. 2017). Three Citrus sinensis constitutive gene promoters confirmed their role in vegetative tissues (Erpen et al. 2018), thus enabling the researchers to use it specifically in vegetative tissues. On the other hand, Jiang et al. 2018 reported that AtSCPL30 promoter could be an alternative for the CaMV35S in terms of reducing transgene silencing and also a good source for the multigene transformation. GAPC2 and EF1 promoter fragments from Citrus sinensis were expressed constitutively in the transgenic tobacco plants (Erpen-Dalla Corte et al. 2020). Description of constitutively expressed plant promoters used for the development of transgenic plants is listed in the table 2.
2.1.2 Tissue-specific promoters
Tissue-specific promoters drive the expression of a targeted gene in a specific tissue(s) or organ at a specific stage(s) of plant growth and development (figure 2b). Since the constitutive promoters cause potential negative effects such as metabolic burden on the transgenics, specific promoters are used to regulate and target gene expressions in an effective manner for improving traits like grain nutritional quality. Due to the demand from biosafety regulators for less intrusive transgene expression, targeted expression of transgenes has become more important for the future development of value-added crops (Bucchini and Goldman 2002).
Schematic depiction of different types of promoters. (a) Stress-inducible promoters can be activated under any biotic or abiotic stressby binding the corresponding transcription factors and cis-acting elements. (b) Tissue-specific promoters are expressed in a particular tissue by binding the corresponding transcription factors and cis-acting elements. (c) Bidirectional promoters drive the transcription of two genes simultaneously that flank the promoter.
2.1.2.1 Promoters that help in biomass production:
Eucalyptus transgenics with AtEXP4pro:CBM2a showing increased plant height, enlargement of xylem, xylem fiber and vessel cells can be an attractive choice for plant biomass improvement (Keadtidumrongkul et al. 2017).
2.1.2.2 Promoters specific to guard cells and vascular tissues:
A partial promoter of KST1 from potato is active in a monocot and the first promoter reported to confer guard cell expression in barley and cucumber (Kelly et al. 2017). Thus, KST1 appears to be a cell specific promoter and highly useful for manipulating proteins in the guard cells. The use of phloem specific promoters (Citrus phloem protein 2-CsPP2, Arabidopsis thalianaphloem protein 2-AtPP2, A. thaliana sucrose transporter 2-AtSUC2, and sucrose synthase 1-SUS1) might increase the chances of producing more bacterial disease resistant transgenic cultivars in many crop plants (Singer et al. 2011; Miyata et al. 2012).
2.1.2.3 Root-specific promoters:
Root-specific promoters are of special interest since they help in understanding the root architecture and in alleviating drought and salt stress conditions. The pNtREL1 promoter can be used to direct root-specific expression of target genes to protect the root from different abiotic stresses (Zhang et al. 2016a, b). Alternatively, Tau Class Glutathione-S-Transferase (SbGSTU) gene promoter of Salicornia brachiata can be used for both constitutive as well as stress-inducible expression of transgenes (Tiwari et al. 2016). More data are needed if such promoters help to mitigate the stress better in comparison with the constitutive promoters with minimum or no yield penalty. A root-preferential promoter GmPRP2 isolated from soybean has been found useful in developing transgenics of novel soybean cultivars (Chen et al. 2014). Rice promoters such as Os03g01700 and Os02g37190 are highly active in the root tissues of rice and can be helpful for the root-specific expression of target gene (Li et al. 2013a, b, c, d, e). Novel nematode-responsive root-specific promoter (NRRS) from Arabidopsis is seen to be expressed high in gall and root of transgenic Arabidopsis plants in defence to the infection of Meloidogyne incognita nematode (Kakrana et al. 2017).
2.1.2.4 Flower-specific promoters:
High-level expression was recorded in a floral-enhanced manner for 1.45 kb promoter of endogenous ubiquitin extension protein (UEP1), although 9-fold lower uidA expression was noticed in the leaves (Annadana et al. 2002). Comparison between CaMV35S and UEP1 promoters revealed that the UEP1 has 40- to 85-fold higher expression in the flower, indicating its high tissue-specificity. The Arabidopsis CER6 gene when transformed into Chrysanthemum resulted in high, specific expression in flower petals due to the CER6 promoter, but had high variability as compared to the UEP1 promoter (Hannoufa et al. 1996).
2.1.2.5 Fruit-specific promoters:
For the fruit-specific expression, promoters related to fruit ripening such as1-aminocyclopropane-1-carboxylate (ACCoxidase), E8 and polygalacturonase (PG) have been characterized from apple (Atkinson et al. 1998) and tomato (Nicholass et al. 1995). A fruit-ripening-induced promoter 1(RIP1) of tomato is perhaps more crucial for controlled manipulation of ripening-related agronomic traits not only in tomato but also in many other fruit crops (Agarwal et al. 2017). In transgenic tomato, the promoter ACC oxidase was able to drive ripening-specific expression of a reporter gene in the fruit only with no detectable activities in other tissues and mature green fruits (Atkinson et al. 1998). Transgenic tomato plants with enhanced sweetness, flavor and nutritive values were developed by expressing the monellin gene under the control of E8 promoter (Reddy et al. 2015). For the expression of target genes and directing expression of the virus-F protein as an oral vaccine to induce systemic immunity in mice, the promoter of the tomato E8 gene was used successfully in a number of instances (Lewinsohn et al. 2001). Although this promoter drives the expression of transgenes in flowers (predominantly anthers), its invariable expression throughout the ripening fruit makes it very valuable. The improved quality of ripened tomatoes in terms of vitamins and micronutrients content makes it valuable for use in transgenic research (Hadley et al. 2002). For the production of biopharmaceuticals, the use of organ-specific promoters are important so as to express the gene of interest in those organs that are able to produce protein(s) in an appropriate manner (Twyman 2003). Deletion analysis showed improvement in the quality and nutritional value of the fruits when the stearoyl-acyl-carrier-protein desaturase (Des) promoter region located between −590 and +10 was used for transforming oil palm and also dicots (Saed et al. 2012). However, comparative studies between constitutive and inducible promoters elaborating the specific merits of inducible promoters especially in the fruits are very meager in the literature. Therefore, such a study is vital to pinpoint the advantages of inducible promoters over that of constitutive (table 3).
2.1.2.6 Seed-specific promoters:
Seed-specific transgene expression has been used in many applications including nutritive value improvement, milled grain quality and production of industrial or pharmaceutical compounds (Ye et al. 2000; Cahoon and Shanklin 2000). Promoter’s specific to seed storage genes is an attractive target for such uses in GM crops. Soybean β-conglycinin gene promoter was used for seed-specific expression, where gene expression was restricted to the embryo during the mid-to-late maturation phase (Lessard et al. 2002). Likewise, groundnut seed promoter (GSP) can potentially be used for modification of seed phenotypes in agronomically important crops (Sunkara et al. 2014). While the sunflower helianthinin full-length promoter was used to drive seed-specific expression in tobacco (Nunberg et al. 1994), the expression of the β-phaseolin promoter in transgenic tobacco plants was restricted to the developing seed only (van der Geest and Hall 1997). Several endosperm-specific promoters like those from hordein in barley (Forde et al. 1985), glutenin in wheat (Lamacchia et al. 2001) and zein in maize (Marzabal et al. 1998) have been studied extensively. The guar MS promoter could manifest its usage for directing endosperm-specific expression of transgenes in legume species (Naoumkina and Dixon 2011). The expression of a 4kb promoter fragment from granule-bound starch synthase1 (gbss1) gene in wheat is restricted to endosperm and developing kernel pericarp tissues only (Kluth et al. 2002).
The WM403 promoter from watermelon may be useful in driving nucellus-specific gene expression in plants inclusive of candidate genes for important nucellus-specific attributes such as apospory or adventitious embryony (Dwivedi et al. 2010). HaFAD2-1 promoter is as strong as the CaMV35S promoter even though it is a tissue-specific promoter and its activity is derived from the embryo, thereby confirming that it can be considered as a strong, highly specific seed promoter useful for genetic manipulation applications in the seed (Zavallo et al. 2010). The Arabidopsis At4g12960 (AtGILT) promoter was employed as a canola seed coat outer integument-specific promoter after the production and selection of desired transformants from many transgenic lines (Wu et al. 2011). Seed coat-specific promoters can also be used to assess the effects of many pathway enzymes/proteins and cell-wall modifying proteins on mucilage structure as has been pointed out by Dean et al. (2017). Maize BD1, Def1 and Def2 promoters were active and reproduced the expression patterns of both Def1 and Def2 genes in transformed immature maize embryos, as well as in developing seeds of transgenic maize (Liu et al. 2016). Thus, an array of tissue-specific promoters has been used in diverse taxa with many beneficial effects. Table 4 highlights a list of tissue-specific promoters that have been used so far in plant genetic engineering studies.
2.1.2.7 Underground storage-tissue-specific promoters:
Promoters like pDJ3S and to a lesser extent pMe1 from yam and casava respectively drive the high and preferential expression of genes in carrot storage roots (Arango et al. 2010). Such promoters help in improving the biomass and nutritional quality of the underground storage tissues.
2.1.3 Abiotic stress inducible
Plant performance is not conditioned to endogenous factors alone, but also to environmental factors and external stimuli. Several promoters are induced by hormones, chemicals, and environmental stresses (figure 2a) (Chakravarthi et al. 2016). Based on the source and type of cells in which they regulate gene expression, hundreds of inducible promoters have been identified (Singh et al. 2002). For induction of promoters under different abiotic stresses, cis-acting elements like dehydration responsive element (DRE) (Yamaguchi-Shinozaki and Shinozaki 2006), an abscisic acid-responsive element (ABRE) (Bonetta and Mccourt 1998), and heat shock element (HSE) have been identified. The necessity of such a system for transgene expression may be vital to develop promoter set-ups where precise temporal regulation of transgene expression is a requisite. Examples include situations where unwanted gene expression is harmful or lethal, especially during the development of the plant (Guo et al. 2003). For example, peanut transgenics overexpressing AtDREB1A transcription factor under the influence of the CaMV35S promoter showed severe stunting that could be overcome by using the rd29A stress-inducible promoter to drive the expression of DREB1A (Bhatnagar-Mathur et al. 2007). Overexpression of a stress-related gene under an inducible promoter may confer better tolerance to stress than when under a constitutive one, while causing minimal or no growth retardation (Nakashima et al. 2007; Li et al. 2013a, b, c, d, e). The growth and development of CaMV35S-TaEXPB23 transgenic tobacco plants were altered under normal conditions, with a faster growth rate at the seedling stage, early flowering and maturation, and a shorter plant height compared to wild-type plants. On the other hand, RD29A-TaEXPB23 transgenic tobacco plants exhibited greater tolerance to water stress than the wild-type. Therefore, the use of stress-inducible promoters, such asrd29A may minimize the negative effects of constitutive transgene expression and improve the water-stress tolerance of plants (Shen et al. 2003; Bhatnagar-Mathur et al. 2007; Li et al. 2013a, b, c, d, e). These studies infer that stress-inducible promoters are superior for enhancing plant productivity under stress conditions in comparison with that of the constitutive promoters. However, it must be ensured that the stress-inducible promoter expression is very tightly controlled in response to the desired stress to avoid any unintended effects on plant growth and development.
Rd29X promoters have been demonstrated to be useful in controlling targeted transgenes to mitigate abiotic stress in soybean (Bihmidine et al. 2013) and peanut (Bhatnagar-Mathur et al. 2007; Rao et al. 2017). However, the usefulness of the endogenous inducible promoters is often limited since they are leaky. There may be a possibility for the activation of other endogenous genes by the same inducer. Srivastava et al. 2014 reported that PsSEOF1 promoter from pea could serve as an important candidate for tissue-specific promoter for engineering plants for both biotic and abiotic stress conditions. HsfB2c and PM19 promoters from rice are highly heat-inducible and further characterization and reorganization of cis-acting elements in their promoters could lead to the development of highly effective, heat-inducible promoters (Rerksiri et al. 2013). It was noticed that PR10 promoter from Erianthus though highly constitutive, was quickly induced upon wounding as well as on treatment with ABA and methyl jasmonate (Chakravarthi et al. 2016). The OsABA2 promoter was shown to drive a low constitutive transgene expression under normal conditions but high induction in response to ABA, salt and drought stresses (Rai et al. 2009). The AlSAP promoter from the halophyte grass Aeluropus littoralis directs a stress-inducible expression pattern in transgenic rice plants making it an interesting candidate for engineering abiotic stress tolerance in cereals (Ben-Saad et al. 2015). Bang et al. 2012 similarly demonstrated that OsNCED3 promoter was stress-inducible in whole rice plant except in the aleurones and endosperm and stably active over three generations. Stress-inducible expression of Hsc70, Lea, Hsp10, Dhn and Apx promoters from Pennisetum glaucum have been shown to confer abiotic stress tolerance in different tissues (leaf, stem and root) of the transgenic tobacco plants when exposed todifferent abiotic stresses (Divya et al. 2019; 2020, Divya et al. unpublished data). Hou et al. 2016 demonstrated that maize Type-II H+-pyrophosphatase promoter has higher expression under drought and salinity conditions compared to the CaMV35S promoter. The AtUSP promoter is highly inducible by phytohormones as well as multiple abiotic stresses that can be exploited as a stress-inducible promoter to develop multi-stress tolerant crops with least adverse effects on other important traits (Bhuria et al. 2016).
Promoter like the AhMTP1 having a variety of cis-acting elements, particularly the MYB-binding sites are involved in the evolution of zinc tolerance (Fasani et al. 2017). The three cadmium-inducible gene promoters such as OsGSTU5, OsGSTU37 and an OsHsp20 from rice could be potentially used for bio-environmental contamination and improving heavy metal tolerance in crops (Qiu et al. 2015). Recent study indicated that pGAL-2kb could be a useful in developing droughttolerant cultivars by driving transgene expression (Conforte et al. 2017). CcHyPRP promoter in Arabidopsis has been shown to be regulated by different stress factors which can be deployed for enhancing abiotic stress tolerance in transgenics (Srinath et al. 2017). ZmPIS and PZ7 promoter fragments in tobacco would be ideal candidates for overexpression of drought- and salinity-responsive genes to improve crop tolerance (Zhang et al. 2016a, b).
Regulated gene expression systems would also be valuable in GE applications such as conditional expression of herbicide, flowering and fruit ripening genes that emphasize the need for developing transgenics by deploying inducible promoter systems (Liu et al. 2013). Myb-related protein-like promoter from rice showed ethanol-inducible promoter activity and could be used to generate transgenic crops with desirable traits as demonstrated by Khanthapok et al. (2018). Afforementioned studies indicate that a wide spectrum of stress-inducible promoters exist, however their effectiveness and relative performance under multiple stress conditions in different taxa needs to be investigated. Recent study by Divya et al. (2019) it’s observed that PgApx and PgDhn promoters are upregulated in drought, heat, cold and salt stresses and PgHsc70 promoter is active in heat and drought stress. In another study, PgHsp10 promoter from same plant i, e., Pennisetum glaucum seen to be expressed in heat and drought stress in transgenic tobacco plants (Divya et al. 2020). Different stress-inducible promoters used in varied studies so far are listed in table 5.
2.1.4 Synthetic
Recent advances in plant promoter engineering are making strides to generate more constitutive, bidirectional or inducible synthetic promoters for a proper transcriptional modulation of transgene expression in plants. Synthetic promoters provide enormous advantages over their counterparts with respect to transgene expression at a specific developmental stage, strength and tissue specificity. Synthetic promoters can be rationally sketched and constructed using specific type, copy number and positioning of motifs upstream of synthetic or native core promoters. To date, most synthetic promoters tested were either hybrids of multiple promoter parts or fusions of specific cis-regulatory elements with a core promoter. Typically, synthetic motif sequence is derived from extant sequences that are multiplied or recombined. The selection, copy number and spacing of cis-elements ascertain the strength, temporal and spatial expression patterns of synthetic promoters. Selection of motifs with known functions can be conducted with the help of previous studies (Banerjee et al. 2015) or from the databases, synthetic motif library screening (Roccaro et al. 2013) and bioinformatics based de novo motif discovery (Tompa et al. 2005). As the deconstructive analysis of plant natural promoters for functional motif discovery has slowly increased and the effectiveness of using systems biology tools for denovo motif discovery has been experimentally demonstrated in Arabidopsis (Koschmann et al. 2012) and soybean (Liu et al. 2014). Once motifs of interest have been selected for synthetic promoter construction, copy number and spacing of motif need to be optimized. Usually, motif copy number often correlates with synthetic promoter strength which has been demonstrated in various plant species like rice (Wu et al. 1998), tobacco (Sawant et al. 2005) and Arabidopsis (Sahoo et al. 2014). The motif dosage effect in synthetic promoters is not surprising, since congruent findings have been observed in native promoters like in Malus (Espley et al. 2009) and Arabidopsis (Cao et al. 2014). When many motifs are accommodated in to a single synthetic promoter, spacing among motifs should be proper that is required for the hierarchical arrangement of their corresponding TFs in order to obtain full synergistic interactions with the RNA polymerase II complex (Sawant et al. 2005). Interestingly, two copies of the ACGT motif in synthetic promoters was shown to result in salicylic acid inducibility when separated by five nucleotides, but were ABA-inducible when separated by 25 nucleotides (Mehrotra and Mehrotra 2010).
The parsley (Petroselinum crispum) protoplast system was used for analyzing MAMP-responsive synthetic promoters which can be used by other plant systems to respond in cases of microbial pathogen attack (Kanofsky et al. 2016). Li et al. (2013a) reported that the synthetic promoter pCL made from Arabidopsis and potato was found optimal use for gene function research in potato tubers in response to low temperature. Altering the structure of CRT/DRE enhanced the CBF-associated transcription complex formation and thus improved the activity of this pCL, synthetic tuber-specific and cold-inducible promoter (Li et al. 2015). Comparative studies amongst the three natural promoters from rice (Rab16A) and two synthetically designed promoters, viz., 4X ABRE (abscisic acid-responsive element) having four tandem repeats of ABRE, and 2X ABRC (abscisic acid-responsive complex) having two tandem repeats of ABRE and two copies of coupling elements showed that 2XABRC make a better salinity/ABA-inducible promoter. The studies of Ganguly et al. (2011) indicated strong GUS expression in the whole seed (both embryo and aleurone layer of endosperm) only by 2X ABRC, while it was localized in the embryo for the other two promoters. Wang et al. (2015) demonstrated the synthesis of rice tissue-specific promoters and also developed a novel, feasible method for screening as well as for functional characterization of tissue-specific cis-acting elements with their flanking sequences at the genome-wide level in rice. This synthetic promoter including the 35S core sequence and two binding sites for cold-inducible CBF transcription factors (PDRE::35S) exhibited transient expression under chilling conditions using synthetic cold-inducible promoter which enhanced the target protein accumulation, and may decrease greenhouse heating expenses (Gerasymenko and Sheludko 2017). Synthetic SynP16 promoter designed from cis-motifs of soybean viz. ABF, ABRE, ABRE-Like, CBF, E2F-VARIANT, G-box, GCC-Box, MYB1, MYB4, RAV1-A, and RAV1-B (in multiple copies and various combination) with a minimal 35s core promoter and a 222 bp synthetic intron sequence. This synp16 promoter induced GUS expression in stress induclible manner and tissues specifically in transgenic soybean and Arabidopsis (Jameel et al. 2020). Thus, synthetic promoters that are rationally designed have become highly efficient components for precise regulation of target gene expression. Different synthetic promoters used thus far are shown in table 6.
2.2 RNA pol III promoters
RNA polymerase III (pol III) has been used for transcription of structural and catalytic RNAs including 5S rRNA, tRNA and most small nuclear RNAs. Promoters for pol III have diverse structural features including reinitiation property and transcribing of at least four different types of genes, such as tRNA, 5S rRNA, U6 sn-RNA, 7SL-Sc-RNA, 7SK RNA. Thus pol III transcribes housekeeping genes required at all times. Different types of pol III promoters such as type 1 (5S RNA), type 2 (tRNA Leu) promoters of the Xenopus laevis, type 3 promoter of the Homo sapiens U6 snRNA gene and the type 4 Sc type of the Saccharomyces cerevisiae promoters were noticed. Other pol III promoters are U3 and U6 which are used in monocots and dicots, respectively (Belhaj et al. 2013). Pol III promoters are generally used to express small RNAs, short hairpin RNA, and guide RNA in the CRISPR/Cas9 system adapted for genome editing. Pol III promoters have not been much characterized excepting in model plants. Also, their relative advantage over that of other promoters need to be explored further.
3 Promoters in genome editing (CRISPR/Cas9 system)
Genome editing is a type of GE where the specific DNA sequence can be inserted, deleted or replaced. This can be accomplished by introducing site-specific double-strand breaks (DSBs) at specific sites in the targeted genome. The induced DSBs are repaired either through non-homologous end joining (NHEJ) or homologous recombination (HR), resulting in specific and targeted mutations. Currently, four families of engineered nucleases are being used: meganucleases (MGNs), zinc finger nucleases (ZFNs), transcription activator-like effector-based nucleases (TALEN), and the clustered regularly interspaced short palindromic repeats (CRISPR). While the MGNs and ZFNs were costly and hard to engineer, TALE nucleases are flexible, specific and relatively complex and CRISPR/Cas9 nucleases are user-friendly and cost-effective (Belhaj et al. 2013). The CRISPR/Cas9 system derived from bacterial immune system and has been widely used for targeted genome editing in diverse organisms including mammalian and most of the plant species (Hsu et al. 2014; Ma et al. 2015; Guo et al. 2018). The CRISPR/Cas9 system uses either single or multi-guide RNA system to induce the specific edits and this efficiency depends on the endonuclease activity of the sgRNA/Cas9 complex and selection of the promoters (Guo et al. 2018). It has been observed that targeted efficiency of the CRISPR/Cas9 depends on the codon optimization of the Cas9 and the promoters. Targeted efficiency increases when expressing Cas9 gene other than CaMV35S promoter such as the dividing cell specific INCURVATA2 (Hyun et al. 2015), egg cell-specific promoters (Wang et al. 2015), the cell division-specific YAO (Yan et al. 2015), and the germ-line-specific SPOROCYTELESS (Mao et al. 2016) and embryo-specific promoter DD45 (Miki et al., 2018). Targeted efficiency increased significantly when ZmUbi1 promoter was used instead of CaMV35S (Feng et al. 2018). Feng et al. (2018) reported that Zmdmc1 promoter could be an alternative to the CaMV35S for expressing the CRISPR/Cas9 system in maize for generating highly efficient targeted genome editing.
The pol III promoters such as U3 and U6 are commonly used to express the small RNAs and guide RNAs of the CRISPR/Cas9 system. Due to their long-term regulation of target genes and their defined sites for transcription initiation and termination, U3 and U6 promoters gained special attention over the others. Characterization of pol III promoters in many organisms was not properly explored and this becomes difficult in choosing the right promoters for CRISPR/Cas9 gene edited targeted mutagenesis. Hence, in most of the studies, pol III promoters such as U3 or U6 from Arabidopsis or rice or maize or wheat are often chosen to drive gRNAs (Nekrasov et al. 2013; Bortesi and Fischer 2015; Feng et al. 2018). Maize ubi1 gene promoter in combination with two rice U6 promoters performed well in maize with mutation efficiency up to 70% (Char et al. 2016). Processing of a pol II transcript into functional gRNA has been found successful using ribozyme or Csy4RNA cleavage systems. Mikami et al. (2017) have shown that functional gRNAs can be efficiently processed using SpCas9 protein and plant RNA cleavage systems without any need for a specific RNA processing. Recent studies have shown that AtU6 promoter from Arabidopsis and Fve U6 promoter of wild strwaberry are equally good for the high-efficiency genome editing (Zhou et al. 2013). Hashimoto et al. (2018) demonstrated increased efficiency and also improved multiplex genome editing using Cas9 gene expression with SlEF1α promoter. Ordon et al. (2020) demonstrated that DD45 and RPS5a promoter-driven Cas9 showed higher mutational frequency. Promoters used for genome editing are shown in the table 5.
4 Bidirectional promoters (BDPs)
A synthetic bidirectional expression module was prepared by placing a computationally designed minimal promoter sequence at the 5′ and 3′ sides of a transcription activation module. BDPs were identified in diverse organisms including plants. Structural features and functional consequences of BDPs received much attention in plant biotechnology due to their effective usage in gene-stacking/pyramiding and targeting the complex traits (Que et al. 2010). However, the underlying mechanisms responsible for the bidirectional transcription and co-expression of BDPs has remained poorly understood in plants where they drive the transcription of the two target genes (Patro et al. 2013). Such promoters play a pivotal role in the transcription of bidirectional gene pairs where the two genes are positioned head-to-head on opposite strands of DNA (figure 2c). The BDPs efficiently regulate two transgenes simultaneously which can be evaluated in multiple model-plant systems. The BDPs representing Rep and coat protein (CP) genes of CLCuBuV were characterized and their efficacy assayed (Khan et al. 2015). They also showed that the strong constitutive CLCuBuV Rep promoter could be of use for higher expression of transgenes in a variety of plant cells. Wang et al. (2016a, b) combined RNA-seq data and cDNA microarray data to discover the potential BDPs in rice genome. Comparative analysis between ZmBD1, ZmDef1 and ZmDef2 promoters revealed that PZmBD1 shared most of the expression characteristics of the two polar promoters, but displayed more stringent embryo specificity, delayed expression initiation, and asymmetric promoter activity (Liu et al. 2016).
The initiation of transcription from both the unidirectional and bidirectional promoters made from the same sequence elements were evaluated by using the uidA and gfp reporter genes. The investigation based on transient and stable transformation of tobacco exhibited that the artificially designed multifactorial activation module activated the transcription simultaneously to proportionate levels in both the directions (Chaturvedi et al. 2006). The transcription regulatory module responded to elicitors like salicylic acid (SA), NaCl and IAA in the forward as well as reverse directions. It implied that constitutive and chemically-inducible bidirectional promoters can be deployed for predictable simultaneous regulation of two genes for genetic engineering in plants (Chaturvedi et al. 2006). Fang et al. 2016 elucidated the unique epigenetic mechanism of BDPs and regulation of the bidirectional gene pairs and eventually used for GE. The bidirectional promoters At4g35985 (P85) and At4g35987 (P87) in both orientations display up-regulation under salt stress. Such regulatory elements of BDPs showing spatial and stress-inducible promoter and functioning in heterologous systems might be an important tool for plant biotechnology and gene stacking applications (Banerjee et al. 2013). Recently, Araceli et al. (2017) have shown that BDPs function can be regulated by degree/intensity of the abiotic stresses like Pi availability. GhZU promoter from Gossypium hirsutum is seen to regulate the expression of gus and gfp genes in both the directions in transgenic Arabiposis plants (Yang et al. 2018). From these studies, it can be stated that BDPs have a potential role to play in plant genetic engineering (Table 7).
5 Conclusions and future perspectives
The selection of a promoter suitable for the targeted transgene expression is one of the most important criteria for developing the GM crops. There are some important implications for producing transgenic plants with higher yield without any compromise on environmental and biosafety concerns. Improved expression of transgenes has been carried by hybrids or combined promoters that are from the constitutive promoters. Constitutive promoters are often seen to be beneficial for a high-level expression of selectable marker genes, necessary for efficient selection and generation of transgenics. Constitutively active promoters are not necessarily always desirable for GE plants as the constitutive overexpression of a transgene may compete for energy and building blocks for synthesis of proteins, RNA and others. Transgenic plants constitutively expressing TFs have been shown to exhibit mild/severe growth retardation in the aerial parts, shorter petioles, rounder leaves, delayed flowering and a dwarf phenotype. In these situations, use of tissue-specific or stress-inducible promoters of moderate strength may be more desirable.
Tissue-specific and stress-inducible promoters can be exploited to minimize the unintended effects of adverse environmental conditions such as heat, cold, drought, and salinity. Inducible promoters are very powerful resources for GE plants, since the expression of genes operably linked to them can be regulated to function at certain stages of growth and development of an organism or a particular tissue which help in reduction of energy expenditure of the plant. Further, inducible promoters will be activated only when required, thereby maintaining yield or productivity even under adverse conditions. On the other hand, synthetic promoters paired with synthetic TFs can be used to provide a coordinated transcriptional control of multiple genes, which would be needed for successful metabolic engineering.
Genome-wide computational prediction and analysis of core promoter elements across plant monocots and dicots will be useful for future work related to genome annotation projects and can inspire research efforts aimed to better understand regulatory mechanisms of transcription. Genome-wide transcriptomic analysis such as RNA-seq will also help in gene regulation as well as in the identification of promoters and cis-acting elements. Advancement in the area of systems biology and the development of new tools would be beneficial in the discovery of novel promoters and their cis-acting elements. Identification and deployment of promoters that can help under diverse stress conditions are desirable, ultimately for the generation of transgenics with better survival and productivity. CRISPR technology has become popular and an alternative to the plant breeding techniques. Genome editing efficiency can be improved by optimizing the diverse range of the promoters. Hence, crop varieties developed by above mentioned technologies are considered as non-GM in USA and other regions. Relaxing in usage of such crop varieties from the scope of the GMO legislation may have a positive impact on the development of the plant biotechnology and breeding sector for the betterment of humans.
References
Acharya S, Ranjan R, Pattanaik S, Maiti IB and Dey N 2014 Efficient chimeric plant promoters derived from plant infecting viral promoter sequences. Planta 239 381–396
Agarwal P, Kumar R, Pareek A and Sharma AK 2017 Fruit preferential activity of the tomato RIP1 gene promoter in transgenic tomato and Arabidopsis. Mol. Genet. Genom. 292 145–156
Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M and Mahfouz MM 2015 CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16 238
Althoff F, Kopischke S, Zobell O, Ide K, Ishizaki K, Kohchi T and Zachgo S 2014 Comparison of the MpEF1alpha and CaMV35 promoters for application in Marchantia polymorpha overexpression studies. Transgenic Res. 23 235–244
An YQ, McDowell JM, Huang S, McKinney EC, Chambliss S and Meagher RB 1996 Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10 107–121
Annadana S, Beekwilder MJ, Kuipers G, Visser PB, Outchkourov N, Pereira A, Udayakumar M, De Jong J and Jongsma MA 2002 Cloning of the chrysanthemum UEP1 promoter and comparative expression in florets and leaves of Dendranthema grandiflora. Transgenic Res. 11 437–445
Araceli OA, Alfredo CR, Javier MM and Luis HE 2017 A phosphate starvation‐driven bidirectional promoter as a potential tool for crop improvement and in vitro plant biotechnology. Plant Biotechnol. J. 15 558–567
Arango J, Salazar B, Welsch R, Sarmiento F, Beyer P and Al-Babili S 2010 Putative storage root specific promoters from cassava and yam: cloning and evaluation in transgenic carrots as a model system. Plant Cell Rep. 29 651–659
Atkinson RG, Bolitho KM, Wright MA, Iturriagagoitia-Bueno T, Reid SJ and Ross GS 1998 Apple ACC-oxidase and polygalacturonase:ripening-specific gene expression and promoter analysis in transgenic tomato. Plant Mol. Biol. 38 449–460
Bang SW, Park SH, Jeong JS, Kim YS, Jung H, Ha SH and Kim JK 2012 Characterization of the stress-inducible OsNCED3 promoter in different transgenic rice organs and over three homozygous generations. Planta 237 211–224.
Banerjee J, Sahoo DK, Dey N, Houtz RL and Maiti IB 2013 An intergenic region shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue specific and stress inducible bidirectional promoter analyzed in transgenic arabidopsis and tobacco plants. PLoS One. 8 e79622
Banerjee J, Sahoo DK, Raha S, Sarkar S, Dey N and Maiti IB 2015 A region containing an as-1 element of Dahlia Mosaic Virus (DaMV) subgenomic transcript promoter plays a key role in green tissue and root-specific expression in plants Plant Mol. Biol. Rep. 33 532–556
Belhaj K, Chaparral-Garcia A, Amount S and Nekrasov V 2013 Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9 1–10
Benn G, Wang CQ, Hicks DR, Stein J, Guthrie C and Dehesh K 2014 A key general stress response motif is regulated non-uniformly by CAMTA transcription factors. Plant J. 80 82–92
Ben-Saad R, Meynard D, Ben-Romdhane W, Mieulet D, Verdeil JL, Al-Doss A, Guiderdoni E and Hassairi A 2015 The promoter of the AlSAP gene from the halophyte grass Aeluropus littoralis directs a stress-inducible expression pattern in transgenic rice plants. Plant Cell Rep. 34 1791–1806
Benfey PN, Ren L and Chua NH 1990 Tissue-specific expression from CaMV35S enhancer subdomains in early stages of plant development. EMBO J. 9 1677–1684
Benfey PN, Takatsuji H, Ren L, Shah DM and Chua NH 1990 Sequence requirements of the 5-enolpyruvylshikimate-3-phosphate synthase 5[prime]-upstream region for tissue-specific expression in flowers and seedlings. Plant Cell 2 849–856
Beringer J, Chen W, Garton R, Sardesai N, Wang PH, Zhou N, Gupta M and Wu H 2017 Comparison of the impact of viral and plant-derived promoters regulating selectable marker gene on maize transformation and transgene expression. Plant Cell Rep. 36 519–528
Bhattacharyya J, Chowdhury AH, Ray S, Jha JK, Das S, Gayen S, Chakraborty A, Mitra J, Maiti MK, Basu A and Sen SK 2011 Native polyubiquitin promoter of rice provides increased constitutive expression in stable transgenic rice plants Plant Cell Rep. 31 271–279
Bhattacharyya S, Dey N and Maiti IB 2002 Analysis of cis-sequence of subgenomic transcript promoter from the Figwort mosaic virus and comparison of promoter activity with the cauliflower mosaic virus promoters in monocot and dicot cells. Virus Res. 90 47–62
Bhattacharyya S, Pattanaik S and Maiti IB 2003 Intron-mediated enhancement of gene expression in transgenic plants using chimeric constructs composed of the Peanut chlorotic streak virus (PClSV) promoter-leader and the antisense orientation of PClSV ORF VII (p7R). Planta 218 115–24
Bhatnagar-Mathur P, Devi MJ, Reddy DS, Lavanya M, Vadez V, Serraj R, Yamaguchi-Shinozaki K and Sharma KK 2007 Stress-inducible expression of At DREB1A in transgenic peanut (Arachis hypogaea L) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep. 26 2071–2082
Bhuria M, Goel P, Kumar S and Singh AK 2016 The Promoter of AtUSPis co-regulated by phytohormones and abiotic stresses in Arabidopsis thaliana. Front Plant Sci. 7 1957
Bihmidine S, Lin J, Stone JM, Awada T, Specht JE and Clemente TE 2013 Activity of the Arabidopsis RD29A and RD29B promoter elements in soybean under water stress. Planta 23 755–64
Biłas R, Szafran K, Hnatuszko-Konka K and Kononowicz AK 2016 Cis-regulatory elements used to control gene expression in plants Plant Cell Tiss. Org. Cul. 127 269–287
Bonetta D and Mccourt P 1998 Genetic analysis ofABA signal transduction pathways. Trends Plant Sci. 3 231–235
Borah BK, Zarreen F, Baruah G and Dasgupta I 2016 Insights into the control of geminiviral promoters. Virology 495 101–111
Bortesi L and Fischer R 2015 The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33 41–52
Bucchini L and Goldman LR 2002 Starlink corn: a risk analysis Environ Health Perspect 110 5
Biłas R, Szafran K, Hnatuszko-Konka K and Kononowicz AK 2016 Cis-regulatory elements used to control gene expression in plants Plant Cell Tiss. Org. Cult. 127 269–287
Cahoon EB and Shanklin J 2000 Substrate-dependent mutant complementation to select fatty acid desaturase variants for metabolic engineering of plant seed oils. Proc Nat Acad Sci. 97 12350–12355
Cao S, Kumimoto RW, Gnesutta N, Calogero AM, Mantovani R and Holt BF 2014 A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26 1009–1017
Cazzonelli CI, McCallum EJ, Lee R and Botella JR 2005 Characterization of a strong constitutive mung bean (Vigna radiata L) promoter with a complex mode of regulation in planta. Transgenic Res. 14 941–967
Chakravarthi M, Syamaladevi DP, Harunipriya P, Augustine SM and Subramonian N 2016 A novel PR10 promoter from Erianthus arundinaceus directs high constitutive transgene expression and is enhanced upon wounding in heterologous plant systems. Mol. Biol. Rep. 43 17–30
Chaturvedi CP, Sawant SV, Kiran K, Mehrotra R, Lodhi N, Ansari SA and Tuli R 2006 Analysis of polarity in the expression from a multifactorial bidirectional promoter designed for high-level expression of transgenes in plants. J. Biotechnol. 123 1231–1312
Char SN, Neelakandan AK, Nahampun H, Frame B, Main M, Spalding M H et al (2017) An Agrobacterium‐delivered CRISPR/Cas9 system for high‐frequency targeted mutagenesis in maize. Plant Biotechnol. J. 15 257–268.
Chen Y, Chen S, Chen F, Li P, Chen L, Guan Z and Chang Q 2012 Functional characterization of a Chrysanthemum dichrum stress-related promoter. Mol. Biotechnol. 52 161–169
Chen L, Jiang B, Wu C, Sun S, Hou W and Han T 2014 GmPRP2 promoter drives root-preferential expression in transgenic Arabidopsis and soybean hairy roots. BMC Plant Biol. 14 245
Christensen AH, Sharrock RA and Quail PH 1992 Maize polyubiquitin genes: structure hermal perturbation of expression and transcript splicing and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18 675–689
Cominelli E, Galbiati M, Albertini A, Fornara F, Conti L, Coupland G and Tonelli C 2011 DOF-binding sites additively contribute to guard cell-specificity of AtMYB60 promoter. BMC Plant Biol. 11 162
Company N, Nadal A, Ruiz C and Pla M 2014 Production of phytotoxic cationic alpha-helical antimicrobial peptides in plant cells using inducible promoters. PLoS One 9 e109990
Conforte AJ, Guimaraes-Dias F, Neves-Borges AC, Bencke-Malato M, Felix-Whipps D and Alves-Ferreira M (2017 Isolation and characterization of a promoter responsive to salt osmotic and dehydration strsses in soybean. Genet. Mol. Biol. 40 226–237
Coussens G, Aesaert S, Verelst W, Demeulenaere M, De Buck S, Njuguna E, Inze D and Van Lijsebettens M 2012 Brachypodium distachyon promoters as efficient building blocks for transgenic research in maize. J. Exp. Bot. 63 4263–4273
Dean GH, Jin Z, Shi L, Esfandiari E, McGee R, Nabata K et al 2017 Identification of a seed coat-specific promoter fragment from the Arabidopsis MUCILAGE-MODIFIED4 gene. Plant Mol. Bio. 95 33–50
Dhankher OP, Li Y, Rosen BP, Shi J, Salt D, Senecoff JF, Sashti NA and Meagher RB 2002 Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and gamma-glutamylcysteine synthetase expression. Nat. Biotechnol. 20 1140–1145
Divya K, Kishor PBK, Bhatnagar-Mathur P, Singam P, Sharma KK, Vadez V and Reddy PS 2019 Isolation and functional characterization of three abiotic stress-inducible (Apx, Dhn and Hsc70) promoters from pearl millet (Pennisetum glaucum L) Mol. Biol.Rep. 46 6039–6052
Divya K, Bhatnagar-Mathur P, Sharma KK, Vadez V and Reddy PS 2020 Functional characterization of the promoter of pearl millet heat shock protein 10 (PgHsp10) in response to abiotic stresses in transgenic tobacco plants. Int. J. Biol. Macromol. 12 103–110.
Du L, Lou Q, Zhang X, Jiao S, Liu Y and Wang Y 2014 Construction of flower-specific chimeric promoters and analysis of their activities in transgenic Torenia. Plant Mol. Biol. Rep. 32 234–245
Dwivedi KK, Roche D and Carman JG 2010 Expression in Arabidopsis of a nucellus-specific promoter from watermelon (Citrullus lanatus). Plant Sci. 179 549–552
Erpen L, Tavano ECR, Harakava R, Dutt M, Grosser JW, Piedade SMS and Mourão Filho FAA 2018 Isolation characterization and evaluation of three Citrus sinensis-derived constitutive gene promoters. Plant Cell Rep. 37 1113–1125
Erpen-Dalla Corte L, Mendes BMJ, Mourão Filho FAA, Grosser JW and Dutt M 2020 Functional characterization of full-length and 5′ deletion fragments of Citrus sinensis-derived constitutive promoters in Nicotiana benthamiana. In Vitro Cell. Dev. Biol. Plant 56 280–289
Espley RV, Brendolise C, Chagne D, Kutty-Amma S, Green S, Volz R, Putterill J, Schouten HJ, Gardiner SE, Hellens RP and Allan AC 2009 Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21 168–183
Fang RX, Nagy F, Sivasubramaniam S and Chua NH 1989 Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. Plant Cell, 1 141–150
Fang Y, Wang L, Wang XY, Ou Q, Pan X, Xiao J and Zhang W 2016 Histone modifications facilitate the coexpression of bidirectional promoters in rice. BMC Genomics 17 768
Fasani E, DalCorso G, Varotto C, Li M, Visioli G, Mattarozzi M and Furini A 2017 The MTP1 promoters from Arabidopsis halleri reveal cis-regulating elements for the evolution of metal tolerance. New Phytol. 214 1614–1630
Fauser F, Schiml S and Puchta H 2014 Both CRISPR/C as‐based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79 348–359
Feki K, Brini F, Ben Amar S, Saibi W and Masmoudi K 2015 Comparative functional analysis of two wheat Na(+)/H(+) antiporter SOS1 promoters in Arabidopsis thaliana under various stress conditions. J. Appl. Genet. 56 15–26.
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P and Zhu JK 2013 Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23 1229
Feng C, Su H, Bai H, Wang R, Liu Y, Guo X and Han F 2018 High‐efficiency genome editing using a dmc1 promoter‐controlled CRISPR/Cas9 system in maize. Plant Biotechnol. J. 16 1848–1857
Forde BG, Heyworth A, Pywell J and Kreis M 1985 Nucleotide sequence of a B1 hordein gene and the identification of possible upstream regulatory elements in endosperm storage protein genes from barley wheat and maize. Nucleic Acids Res. 13 7327–7339
Francia P, Simoni L, Cominelli E, Tonelli C and Galbiati M 2008 Gene trap-based identification of a guard cell promoter in Arabidopsis. Plant Signal. Beh. 3 684–686
Fromm ME, Morrish F, Armstrong C, Williams R, Thomas J and Klein TM 1990 Inheritance and expression of chimeric genes in the progeny of transgenic maize plants. Biotechnology (NY) 8 833–839
Gago J, Grima-Pettenati J and Gallego PP 2011 Vascular-specific expression of GUS and GFP reporter genes in transgenic grapevine (Vitis vinifera L cv Albarino) conferred by the EgCCR promoter of Eucalyptus gunnii. Plant Physiol. Biochem. 49 413–419
Ganguly M, Roychoudhury A, Sarkar SN, Sengupta DN, Datta SK and Datta K 2011 Inducibility of three salinity/abscisic acid-regulated promoters in transgenic rice with gusA reporter gene Plant Cell Rep. 30 1617–1625
Gerasymenko IM and Sheludko YV 2017 Synthetic cold-inducible promoter enhances recombinant protein accumulation during Agrobacterium-mediated transient expression in Nicotiana excelsior at chilling temperatures Biotechnol. Lett. 39 1059–1067
Govindarajulu M, Elmore JM, Fester T and Taylor CG 2008 Evaluation of constitutive viral promoters in transgenic soybean roots and nodules. Mol. Plant Microbe Interact. 21 1027–1035
Guo HS, Fei JF, Xie Q and Chua NH 2003 A chemical-regulated inducible RNAi system in plants. Plant J. 34 383–392
Guo J, Li K, Jin L, Xu R, Miao K, Yang F and Miao Y 2018 A simple and cost-effective method for screening of CRISPR/Cas9-induced homozygous/biallelic mutants. Plant Methods 14 40
Gupta P, Raghuvanshi S and Tyagi AK 2001 Assessment of the efficiency of various gene promoters via biolistics in leaf and regenerating seed callus of millets Eleusine coracana and Echinochlo acrusgalli. Plant Biotech. 18 275–282
Hadley CW, Miller EC, Schwartz SJ and Clinton SK 2002 Tomatoes lycopene and prostate cancer: progress and promise. Exp. Biol. Med. 227 869–880
Han YJ, Kim YM, Hwang OJ and Kim JI 2015 Characterization of a small constitutive promoter from Arabidopsis translationally controlled tumor protein (AtTCTP) gene for plant transformation. Plant Cell Rep. 34 265–275
Hannoufa A, Negruk V, Eisner G and Lemieux B 1996 The CER3 gene of Arabidopsis thaliana is expressed in leaves stems roots flowers and apical meristems. Plant J. 10 459–467
Hashimoto R, Ueta R, Abe C, Osakabe Y and Osakabe K 2018 Efficient multiplex genome editing induces precise and self-ligated type mutations in tomato plants. Front. Plant Sci. 9 916
Hernandez-Garcia CM and Finer JJ 2014 Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 217–218 109–119
Hernandez-Garcia CM and Finer JJ 2016 A novel cis-acting element in the GmERF3 promoter contributes to inducible gene expression in soybean and tobacco after wounding. Plant Cell Rep. 35 303–316
Hernandez-Garcia CM, Martinelli AP, Bouchard RA and Finer JJ 2009 A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep. 28 837–849
Hou J, Jiang P, Qi S, Zhang K, He Q, Xu C, Ding Z, Zhang K and Li K 2016 Isolation and functional validation of salinity and osmotic stress inducible promoter from the maize Type-II H+-Pyrophosphatase gene by deletion analysis in transgenic tobacco Plants. PLoS One 11 e0154041
Hsu PD, Lander ES and Zhang F 2014 Development and applications of CRISPR-Cas9 for genome engineering. Cell 157 1262–1278
Hyun Y, Kim J, Cho SW, Choi Y, Kim JS and Coupland G 2015 Site-directed mutagenesis in Arabidopsis thaliana using dividing tissue-targeted RGEN of the CRISPR/Cas system to generate heritable null alleles. Planta 241 271–284
Jameel A, Noman M, Liu W, Ahmad N, Wang F, Li X and Li H 2020 Tinkering cis motifs jigsaw puzzle led to root-specific drought-inducible novel synthetic promoters. Int. J. Mol. Sci. 21 1357
Jiang P, Zhang K, Ding Z, He Q, Li W, Zhu S and Li K 2018 Characterization of a strong and constitutive promoter from the Arabidopsis serine carboxypeptidase-like gene AtSCPL30 as a potential tool for crop transgenic breeding. BMC Biotech. 18 59
Johnson JA, Bragg JN, Lawrence DM and Jackson AO 2003 Sequence elements controlling expression of Barley stripe mosaic virus subgenomic RNAs in vivo. Virology 313 66–80
Kakrana A, Kumar A, Satheesh V, Abdin MZ, Subramaniam K, Bhattacharya RC and Jain PK 2017 Identification validation and utilization of novel nematode-responsive root-specific promoters in Arabidopsis for inducing host-delivered RNAi mediated root-knot nematode resistance. Front Plant Sci. 8 2049
Kanofsky K, Lehmeyer M, Schulze J and Hehl R 2016 Analysis of microbe-associated molecular pattern-responsive synthetic promoters with the parsley protoplast system. Methods Mol. Biol. 1482 163–174
Khanthapok P, Sang-Awut N, Chakhonkaen S, Pitngam K, Osadcenco A, Sukrong S and Muangprom A 2018 Identification of ethanol-inducible genes and isolation of the myb-related protein-like promoter in Oryza sativa L. J. Plant Growth Regul. 37 452–470
Keadtidumrongkul P, Suttangkakul A, Pinmanee P, Pattana K, Kittiwongwattana C, Apisitwanich S and Vuttipongchaikij S 2017 Growth modulation effects of CBM2a under the control of AtEXP4 and CaMV35S promoters in Arabidopsis thaliana, Nicotiana tabacum and Eucalyptus camaldulensis. Transgenic Res. 26 447–463
Kelly G, Lugassi N, Belausov E, Wolf D, Khamaisi B, Brandsma D, Kottapalli J, Fidel L, Ben-Zvi B, Egbaria A, Acheampong AK, Zheng C, Or E, Distelfeld A, David-Schwartz R, Carmi N and Granot D 2017 The Solanum tuberosum KST1 partial promoter as a tool for guard cell expression in multiple plant species. J. Exp. Bot. 68 2885–2897
Khan ZA, Abdin MZ and Khan JA 2015 Functional characterization of a strong bi-directional constitutive plant promoter isolated from cotton leaf curl Burewala virus. PLoS One 10 e0121656
Kevin E, Mc Bride and Kristin R 1989 Improved binary vectors for Agrobacterium-mediated plant transformation. SummerfeltCalgene Inc. 1920 Fifth St., Davis, CA 95616, USA
Kluth A, Sprunck S, Becker D, Lorz H and Lutticke S 2002 5’ deletion of a gbss1 promoter region from wheat leads to changes in tissue and developmental specificities. Plant Mol. Biol. 49 669–682
Koia J, Moyle R, Hendry C, Lim L and Botella JR 2013 Pineapple translation factor SUI1 and ribosomal protein L36 promoters drive constitutive transgene expression patterns in Arabidopsis thaliana. Plant Mol. Biol. 81 327–336
Koschmann J, Machens F, Becker M, Niemeyer J, Schulze J, Bulow L, Stahl DJ and Hehl R 2012 Integration of bioinformatics and synthetic promoters leads to the discovery of novel elicitor-responsive cis-regulatory sequences in Arabidopsis. Plant Physiol. 160 178–191
Kyozuka J, Fujimoto H, Izawa T and Shimamoto K 1991 Anaerobic induction and tissue-specific expression of maize Adh1 promoter in transgenic rice plants and their progeny. Mol. Gen. Genet. 228 40–48
Lamacchia C, Shewry PR, Di Fonzo N, Forsyth JL, Harris N, Lazzeri PA, Napier JA, Halford NG and Barcelo P 2001 Endosperm-specific activity of a storage protein gene promoter in transgenic wheat seed. J. Exp. Bot. 52 243–250
Lessard PA, Kulaveerasingam H, York GM, Strong A and Sinskey AJ 2002 Manipulating gene expression for the metabolic engineering of plants. Metab. Eng. 4 467–479
Lewinsohn E, Schalechet F, Wilkinson J, Matsui K, Tadmor Y, Nam KH, Amar O, Lastochkin E, Larkov O, Ravid U, Hiatt W, Gepstein S and Pichersky E 2001 Enhanced levels of the aroma and flavor compound S-linalool by metabolic engineering of the terpenoid pathway in tomato fruits. Plant Physiol. 127 1256–1265
Li F, Han Y, Feng Y, Xing S, Zhao M, Chen Y and Wang W 2013 Expression of wheat expansin driven by the RD29 promoter in tobacco confers water-stress tolerance without impacting growth and development. J. Biotechnol. 163 281–291
Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM and Sheen J 2013 Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31 688–691
Li M, Wang X, Cao Y, Liu X, Lin Y, Ou Y, Zhang H and Liu J 2013a Strength comparison between cold-inducible promoters of Arabidopsiscor15a and cor15b genes in potato and tobacco. Plant Physiol. Biochem. 71 77–86
Li M, Xie C, Song B, Ou Y, Lin Y, Liu X, Zhang H and Liu J 2015 Construction of efficient tuber-specific and cold-inducible promoters in potato. Plant Sci. 23 514–524
Li T, Huang S, Zhou J and Yang B 2013 Designer TAL effectors induce disease susceptibility and resistance to Xanthomonas oryzaepv oryzae in rice. Mol. Plant 6 781–789
Li Y, Liu S, Yu Z, Liu Y and Wu P 2013b Isolation and characterization of two novel root-pecific promoters in rice (Oryza sativa L). Plant Sci. 207 37–44
Li J, Xu RF, Qin RY, Ma H, Li H, Zhang YP, Li L, Wei PC and Yang JB 2014 Isolation and functional characterization of a novel rice constitutive promoter. Plant Cell Rep. 33 1651–60
Li Z, Jayasankar S and Gray DJ 2001 Expression of a bifunctional green fluorescent protein (GFP) fusion marker under the control of three constitutive promoters and enhanced derivatives in transgenic grape (Vitis vinifera). Plant Sci. 160 877–887
Liu C, Fukumoto T, Matsumoto T, Gena P, Frascaria D, Kaneko T et al 2013 Aquaporin OsPIP1; 1 promotes rice salt resistance and seed germination. Plant Physiol. Biochem. 63 151–158
Liu W, Mazarei M, Peng Y, Fethe MH, Rudis MR, Lin J, Millwood RJ, Arelli PR and Stewart CN 2014 Computational discovery of soybean promoter cis-regulatory elements for the construction of soybean cyst nematode-inducible synthetic promoters. Plant Biotechnol. J. 12 1015–1026
Liu YG and Whittier RF 1995 Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25 674–681
Liu X, Yang W, Li Y, Li S, Zhou X, Zhao Q, Fan Y, Lin M and Chen R 2016 The intergenic region of the maize defensin-like protein genes Def1 and Def2 functions as an embryo-specific asymmetric bidirectional promoter. J. Exp. Bot. 67 4403–4413
Liu Z, Luo QH, Wang JM, Li XF and Yang Y 2015 Functional characterization and analysis of the Arabidopsis UGT71C5 promoter region. Genet. Mol. Res. 14 191 73–83
Long L, Guo DD, Gao W, Yang WW, Hou LP, Ma XN and Song CP 2018 Optimization of CRISPR/Cas9 genome editing in cotton by improved sgRNA expression. Plant Methods 14 85
Lota F, Wegmüller S, Buer B, Sato S, Bräutigam A, Hanf B and Bucher M 2013 The cis-acting CTTC-P1BS module is indicative for gene function of LjVTI12 a Qb-SNARE protein gene that is required for arbuscule formation in Lotus japonicas. Plant J. 74 280–293
Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R et al. 2015 A robust CRISPR/Cas9 system for convenient high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8 1274–1284
Ma F, Qi H, Hu Y, Jiang Q, Zhang LG, Xue P and Zhang Q 2018 The mitochondrial endonuclease M20 Participates in the down-regulation of mitochondrial DNA in pollen cells. Plant Physiol. 178 1537–1550
Maiti IB and Shepherd RJ 1998 Isolation and expression analysis of peanut chlorotic streak caulimovirus (PClSV) full-length transcript (FLt) promoter in transgenic plants. Biochem. Biophys. Res. Commun. 244 440–444
Malik K, Wu K, Li XQ, Martin-Heller T, Hu M, Foster E, Tian L, Wang C, Ward K, Jordan M, Brown D, Gleddie S, Simmonds D, Zheng S, Simmonds J and Miki B 2002 A constitutive gene expression system derived from the tCUP cryptic promoter elements. Theor. Appl. Genet. 105 505–514
Mann DG, King ZR, Liu W, Joyce BL, Percifield RJ, Hawkins JS, LaFayette PR, Artelt BJ, Burris JN, Mazarei M, Bennetzen JL, Parrott WA and Stewart CN 2011 Switchgrass (Panicum virgatum L) polyubiquitin gene (PvUbi1 and PvUbi2) promoters for use in plant transformation. BMC Biotechnol. 11 74
Mao Y, Zhang Z, Feng Z, Wei P, Zhang H, Botella JR and Zhu JK 2016 Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 14 519–532
Marshallsay C, Connelly S and Filipowicz W 1992 Characterization of the U3 and U6 snRNA genes from wheat: U3 snRNA genes in monocot plants are transcribed by RNA polymerase III. Plant Mol. Biol. 19 973–983
Marzabal P, Busk PK, Ludevid MD and Torrent M 1998 The bifactorial endosperm box of gamma-zein gene: characterisation and function of the Pb3 and GZM cis-acting elements. Plant J. 16 41–52
McElroy D, Blowers AD, Jenes B and Wu R 1991 Construction of expression vectors based on the rice actin 1 (Act1) 5’ region for use in monocot transformation. Mol. Gen. Genet. 231 150–160
Mehrotra R and Mehrotra S 2010 Promoter activation by ACGT in response to salicylic and abscisic acids is differentially regulated by the spacing between two copies of the motif. J. Plant Physiol. 167 1214–1218
Meyer S, Mumm P, Imes D, Endler A, Weder B, Al‐Rasheid KA and Hedrich R 2010 AtALMT12 represents an R‐type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 63 1054–1062
Miki D, Zhang W, Zeng W, Feng Z and Zhu JK 2018 CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat. Commun. 9 1967
Mikami M, Toki S, and Endo M 2017 In Planta processing of the SpCas9–gRNA complex. Plant Cell Physiol 58 1857–1867
Miyata LY, Harakava R, Stipp LC, Mendes BM, Appezzato-da-Glória B, de Assis Alves Mourão and Filho F 2012 GUS expression in sweet oranges (Citrus sinensisL Osbeck) driven by three different phloem-specific promoters. Plant Cell Rep. 31 2005–2013
Mittler R and Blumwald E 2010 Genetic engineering for modern agriculture: challenges and perspectives. Annu. Rev. Plant. Biol. 61 443–462
Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K and Yamaguchi-Shinozaki K 2007 Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51 617–630
Naoumkina M and Dixon RA 2011 Characterization of the mannan synthase promoter from guar (Cyamopsis tetragonoloba). Plant Cell Rep. 30 997–1006
Nekrasov V, Staskawicz B, Weigel D, Jones JD and Kamoun S 2013 Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotech. 31 691
Nicholass FJ, Smith CJ, Schuch W, Bird CR and Grierson D 1995 High levels of ripening-specific reporter gene expression directed by tomato fruit polygalacturonase gene-flanking regions. Plant Mol. Biol. 28 423–435
Nunberg AN, Li Z, Bogue MA, Vivekananda J, Reddy AS and Thomas TL 1994 Developmental and hormonal regulation of sunflower helianthinin genes:proximal promoter sequences confer regionalized seed expression. Plant Cell 6 473–486
Obertello M, Santi C, Sy MO, Laplaze L, Auguy F, Bogusz D and Franche C 2005 Comparison of four constitutive promoters for the expression of transgenes in the tropical nitrogen-fixing tree Allocasuarina verticillata. Plant Cell Rep. 24 540–548
Odell JT, Nagy F and Chua NH 1985 Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313 810–812
Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, Kim M and Kim JK 2005 Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol. 138 341–351
Ordon J, Bressan M, Kretschmer C, Dall’Osto L, Marillonnet S, Bassi R and Stuttmann J 2020 Optimized Cas9 expression systems for highly efficient Arabidopsis genome editing facilitate isolation of complex alleles in a single generation. Funct. Integr. Genomics 20 151–162
Osakabe Y, Osakabe K and Chiang VL 2009 Characterization of the tissue-specific expression of phenylalanine ammonia-lyase gene promoter from loblolly pine (Pinus taeda) in Nicotiana tabacum. Plant Cell Rep. 28 1309–1317
Park SH, Yi N, Kim YS, Jeong MH, Bang SW, Choi YD and Kim JK 2010 Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J. Exp. Bot. 61 2459–2467
Patro S, Maiti IB and Dey N 2013 Development of an efficient bi-directional promoter with tripartite enhancer employing three viral promoters. J. Biotechnol. 163 311–317
Porto MS, Pinheiro MP, Batista VG, dos Santos RC, Filho Pde A and de Lima LM 2014 Plant promoters: an approach of structure and function. Mol. Biotechnol. 56 38–49
Potenza C, Aleman L and Gopalan CS 2004 Targeting transgene expression in research agricultural and environmental applications: promoters used in plant transformation. InVitro Cell Dev. Biol. Plant 40 1–22
Que Q, Chilton M-DM, de Fontes CM, He C, Nuccio M, Zhu T, Wu Y, Chen JS, Shi L 2010 Trait stacking in transgenic crops: challenges and opportunities. GM Crops 1 220–229
Qiu CH, Li H, Li J, Qin RY, Xu RF, Yang YC, Ma H, Song FS, Li L, Wei PC and Yang JB 2015 Isolation and characterization of three cadmium-inducible promoters from Oryza sativa. J. Biotechnol. 216 1–19
Rai M, He C and Wu R 2009 Comparative functional analysis of three abiotic stress-inducible promoters in transgenic rice. Transgenic Res. 187 87–99
Rao TSRB, Naresh JV, Reddy PS, Reddy MK and Mallikarjuna G 2017 Expression of Pennisetum glaucum eukaryotic translational initiation factor 4A (PgeIF4A) confers improved drought salinity and oxidative stress tolerance in groundnut. Front. Plant Sci. 8 453
Reddy CS, Vijayalakshmi M, Kaul T, Islam T and Reddy MK 2015 Improving flavour and quality of tomatoes by expression of synthetic gene encoding sweet protein monellin. Mol. Biotechnol. 57 448–53
Reddy PS, Mahanty S, Kaul T, Nair S, Sopory SK and Reddy MK 2008 A high-throughput genome-walking method and its use for cloning unknown flanking sequences. Anal. Biochem. 381 248–253
Rerksiri W, Zhang X, Xiong H and Chen X 2013 Expression and promoter analysis of six heat stress-inducible genes in rice. Sci. World J. 397401
Roccaro M, Ahmadinejad N, Colby T and Somssich IE 2013 Identification of functional cis-regulatory elements by sequential enrichment from a randomized synthetic DNA library. BMC Plant Biol. 13 164
Rusconi F, Simeoni F, Francia P, Cominelli E, Conti L, Riboni M and Galbiati M 2013 The Arabidopsis thaliana MYB60 promoter provides a tool for the spatio-temporal control of gene expression in stomatal guard cells. J. Exp. Bot. 64 3361–3371
Saed TR, Ismail I, Zainal Z and Abdullah SN 2012 The stearoyl-acyl-carrier-protein desaturase promoter (Des) from oil palm confers fruit-specific GUS expression in transgenic tomato. J. Plant Physiol. 1691 290–300
Sahoo DK, Sarkar S, Raha S, Maiti IB and Dey N 2014 Comparative analysis of synthetic DNA promoters for high-level gene expression in plants. Planta 240 855–875
Sakaue K, Harada H and Matsuda Y 2008 Development of gene expression system in a marine diatom using viral promoters of a wide variety of origin. Physiol. Plant. 133 59–67
Sawant SV, Kiran K, Mehrotra R, Chaturvedi CP, Ansari SA, Singh P, Lodhi N and Tuli R 2005 A variety of synergistic and antagonistic interactions mediated by cis-acting DNA motifs regulate gene expression in plant cells and modulate stability of the transcription complex formed on a basal promoter. J. Exp. Bot. 56 2345–2353
Schiml S, Fauser F and Puchta H 2014 The CRISPR/C as system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in A rabidopsis resulting in heritable progeny. Plant J. 80 1139–1150
Schmulling T, Schell J and Spena A 1989 Promoters of the rolA B and C genes of Agrobacterium rhizogenes are differentially regulated in transgenic plants. Plant Cell 1 665–670
Seemanpillai M, Dry I, Randles J and Rezaian A 2003 Transcriptional silencing of geminiviral promoter-driven transgenes following homologous virus infection. Mol. Plant Microbe. Interact. 16 429–438
Shah SH, Jan SA, Ahmad N, Khan SU, Kumar T, Iqbal A and Nasir F 2015 Use of different promoters in transgenic plant development: current challenges and future perspectives. Am Eurasian J. Agric. Environ. Sci. 15 664–675
Shahmuradov IA, Umarov RK and Solovyev VV 2017 TSSPlant: a new tool for prediction of plant Pol II promoters. Nucleic Acids Res. 45 e65
Shan Q, Wang Y, Chen K, Liang Z, Li J, Zhang Y et al 2013 Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant 6 1365–1368
Sharma KK, Lavanya M and Anjaiah V 2006 Agrobacterium-mediated production of transgenic pigeonpea (Cajanus cajan L. Millsp.) expressing the synthetic Bt cry1Ab gene. In Vitro Cell. Dev. Biol.-Plant 42 165–173
Shen YG, Zhang WK, Yan DQ, Du BX, Zhang JS, Liu Q and Chen SY 2003 Characterization of a DRE-binding transcription factor from a halophyte Atriplex hortensis. Theor. Appl. Genet. 107 155–161
Singer SD, Hily JM and Cox KD 2011 The sucrose synthase-1 promoter from Citrus sinensis directs expression of the β-glucuronidase reporter gene in phloem tissue and in response to wounding in transgenic plants. Planta 234 623–637
Smale ST and Kadonaga JT 2003 The RNA polymerase II core promoter Annu. Rev. Biochem 72 449–479
Singh KB, Foley RC and Onate-Sanchez L 2002 Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5 430–436
Srinath T, Reddy VD and Rao KV 2017 Isolation and functional characterization of a novel stress inducible promoter from pigeonpea (Cajanus cajan L). Plant Cell Tiss. Org. Cult. 128 457–468
Srivastava VK, Raikwar S and Tuteja N 2014 Cloning and functional characterization of the promoter of PsSEOF1 gene from Pisum sativum under different stress conditions using Agrobacterium-mediated transient assay. Plant Signal Behav. 9 e29626
Sunkara S, Bhatnagar-Mathur P and Sharma KK 2014 Isolation and functional characterization of a novel seed-specific promoter region from peanut. Appl Biochem Biotechnol. 172 325–339
Tao YB, He LL, Niu LJ and Xu ZF 2015 Isolation and characterization of an ubiquitin extension protein gene (JcUEP) promoter from Jatropha curcas. Planta 241 823–836
Tashkandi M, Ali Z, Aljedaani F, Shami A and Mahfouz MM 2018 Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal. Behav. 13 e1525996
Tiwari V, Patel MK, Chaturvedi AK, Mishra A and Jha B 2016 Functional characterization of the tau class glutathione-S-transferases gene (SbGSTU) promoter of Salicornia brachiata under salinity and osmotic stress. PLoS One 11 e0148494
Tompa M, Li N, Bailey TL, Church GM, et al. 2005 Assessing computational tools for the discovery of transcription factor binding sites Nat. Biotechnol. 23 137–144
Twyman RM 2003 Growth and development: control of gene expression regulation of transcription; In Encyclopedia of applied plant sciences (Eds) Thomas B, Murphy DJ and Murray GB (Elsevier Science, London UK) pp 558–567
van der Geest AH and Hall TC 1997 The β-phaseolin5 matrix attachment region acts as an enhancer facilitator. Plant Mol. Bio. 33 553–557
Van der Does D, Leon-Reyes A, Koornneef A, Van Verk MC, Rodenburg N, Pauwels L, Goossens A, Körbes AP, Memelink J, Ritsema T, Van Wees SC and Pieterse CM 2013 Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25 744–761
Viana AA, Fragoso RR, Guimarães LM, Pontes N, Oliveira-Neto OB, Artico S et al 2011 Isolation and functional characterization of a cotton ubiquitination-related promoter and 5'UTR that drives high levels of expression in root and flower tissues. BMC Biotechnol. 11 115
Wang JY, Wang JP and Yang HF 2016a Identification and functional characterization of the NAC gene promoter from Populus euphratica. Planta 244 417–427
Wang P, Du Y, Zhao X, Miao Y and Song CP 2013 The MPK6-ERF6-ROS-responsive cis-acting Element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol. 161 1392–1408
Wang R, Yan Y, Zhu M, Yang M, Zhou F, Chen H and Lin Y 2016b Isolation and Functional Characterization of bidirectional promoters in rice. Front. Plant Sci. 77 66
Wang R, Zhu M, Ye R, Liu Z, Zhou F, Chen H and Lin Y 2015 Novel green tissue-specific synthetic promoters and cis-regulatory elements in rice. Sci. Rep. 51 825
Wang J and Oard JH 2003 Rice ubiquitin promoters: deletion analysis and potential usefulnessin plant transformation systems:Genetic transformation and hybridization. Plant Cell Rep. 221 29–34
Wang Y, Lin S, Song Q, Li K, Tao H, Huang J et al 2014 Genome-wide identification of heat shock proteins (Hsps) and Hsp interactors in rice: Hsp70s as a case study. BMC Genom. 15 344
Wang X, Wang L, Wang Y, Liu H, Hu D. Zhang N et al 2018 Arabidopsis PCaP2 plays an important role in chilling tolerance and ABA response by activating CBF-and SnRK2-mediated transcriptional regulatory network. Frontiers in plant science 9 215
Wu CY, Suzuki A, Washida H and Takaiwa F 1998 The GCN4 motif in a rice glutelin gene is essential for endosperm-specific gene expression and is activated by Opaque-2 in transgenic rice plants. Plant J. 14 673–683
Wu L, E-Mezawy A and Shah S 2011 A seed coat outer integument-specific promoter for Brassica napus. Plant Cell Rep. 30 75–80
Xiao K, Zhang C, Harrison M and Zeng YW 2005 Isolation and characterization of novel plant promoter that directs strong constitutive expression of transgene in plants. Mol. Breed. 15 221–231
Xu D, Duan X, Wang B, Hong B, Ho T and Wu R 1996 Expression of a late embryogenesis abundant protein gene HVA1 from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110 249–257
Xu F, Huang XH, Li LL, Deng G, Cheng H, Rong XF, Li JB and Cheng SY 2013 Molecular cloning and characterization of GbDXS and GbGGPPS gene promoters from Ginkgo biloba. Genet. Mol. Res. 4 293–301
Yamaguchi-Shinozaki K and Shinozaki K 2006 Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803
Yan L, Wei S, Wu Y, Hu R, Li H, Yang W and Xie Q 2015 High-efficiency genome editing in Arabidopsisusing YAO promoter-driven CRISPR/Cas9 system. Mol. Plant 8 1820–1823
Yang IC, Iommarini JP, Becker DK, Hafner GJ, Dale JL and Harding RM 2003 A promoter derived from taro bacilliform badnavirus drives strong expression in transgenic banana and tobacco plants. Plant Cell Rep. 21 1199–1206
Yang J, Wang X, Hasi A and Wang Z 2018 Structural and functional analysis of a bidirectional promoter from Gossypium hirsutum in Arabidopsis. Int. J. Mol. Sci. 19 3291
Ye X, Al-Babili S, Kloti A, Zhang J, Lucca P, Beyer P and Potrykus I 2000 Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287 303–305
Ye R, Zhou F and Lin Y 2012 Two novel positive cis-regulatory elements involved in green tissue-specific promoter activity in rice (Oryza sativa L ssp). Plant Cell Rep. 31 1159–72
Zavallo D, Lopez Bilbao M, Hopp HE and Heinz R 2010 Isolation and functional characterization of two novel seed-specific promoters from sunflower (Helianthus annuus L). Plant Cell Rep. 29 239–248
Zhang C, Pan S, Chen H, Cai T, Zhuang C, Deng Y, Zhuang Y, Zeng Y, Chen S and Zhuang W 2016 Characterization of NtREL1 a novel root-specific gene from tobacco and upstream promoter activity analysis in homologous and heterologous hosts. Plant Cell Rep. 35 757–769
Zhang H, Hou J, Jiang P, Qi S, Xu C, He Q, Ding Z, Wang Z, Zhang K and Li K 2016 Identification of a 467 bp promoter of maize phosphatidylinositol synthase gene (ZmPIS) which confers high-level gene expression and salinity or osmotic stress inducibility in transgenic tobacco. Front. Plant Sci. 7 42
Zhou J, Yang Y, Wang X, Yu F, Yu C, Chen J, Cheng Y, Yan C and Chen J 2013 Enhanced transgene expression in rice following selection controlled by weak promoters. BMC Biotechnol. 27 13–29
Acknowledgements
PSR acknowledges financial support from the Department of Science and Technology, Govt. of India, New Delhi, for a fellowship and research grant through the INSPIRE Faculty Award No. IFALSPA-06 and Young Scientist Award SB/YS/LS-12/2013. This work was undertaken as part of the CGIAR Research Program on Grain Legumes & Dryland Cereals (CRP-GLDC).
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Topical Collection: Genetic Intervention in Plants: Mechanisms and Benefits.
Rights and permissions
About this article
Cite this article
Kummari, D., Palakolanu, S.R., Kishor, P.B.K. et al. An update and perspectives on the use of promoters in plant genetic engineering. J Biosci 45, 119 (2020). https://doi.org/10.1007/s12038-020-00087-6
Published:
DOI: https://doi.org/10.1007/s12038-020-00087-6